Probing substrate binding to the metal binding sites in metallo-β-lactamase L1 during catalysis†‡
Mahesh
Aitha
,
Sameer
Al-Adbul-Wahid
,
David L.
Tierney
* and
Michael W.
Crowder
*
Department of Chemistry and Biochemistry, Miami University, 650 East High Street, Oxford, Ohio 45056, USA. E-mail: tiernedl@miamioh.edu; crowdemw@MiamiOH.edu; Fax: (513) 529 5715; Tel: (513) 529 2813
First published on 4th January 2016
Abstract
Metal ions in metallo-β-lactamases (MBLs) play a major role in catalysis. In this study we investigated the role of the metal ions in the Zn1 and Zn2 sites of MBL L1 during catalysis. A ZnCo (with Zn(II) in the invariant Zn1 site and Co(II) in the Zn2 site) analog of MBL L1 was prepared by using a biological incorporation method. Extended X-ray absorption fine structure (EXAFS) spectroscopic studies were used to confirm that the ZnCo analog was prepared. To study the roles of the Zn(II) and Co(II) ions during catalysis, rapid freeze quench (RFQ)-EXAFS studies were used to probe the reaction of the ZnCo-L1 analog with chromacef when quenched at 10 ms, 50 ms, and 100 ms. The L1-product complex was also analyzed with EXAFS spectroscopy. The data show that the Zn–Co distance is 3.49 Å in the resting enzyme and that this distance increases by 0.3 Å in the sample that was quenched at 10 ms. The average Zn–Co distance decreases at the other time points until reaching a distance of 3.58 Å in the L1-product complex. The data also show that a Co–S interaction is present in the 100 ms quenched sample and in the L1-product complex, which suggests that there is a significant rearrangement of product in the active site.
1. Introduction
The β-lactamases inactivate β-lactam-containing antibiotics, inhibitors of bacterial transpeptidases.1,2 As of late 2013, over 1300 β-lactamases have been identified.3 They have been sub-classified into 4 major groups, A–D, based on their sequence homologies.3–6 Class A, C, and D enzymes are called serine β-lactamases (SBLs), because they use an active site serine as the nucleophile to catalyze β-lactam hydrolysis.7 Several clinical inhibitors, such as clavulanic acid, sulbactam, tazobactam, and avibactam, are available to combat SBL-producing bacteria. In contrast, the class B enzymes, called metallo-β-lactamases (MBLs), require one or two Zn(II) ions for full catalytic activity.8 Several bacterial strains have been identified that produce chromosomally-encoded MBLs, such as Bacillus cereus (BcII),9Bacteroides fragilis (CcrA),10Elizabethkingia meningoseptica (BlaB),11 and Stenotrophomonas maltophilia (L1),12 and plasmid-encoded MBLs, such as IMP,13 VIM,14 and NDM15 variants, have shown remarkable horizontal transferability. The MBLs hydrolyze all known penicillins, carbapenems, and cephalosporins, and to date, there are no clinically-available inhibitors towards these enzymes.The MBLs have been further classified into three subgroups (B1–B3) based on their sequence homologies and the number of metal ions in their active sites.8 Most of the B1 and B3 enzymes bind 2 equivalents of Zn(II) at what are termed the Zn1 and Zn2 sites, with the major difference being the replacement of a Cys ligand at the Zn2 site (B1) with a His ligand (B3) (Fig. 1). A crystal structure of hydrolyzed moxalactam bound to the B3 MBL L1 provided the first glimpse of product binding at the metal site.16 In this structure, the Zn1 metal ion was five-coordinate (three His nitrogens, one oxygen from the newly-generated carboxylate, and a solvent in the bridging position), while the Zn2 ion was six-coordinate (two His nitrogens, an oxygen from Asp120, one oxygen from the invariant carboxylate at the substrate 2-position, the β-lactam nitrogen, and the bridging solvent). The metal–metal distance in this L1-product complex was 3.68 Å (0.18 Å greater than in the resting enzyme).
