6-O-Nucleotidyltransferase: an aminoglycoside-modifying enzyme specific for streptomycin/streptidine†
Montserrat
Latorre
,
Julia
Revuelta
,
Eduardo
García-Junceda
* and
Agatha
Bastida
*
Departamento de Química Orgánica Biológica, Instituto de Química Orgánica General, CSIC, Madrid, 28006, Spain. E-mail: agatha.bastida@.csic.es; eduardo.junceda@csic.es
First published on 16th November 2015
Abstract
Aminoglycosides are especially useful for the treatment of hospital-acquired infections. The main problem for the application of these antibiotics is the presence of bacterial resistance enzymes, in particular, nucleotidyltransferases (ANTs). These enzymes catalyze the transfer of an adenylyl group from the MgATP complex to different positions of the antibiotic. To understand the mechanisms that lead to antibiotic inactivation, we have performed a comprehensive experimental analysis of one of those enzymes. The 6-O- nucleotidyltransferase enzyme (ANT(6)) from Bacillus subtilis was cloned, overexpressed and purified in E. coli. The kinetic parameters revealed a narrow specificity of the ANT(6) for MgATP/streptomycin as substrates. The binding epitope of the streptomycin recognized by the ANT(6) is the streptidine moiety. Therefore, the use of streptidine as a “decoy acceptor” allows the recovery of the antibiotic activity of streptomycin E. coli cells that are overexpressing the ANT(6).
Introduction
Aminoglycoside antibiotics are of special interest in medicine for the treatment of severe bacterial infections.1–3 These antibiotics are active against either Gram + or Gram − bacteria, since their primary mechanism of action is the inhibition of bacterial protein synthesis by binding to the tRNA acceptor A site in the small ribosomal subunit that is causing a misreading and/or hindering the translocation step.4–8 For most of the aminoglycosides, the aminocyclitol moiety is a 2-deoxystreptamine ring (2-DOS) and can be substituted at positions 4 and 6 (kanamycin family 3) or at 4 and 5 (neomycin 2 family). However, a number of active aminoglycosides are structurally atypical, such as streptomycin (ST) 1 (Fig. 1).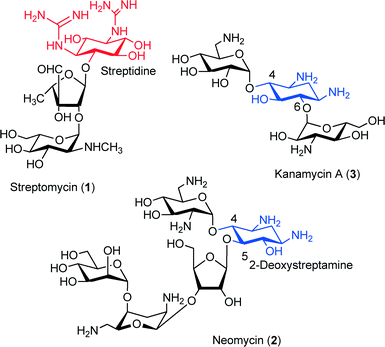 | ||
| Fig. 1 Structures of aminoglycoside antibiotics: streptomycin (1), neomycin (2) and kanamycin A (3). | ||
The emergence of bacterial resistance for all classes of antipathogenic agents has become a serious problem over recent years.9–11 Aminoglycosides were one of the first groups of antibiotics to meet the challenge of resistance.12 The most prevalent source of clinically relevant resistance to aminoglycoside antibiotics is conferred by the enzymatic inactivation of the drugs by the aminoglycoside-modifying enzymes (AMEs).13,14
The AMEs can be classified as N-acetyltransferases (AACs), O-adenyltransferases (ANTs) or O-phosphotransferases (APHs). In each of these families, there are several enzymes that catalyzed the reactions with different regioselectivities and substrate specificities.
The ANT' family is the smallest of the three groups with only a few enzymes identified to date [ANT(6)], [ANT(9)], [ANT(4′)] and [ANT(2′′)].15–18 One of the most prevalent ANT enzymes is the streptomycin adenylyltransferase (ANT (6)).19–21 This enzyme catalyzes the transfer of an adenyl group from the [MgATP]− complex to the OH at the 6 position of streptidine unit in the streptomycin (1), thus leading to a sharp decrease in the drug's affinity for its RNA target (Scheme 1). Streptomycin (1) was the first aminoglycoside antibiotic to be discovered in 1943. It has a three-ring structure (Scheme 1), comprising a substituted aminocyclitol (streptidine moiety), linked to a modified ribose (α-L-streptose), which is linked in turn to an N-methyl-α-L-glucosamine residue.
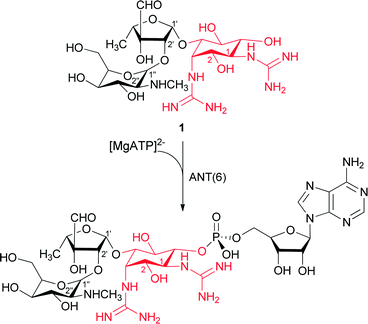 | ||
| Scheme 1 ANT(6) catalyzes the adenylation of streptidine moiety (red) in streptomycin (1) antibiotic, giving AMP-streptomycin inactive. | ||
Some years ago, our group reported that the binding epitope of the streptomycin recognized by this enzyme was the streptidine moiety22 we also noted its use as a “decoy acceptor”‡23,24 by the ANT(6) enzyme to rescue the antibiotic activity of streptomycin (1) from bacterial strains overexpressing this aminoglycoside-modifying enzyme. We showed that the streptidine was a substrate for the aminoglycoside-inactivating enzyme ANT(6), competing with the streptomycin (1) for the active center of the enzyme. Thus, the addition of this molecule in a cell culture restored the activity of the streptomycin antibiotic normally rendered inactive by the ANT(6) enzyme, because the streptidine was acting as a “decoy acceptor” of the ANT(6).
Therefore, we demonstrated that the use of decoy acceptors can be an effective strategy for combating the phenomenon of resistance promoted by AMEs. However, for the design of more efficient decoy acceptors, a deeper knowledge of the molecular mechanism of the AMEs, their structures and interactions with the drugs is needed. Herein, we report the biochemical characterization of the ANT(6) from Bacillus subtilis. To this end and we cloned, overexpressed the enzyme in E. coli. The kinetic parameters revealed a narrow specificity of the ANT(6) for MgATP/streptomycin as substrates. Finally, we assayed the use of 2-deoxystreptamine as a decoy acceptor, and its ability to restore the antibiotic activity of streptomycin against resistant bacterial strains.
Results and discussion
Cloning, over-expression, purification and characterization of ANT(6)
The aadK gene from B. subtilis was amplified by PCR, using primers designed specifically to complement 15 bp at the 5′ ends of codifying and complementary DNA strains. The recognition sequence for the restriction enzymes EcoRI and HindIII was introduced in the amplification product during the PCR process. The double-digested PCR product was cloned into the vector pET-28b(+) to yield the recombinant plasmid pET-aadk NHis6-tag.The aadK gene was expressed in E. coli BL21(DE3) at 30 °C. SDS-PAGE analysis of the gene expression showed an IPTG-inducible protein of 37.4 kDa of the ANT(6) with an NHis6-tag. After addition of the thrombin enzyme, the ANT(6)ΔHis enzyme was obtained with 35.5 kDa of weight (Fig. S1†). The two-step purification of the ANT(6) yielded well over 40 mg of protein per liter of the cell culture, with a purity degree of at least 95% (Fig. S1†). The ANT(6) was aggregated without MgCl2, so the use of that cation is essential to avoid the formation of aggregations (Fig. S2a†). The purified ANT(6)ΔHis is a dimer based on an ultracentrifugation experiment (Fig. S2b†) and its mobility over Superdex 200 in standardized analytical-gel filtration experiments.
The activity of the pure recombinant enzyme was followed by HPLC and reaction products identified by 1H-NMR.25 The recombinant ANT(6) exhibited activity between pH 6.5 and pH 8.5, with the maximum activity at pH 7.5. At pH values over 7.5, the activity level drops drastically to virtually zero (Fig. S3†).