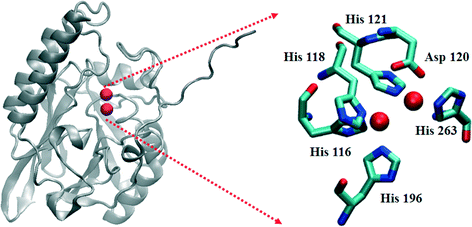 | ||
| Fig. 1 Structure of the L1 monomer. The active site is shown on the right. The spheres represent Zn(II) ions. | ||
Previously, we used rapid freezing techniques to examine MBL catalytic intermediates spectroscopically on the millisecond timescale.17–22 RFQ-EXAFS and RFQ-EPR studies showed different coordination numbers for the metal ions in L1 samples freeze quenched at 10 ms.18,20 RFQ-EPR studies on ZnCo-L1 demonstrated that Co(II) (in the Zn2 site) is five-coordinate in the resting state, proceeds through a four-coordinate species during the reaction, and is five-coordinate in the enzyme product complex.20 In contrast, RFQ-EXAFS studies on ZnZn-L1 indicated an increase in the average coordination number of Zn(II) in the sample freeze quenched at 10 ms and in the product complex, as compared to the resting enzyme.18 The EXAFS studies indicated an increase in the metal–metal distance in L1 after 10 ms of reaction with nitrocefin. The EXAFS studies indicated formation of a Zn–S interaction in the product complex,18,23 afforded by rotation around the C6–C7 bond after hydrolysis. As EXAFS is a bulk technique, it was not possible to explore the metal sites independently in the ZnZn-L1 analog or to determine if the enzyme-product complex was catalytically-competent (i.e., if the rotation occurred while the product was still in the active site or if it released, rotated, and rebound). To more closely examine the formation of this product complex, we report here RFQ-EXAFS studies of a heterobimetallic analog of L1 (ZnCo, with Zn(II) in the Zn1 site and Co(II) in the Zn2 site)20 that allowed us to probe each metal site independently.
2. Results and discussion
2.1 Over-expression and purification
The heterobimetallic (ZnCo) analog of L1 was prepared as previously described.20,24 The purified enzyme exhibited a kcat of 23 ± 1 s−1 and a Km of 2.5 ± 0.1 μM when using chromacef as substrate, and ICP-MS showed it bound 1.0 ± 0.1 equivalents of Zn(II) and 0.9 ± 0.1 equivalents of Co(II). The steady-state kinetic constants are similar to those previously reported for wild-type L1 (kcat = 25 ± 1 s−1; Km = 2.9 ± 0.1 μM).25 Stopped-flow kinetic studies were conducted on the reaction of 40 μM ZnCo-L1 and 40 μM chromacef (Fig. 2). The substrate decayed quickly over the first 100 ms of the reaction, and the concentration of the ring-opened, anionic intermediate maximized during the first 20 ms of the reaction. Under these conditions, the intermediate persisted at relatively high concentrations (around 30–35 μM) for over 50 ms. These progress curves are similar to those of NDM-1 when using chromacef as substrate22 and to those of L1 when using nitrocefin as substrate.20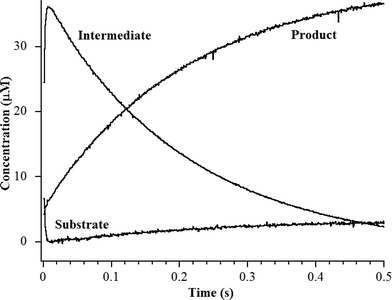 | ||
| Fig. 2 Stopped-flow kinetic traces of the reaction of 40 μM L1 with 40 μM chromacef in 30 mM Tris, pH 7.6, at 10 °C. Absorbance data were converted to concentration data using published extinction coefficients for substrate, intermediate, and product (see Materials and methods).22 | ||
2.2 EXAFS of unreacted ZnCo-L1
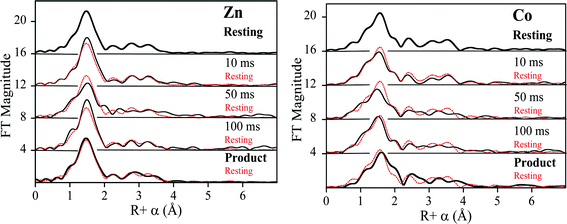