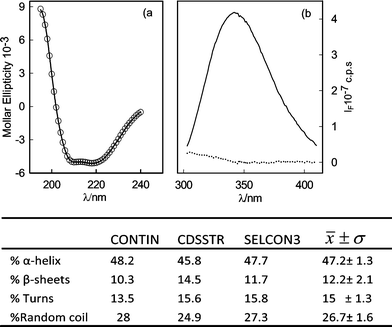
Regarding the secondary structure of the recombinant enzyme, the far-UV CD spectrum of purified ANT(6) shows two minima in ellipticity at ~210 and ~222 nm, indicative of a main α-helical secondary structure (Fig. 2a). The percentages of the different secondary structure elements were estimated using the CDPro26 software package ~48.2% α-helix, ~8.7% β-sheet, ~15.9% turns, and ~27.3% of unordered structure were thereby obtained. The tertiary structure of the protein in the microenvironment of the protein fluorophores was analyzed using fluorescence-emission spectroscopy of 19 tyrosines and 7 tryptophan present in the ANT(6) sequence (Fig. 2b). After excitation at 275 nm, the fluorescence spectrum exhibited an emission maximum centered at 345 nm, close to the expected emission of free tryptophans exposed to an aqueous solution (348–350 nm). The emission spectrum obtained upon excitation at 295 nm, was almost identical to the previous spectrum, indicating an almost negligible contribution of tyrosines to the fluorescence spectrum of the recombinant ANT(6) enzyme (Fig. 2b).
The thermal stability of the enzyme was initially assessed by using CD spectroscopic analysis, in which the temperature dependence of the dichroism signal at λ = 222 nm was monitored (Fig. 3). The curve following the thermal unfolding of the ANT(6) could be fitted to a simple thermodynamic unfolding model. The melting temperature of the recombinant ANT(6) was determined to be 56 ± 0.2 °C. The transition region was sharp, thus indicating that the protein existed initially as a compact, well-folded structure and that the unfolding reaction was highly cooperative.
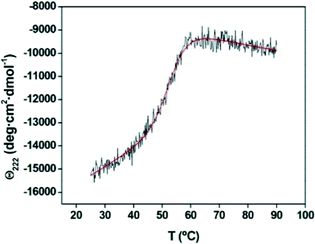 | ||
| Fig. 3 Thermal denaturation profile of the ANT(6) enzyme. Melting temperature (Tm) was assessed using CD by following the temperature dependence of the dichroism signal at 222 nm. | ||
The influence of divalent cations in the activity and stability of recombinant ANT(6)
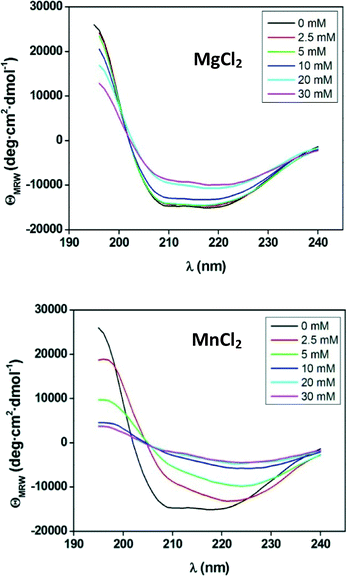
The quaternary structure of the enzyme determined by sedimentation velocity experiments (Fig. S1a†) showed that the presence of Mg2+ strongly affected the aggregation state of the enzyme. The magnesium ion is by far the most frequently found metal ion cofactor in enzymatic systems. This is most likely related to the ability of Mg2+ to form stable complexes with ATP, which is normally associated with the metal ion when acting in a physiological environment. On the other hand, Mn2+ is the required cofactor in many other transferase enzymes, such as glycosyltransferases of the Leloir patway.27 Therefore, we decided to study the influence of Mg2+ or Mn2+ ion on the 2D-structure (Fig. 4).The ANT(6) showed a similar activity in the presences of MnCl2 and MgCl2 (0.07 and 0.08 U mg−1, respectively) but was much more stable in the presence of Mg2+ (Fig. 4). Concentrations of MgCl2 above 20 mM promote a conformational change of the protein that is reflected in the decrease of the peaks ≈195, ≈208 and ≈222 nm, which corresponds to an increase in β-sheet structure of about 7% but which maintains a relatively high degree of structure. However, manganese promotes a higher loss of the protein's secondary structure. Thus, from a concentration of 10 mM, the CD spectrum gives almost no signal indicating that the protein has lost its secondary structure and, therefore, its chirality. This effect could be due to the higher flexibility of Mn2+ ligand bonds in both length and angle, which could promote the conformational change of the recombinant protein.28
ANT(6)-substrate specificity
The main kinetic parameters for ANT(6) from B. subtilis are displayed in Table 1.| Substrate | K M (M) | k cat (s−1) | k cat/KM (s−1 M−1) |
|---|---|---|---|
| a 0.5 mM of streptomycin, 2 mM [ATPMgCl2], pH 7.5. | |||
| Streptomycin | 3.8 × 10−5 | 0.03 | 9.2 × 102 |
| ATPMg | 8.4 × 10−5 | 0.018 | 2.1 × 102 |
| Streptidine | 6 × 10−4 | 0.0006 | 1.2 |
To the best of our knowledge, the kinetic parameters of only three other ANTs have been described: the bifunctional enzyme ANT(3′′)-Ii/AAC(6′)-IId from Serratia marcescens,28 the ANT(2′′)-I from Klebsiella pneumoniae29,30 and the ANT(4′) from S. aureus.31,32 The ANT(3′′)-Ii domain carried out the adenylation of streptomycin with a higher catalytic efficiency (kcat/KM 3.2 105) than that of the ANT(6) from B. subtilis (Table 1). This domain was highly specific for streptomycin; that is, it did not adenylate 4,5- or 4,6-aminoglycosides, thus showing a similar behaviour to that of the ANT(6). In the case of the ANT(2′′)-I/ANT(4′), its catalytic efficiency is similar to that of our enzyme but it does not recognize streptomycin as an aminoglycoside.
In addition, we studied the effect of MgCl2 concentration on the enzyme activity as a function of the relationship [MgCl2]/[ATP]. The maximum activity was reached at a 17 mM concentration of free Mg2+ (Fig. 5). Above this concentration, and with a free-magnesium concentration of 37 mM, neither increased activity nor an inhibitory effect was detected. The concentration of magnesium required to reach maximum activity was higher than that required to saturate the ATP. Some authors suggest that this may be because the enzyme requires two molecules of the divalent ion to carry out its activity: one that would bind to the ATP and the other that would bind directly to the enzyme,15 which has been observed in the 3D-structure of ANT(4′)/Kanamycin Complex (pdb 1kny).
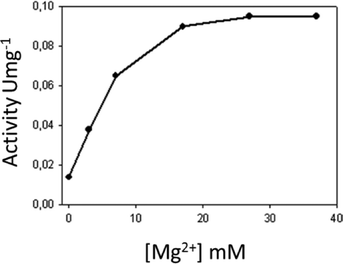 | ||
| Fig. 5 The effect of the magnesium free-ion in the adenylated activity of the streptomycin with the ANT(6). | ||
We have already showed that streptidine is the minimum moiety of streptomycin that is recognized by the enzyme and that can be adenylated by the ANT(6) (Fig. S5†).22 In order to study the ANT(6) specificity, the enzyme activity was tested using different aminoglycosides, such as streptomycin, neomycin, kanamycin, spectinomycin, ribostamycin, paromomycin, neamine and the 2-deoxystreptamine (the common moiety of the 4,5- and 4,6-aminoglycosides). The enzyme was highly specific for streptomycin (Table S1†) and did not recognize 4,5- or 4,6-aminoglycosides (no activity was detected with kanamycin or ribostamycin). In fact, the enzyme only provides acceptor groups for hydrogen-bond interactions with aminoglycoside substrates. The abundance of acidic residues in the binding pocket of the enzyme can be readily explained by reference to the fact that aminoglycosides are invariably positively charged molecules. The other ANT enzymes described, such as ANT(4′) and ANT(2′′), accept different aminoglycosides as substrates, thus showing high rates of substrate promiscuity.31,32 This differing behavior can be explained by reference to the fact that, in the former case, the natural substrates are 4,5 or 4,6-aminoglycosides, both of which share the 2-deoxystreptamine moiety (Fig. 1).
We have also studied the specificity of the nucleoside triphosphate (ATP, GTP, CTP and UTP). The enzyme recognizes the nucleotides with similar affinities25 but only showed activity with ATP/GTP (Table S2†). According to these data, ANT(6) shows a clear preference for nucleotide triphosphates incorporating purine aromatic systems. On the other hand, modifications of the inorganic fragment led to significant changes in activity (Table S2†). Thus, replacement of a single oxygen atom by a methylene group (ATP versus AMPCPP) and the suppression of the terminal γ-phosphate group (ATP versus ADP, AMP) lead to a total loss of activity. Recent results obtained by our research group have demonstrated that ANT(4′) from S. aureus can employ inorganic triphosphate (P3) as a substrate to promote the regioselective phosphorylation of aminoglycosides.33 However, in the case of the ANT(6), no reaction of phosphorylation with streptomycin/P3 was detected.
In vivo activity of ANT(6)
Streptomycin is powerful antibiotic with minimal inhibitory concentrations (MICs) of 5 μg ml−1 for E. coli BL21(DE3) strain. However, when the ANT(6) enzyme is present, it lost its antibiotic activity (MIC > 200 μg ml−1). On the other hand, streptidine and 2-deoxystreptamine (2-DOS) (common moieties of the streptomycin and 4,5- 4,6-aminoglycosides, respectively) did not show any antibiotic activity in the absence of ANT(6) (MIC >100 μg ml−1) (Table 2). In the case of 4,5-aminoglycosides, binding to neomycin was detected but no reaction was observed (Table S2†).25 As we have described, the streptidine is a substrate of the ANT(6), so when it is added together with streptomycin, it works as a “decoy acceptor”.22 With these previous results in mind, we assayed the 2-DOS34 as a “decoy acceptor” of the ANT(6) in the presence of streptomycin. Meanwhile, streptidine is able to reduce the MIC for streptomycine to 10 μg ml−1 when ANT(6) is expressed, but 2-DOS did not show any affect (Table 2). This result could indicate that ANT(6) requires the presence of a guanidine group in the aminocyclitol, since it is so specific for streptomycin, unlike the other ANTs hitherto described. This was verified because AMP-2-DOS product was not detected by ESI-MS or HPLC.| Amynoglycoside | E. coli | MICaE. coli (pET) | E. colipET-aadk |
|---|---|---|---|
| a MIC: minimum inhibitory concentration, values (μg mL−1). b Data taken from reference.22 c 2-DOS (2-deoxystreptamine). | |||
| Streptomycinb | 5 | 5 | >200 |
| Streptidine | >100 | >100 | >200 |
| Streptomycin + streptidineb | 5 | 5 | 10 |
| 2-DOSc | >100 | >100 | >100 |
| Streptomycin + 2-DOS | 2 | 100 | 100 |
Experimental
Materials
pET-28b(+) vector was obtained from Novagen. Taq DNA polymerase was purchased from Ecogen. T4 DNA ligase was obtained from MBI Fermentas. Restriction enzymes EcoRI and HindIII were purchased from Boehringer Manheim. Isopropyl-L-thio-β-D-galactopyranoside (IPTG) was purchased from Applichem. E. coli competent cells BL21 (DE3) was purchased from Stratagene Co. (San Diego, CA). Kanamicyn, ATP and other aminoglycoside antibiotics were obtained from Sigma. Nickel-iminodiacetic acid (Ni2+IDA) agarose was generous donated by ABT. Bacillus subtilis was obtained from CECT.Cloning, expression and purification the ANT(6)
PCR amplification was performed in a 100 μL reaction mixture containing DNA (3 μL) of Bacillus subtilis as template, water (70.5 μL), buffer (100 mm Tris-HCl, 500 mm KCl, pH 8.0, 10 μL), MgCl2 (2 mm), dNTPs (200 μm), primers (1 μm) ANT6-Ct (5′cgaagctttcactttactgagcaataaa3′) and ANT6-Nt (5′gccgcgaattccatgcgaagtgagcaggaaat3′) and Taq DNA polymerase (2.5 U). The reaction was subjected to 25 cycles of amplification. The cycle conditions were set as follows: denaturation at 94 °C for 1 min, annealing at 55 °C for 1.0 min, and elongation at 72 °C for 1.5 min. The PCR product was introduces into the pGem-T easy vector. The sequencing of the DNA insert was carried out with the promotors T7 and SP6. Then aadK insert was digested with EcoRI and HindIII enzymes and inserted into the pET-28b(+) vector that the resulting plasmid pET-ANT(6) was transformed into E. coli strain BL21 (DE3) which then showed the expected resistance pattern to the streptomycin.The selected clone was grown up on 100 mL of LB medium containing 26 μg mL−1 kanamycin at 37 °C with shaking. When the cell growth reached an optical density at 600 nm (O.D.600) of 0.5, the temperature was switched to 30 °C and the culture was induced with 0.5 mM IPTG. After 24 hours of induction the expression level was analysed by SDS-PAGE using gels with 13% of polyacrylamide in the separation zone. The culture broth was centrifuged (5000 × g, 20 min, 4 °C), and the cells were suspending in Tris buffer (8 mL g−1 cells, 50 mm pH 8.0) and was added EDTA (50 mm, pH 8.2) and lysozyme (2 mg g−1 cells). The suspension was gently stirred at room temperature during 1 hour, and the suspension was kept at 4 °C overnight. The preparation was gently sonicated for 40 s and cooled down in ice (4 times) to decrease viscosity. DNase (10 μg g−1 cells) and MgCl2 (0.95 μg mL−1 of preparation) were added, and the mixture was refrigerated for 20 min. The mixture was then centrifuged for 30 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g to separate the soluble proteins from the insoluble ones. To the soluble fraction 1% of streptomycin was added and stirred for 20 min at 4 °C. Then the cell free extract (CFE) was recovered by centrifugation.
000 × g to separate the soluble proteins from the insoluble ones. To the soluble fraction 1% of streptomycin was added and stirred for 20 min at 4 °C. Then the cell free extract (CFE) was recovered by centrifugation.
1.5 mL of CFE was applied to 0.5 gr of Ni2+IDA-agarose, previously equilibrated with phosphate buffer, pH 7.0. The proteins non-specifically retained were washed with the same buffer. ANT(6) was eluted with the same buffer containing 0.5 M of imidazole. After that, the solution was dialyzed against phosphate buffer 5 mM, pH = 7.0. After this step, the recombinant enzyme was found to be more than 90% pure as assessed by SDS-polyacrylamide gel electrophoresis. Further purification of the recombinant ANT(6) was accomplished by size-exclusion chromatography on HiLoad 26/60 Superdex 200 PG column controlled using the AKTA-FPLC system. The column was developed in 20 mM phosphate buffer pH 7.5 containing 0.15 M NaCl at a constant rate of 1 ml min−1. Fractions showing activity were pooled and dialyzed with water and concentrated by lyophilisation. Protein concentrations were estimated by the Bio-Rad protein assay method using bovine serum albumin as a standard.
Circular dichroism spectroscopy
Far-UV circular dichroism (CD) spectra were recorded in the wavelength range of 195–240 nm using a Jasco J-810 spectropolarimeter equipped with a constant temperature cell holder Jasco PTC423-S Peltier. The protein concentration was 5.3 μM and the optical path length 0.1 cm. The contribution of the buffer was always subtracted. For each sample, three spectra were accumulated at a scan speed of 20 nm min−1 with a bandwidth of 0.2 nm and averaged automatically. All spectra were background-corrected, smoothed and transformed into mean residue ellipticity (θ) in the JASCO spectra manager. A value of 110 g mol−1 was used as a mean residue weight for ANT(6).The secondary structure of the protein was evaluated by computer fit of the dichroism spectrum into four simple components (α-helix, β-sheet, turns and random coil) using the CDPro software package containing three commonly used programs: SELCON3, CONTIN/LL and CDSSTR.26Ultracentrifugation
The quaternary structure of recombinant ANT(6) from B. subtillis was assessed using sedimentation equilibrium experiments. These experiments were performed at the Department of Chemical Physics of Biological Macromolecules of the IQFR-CSIC. The initial concentration of the protein used in these experiments was 0.96 mg mL−1.Fluorescence experiments
The experiments were conducted on Perkin Elmer LS50B luminescence spectrometer with the program FLWin. A quartz cell with a 1 cm path length in both the excitation (275 nm and 295 nm) and emission (295 to 400 nm) directions was used in all the experiments that were performed at 25 °C. The ANT(6) protein concentration was 30 μM in phosphate buffer 5 mM, pH = 7.5. The spectres were normalized by origin 7.0 program.Measurement of enzyme activity
The activity of the pure recombinant enzyme was follow by HPLC and the identity of the adenylated products were confirmed by ESI-MS. The formation of the product was followed by HPLC at 260 nm (Vydac column eluted with TFA pH 3.0 buffer). Enzyme concentration was calculated using a molar absortion coefficient at 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 M−1 cm−1 at 280 nm. The reaction mixture contained 50 mM Hepes buffer, pH (6.5–8), nucleoside derivatives (3 mM), aminoglicoside (0–2.5 mM), MgCl2 (0–35 mM) and the reaction were started by the addition of ANT(6) enzyme (0.5 μM) at room temperature in a volume of 1 ml.
400 M−1 cm−1 at 280 nm. The reaction mixture contained 50 mM Hepes buffer, pH (6.5–8), nucleoside derivatives (3 mM), aminoglicoside (0–2.5 mM), MgCl2 (0–35 mM) and the reaction were started by the addition of ANT(6) enzyme (0.5 μM) at room temperature in a volume of 1 ml.
Minimal inhibitory concentrations (MIC)
The different strains were grown in 1 ml tubes and Mueller–Hinton broth to an optical density OD600 of 0.4 units, then ANT(6) was expressed by induction with 0.5 mM of IPTG until 0D600 0.6 when. Then, the desired concentrations of antibiotics were added from stock solutions. The samples were incubated at 37 °C for 12 h, when the control culture (with no antibiotic) had an OD600 of 1.2–1.6. The OD600 of each sample was read and the MIC was taken as the lowest antibiotic concentration inhibiting bacterial growth greater than 90%.Conclusions
In conclusion, we have described the overexpression and biochemical characterization of the ANT (6) from B. subtilis. The results reported throughout this manuscript highlight the high specificity of the ANT(6) for streptomycin-ATP/GTP-Mg of an enzyme that is involved in bacterial resistance against aminoglycoside antibiotics.From a medicinal chemistry perspective, the combination of the features present in the ANT(6) give the enzyme a narrow tolerance to chemical variations in the aminoglycoside/nucleotide, making it very useful in the design of non-inactivable derivatives in order to avoid bacterial resistance. Moreover, the design of new inhibitors or decoy acceptors for this enzyme must considerer the minimum unit of streptidine that is essential, and the fact that only nucleotide derivatives of purine bases will be recognized.
Acknowledgements
This investigation was supported by a research grant of the Spanish research “Dirección General de Investigación,” CTQ2013-45538-P”.Notes and references
- J. Davies, Can. J. Infect. Dis. Med. Microbiol., 2006, 17, 287–290 Search PubMed.
- D. Greenwood, Antimicrobial Chemotherapy, ed. D. Greenwood, Oxford University Press, Oxford, 1995, pp. 32–48 Search PubMed.
- L. Leibovici, L. Vidal and P. Mical, J. Antimicrob. Chemother., 2009, 63, 246–251 CrossRef CAS PubMed.
- D. Moazed and H. F. Noller, Cell, 1986, 47, 985–994 CrossRef CAS PubMed.
- B. Becker and M. A. Cooper, ACS Chem. Biol., 2013, 8, 105–115 CrossRef CAS PubMed.
- D. Moazed and H. F. Noller, Nature, 1987, 327, 389–394 CrossRef CAS PubMed.
- D. Moazed and H. F. Noller, J. Mol. Biol., 1990, 211, 135–145 CrossRef CAS PubMed.
- E. F. Gale, E. Cundliffe, P. E. Reynolds, M. H. Richmond and M. J. Waring, The Molecular Basis of Antibiotic Action, ed. M. J. Waring, Wiley, London, 1981 Search PubMed.
- S. Magnet and J. S. Blanchard, Chem. Rev., 2005, 105, 477–498 CrossRef CAS PubMed.
- C. A. Smith and E. N. Baker, Curr. Drug Targets: Infect. Disord., 2002, 2, 143 CrossRef CAS.
- J. N. Pendleton, S. P. Gorman and B. F. Gilmore, Expert Rev. Anti-Infect. Ther., 2013, 11, 29–308 CrossRef PubMed.
- S. B. Vakulenko and S. Mobashery, Clin. Microbiol. Rev., 2003, 16, 430–450 CrossRef CAS PubMed.
- G. D. Wright, Curr. Opin. Microbiol., 1999, 2, 499–503 CrossRef CAS PubMed.
- P. Keith, Antimicrob. Agents Chemother., 2005, 49, 479–487 CrossRef PubMed.
- L. C. Pedersen, M. M. Benning and H. M. Holden, Biochemistry, 1995, 34, 13305–13311 CrossRef CAS PubMed.
- M. S. Ramirez and M. E. Tolmasky, Drug Resist. Updates, 2010, 13(6), 151–171 CrossRef CAS PubMed.
- J. E. Van Pelt and D. B. Northrop, Arch. Biochem. Biophys., 1984, 230, 250–263 CrossRef CAS PubMed.
- C. Kim, D. Hesek, J. Zajicek, S. B. Vakulenko and S. Mobashery, Biochemistry, 2006, 45, 8368–8377 CrossRef CAS PubMed.
- G. Werner, B. Hildebrandt and W. Witte, Microb. Drug Resist., 2003, 9, 9–16 CrossRef PubMed.
- S. Jana and J. K. Deb, Biotechnol. Lett., 2005, 27, 519–524 CrossRef CAS PubMed.
- M. Tolmasky, in Enzyme-Mediated Resistance to Antibiotics, ed. R. Bonomo and M. Tolmasky, ASM Press, Washington, DC, 2007, pp. 35–52 Search PubMed.
- M. Latorre, P. Peñalver, J. Revuelta, J. L. Asensio, E. García-Junceda and A. Bastida, Chem. Commun., 2007, 2829 RSC.
- M. Okayama, K. Kimata and S. Suzuki, J. Biochem., 1973, 74, 1069–1073 CAS.
- N. B. Schwartz, L. Galligani, P. L. Ho and A. Dorfman, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 4047–4051 CrossRef CAS.
- F. Corzana, I. Cuesta, A. Bastida, A. Hidalgo, M. Latorre, C. González, E. García-Junceda, J. Jiménez-Barbero and J. L. Asensio, Chem. – Eur. J., 2005, 11, 5102–5113 CrossRef CAS PubMed.
- N. Sreerama and R. W. Woody, Anal. Biochem., 2000, 287, 252–260 CrossRef CAS PubMed.
- S. Chang, G. N. Singh, J. R. Phillips and J. S. Thorson, Curr. Opin. Biotechnol., 2011, 22, 800–808 CrossRef PubMed.
- K. Choonkeun, H. Dusan, Z. Jaroslav, S. B. Vakulenko and S. Mobashery, Biochemistry, 2006, 45, 8368–8377 CrossRef PubMed.
- C. A. Gates and D. B. Norton, Biochemistry, 1988, 27, 3820–3825 CrossRef CAS PubMed.
- E. Wright and E. H. Serpersu, Biochemistry, 2005, 44, 11581–11591 CrossRef CAS PubMed.
- J. Revuelta, T. Vacas, M. Torrado, F. Corzana, C. González, J. Jiménez-Barbero, M. Menéndez, A. Bastida and J. L. Asensio, J. Am. Chem. Soc., 2008, 16, 5086–5103 CrossRef PubMed.
- R. Matesanz, J. F. Diaz, F. Corzana, A. G. Santana, A. Bastida and J. L. Asensio, Chem. – Eur. J., 2012, 18, 2875–2889 CrossRef CAS PubMed.
- J. Revuelta, F. Corzana, A. Bastida and J. L. Asensio, Chem. – Eur. J., 2010, 16, 8635–8640 CrossRef CAS PubMed.
- F. R. Pinsetta, D. F. Kawano, M. Rodrigues de Carvalho, J. A. A. de Oliveira, A. P. Corrado, M. Hyppolito and I. Carvalho, Carbohydr. Res., 2013, 373(24), 97–102 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5md00496a |
| ‡ The term “decoy acceptors” was introduced by M. Okayama, et al.23 and N. B. Schwartz24 to designate those compounds, that being substrates for a particular enzyme are able to inhibit a metabolic pathway, to distinguish them from inhibitors, which are compounds that block the enzyme without being modified by it. |
| This journal is © The Royal Society of Chemistry 2016 |