Previous EXAFS studies on the resting di-Zn enzyme yielded metal site metrics similar to those from available crystal structures,12 including an average of 4.5 N/O donors per Zn at 2.02 Å, 2.5 His ligands per Zn and a metal–metal separation of 3.42 Å.23 EXAFS of the ZnCo-L1 used for RFQ studies here were consistent with Zn(II) occupying the Zn1 site and Co occupying the Zn2 site. Fourier-transformed Zn K- and Co K-edge EXAFS spectra of resting ZnCo-L1 are shown in Fig. S1 and S2,† and fitting data are summarized in Tables S1 and S2.† Nonlinear least square regression analyses on the resting ZnCo-L1 sample showed a Zn K-edge first shell consistent with 4 N/O ligands at 2.01 Å and a second shell coordinated by 3 His; these values are consistent with the published ligand environment of the metal ion in the Zn1 site (Table 1).12 Inclusion of a metal–metal (Zn–Co) interaction at 3.48 Å in the model resulted in a 17% improvement in the fit residual (Fig. S1 and Table S1†). Analyses of the Co K-edge of the resting ZnCo-L1 sample were consistent with a first shell coordinated by 5 N/O ligands at 2.08 Å and a second shell coordinated by 2 His and 1 carbon at 2.41 Å, possibly from Asp120.12 The inclusion of a metal–metal interaction at 3.50 Å (Co–Zn) resulted in an improvement of the fit residual by 30% (Fig. S2 and Table S2†). The Zn K-edge and Co-K-edge EXAFS results showed a metal–metal distance of 3.48 Å (Zn–Co) and 3.50 Å (Co–Zn), which are the same within experimental error. The mean distance of 3.49 Å is considered the metal–metal distance in resting ZnCo-L1, which is 0.8 Å longer than in the ZnZn-analog. This result was not surprising because cobalt-substituted enzymes often show slightly increased metal–metal distances.26,27| Sample (ms) | Zn–N/O | Co–N/O | M–M (Zn) | M–M (Co) |
|---|---|---|---|---|
| 0 | 2.01 | 2.10 | 3.48 | 3.50 |
| 10 | 2.01 | 2.06 | 3.82 | 3.81 |
| 50 | 2.00 | 2.10 | 3.70 | 3.68 |
| 100 | 2.01 | 2.07 | 3.59 | 3.58 |
| Prod. | 2.02 | 2.08 | 3.59 | 3.57 |
EXAFS results on the resting ZnCo analog of L1 were consistent with data from previously-reported crystal structures;12 Zn(II) was present in the Zn1 site and Co(II) was present in the Zn2 site.20 Zn(II) was coordinated by 4 N/O scatterers in the first shell, which is consistent with 3 His and a hydroxyl bridge. Cobalt was coordinated by 5 N/O scatterers in the first shell, which is consistent with 2 His, 1 Asp, 1 terminally-bound water, and the hydroxyl bridge as ligands. Our EXAFS results confirm the fidelity of the heterobimetallic (ZnCo) analog of L1 and suggest that information gleaned using RFQ studies on this analog can be extrapolated to the naturally-occurring ZnZn analog. This analog also allowed us to address some finer points of the reaction mechanism catalyzed by L1.
2.3 RFQ-EXAFS studies on the reaction of ZnCo-L1 with chromacef
To probe the hydrolysis reaction, ZnCo-L1 (ca. 1 mM) was rapidly mixed with chromacef (ca. 1 mM), and the reaction was freeze-quenched at 10 ms, 50 ms, and 100 ms. Each sample was analyzed with EXAFS spectroscopy (Fig. 3), and fitting data are summarized in Table 1. Best fit results of the ZnCo-L1/chromacef sample freeze-quenched at 10 ms indicated a Zn K-edge first shell with 4 N/O ligands at 2.02 Å and a second shell coordinated by 3 His ligands. Inclusion of a metal–metal interaction at 3.82 Å improved the fit residual by 14% (Fig. S3 and Table S3†). Co K-edge fit results of the 10 ms sample revealed a first shell coordinated by 5 N/O ligands at 2.06 Å and a second shell coordinated by 2 His. While the inclusion of a Co–C interaction in the spectrum of resting ZnCo-L1 improved the fit residual, the inclusion of this same interaction in the spectrum of ZnCo-L1/chromacef quenched at 10 ms did not improve the fit and was not included in the final model. Inclusion of a metal–metal interaction at 3.81 Å to the model improved the fit residual by 34% (Fig. S4 and Table S4†). A similar increased metal–metal distance was previously observed in the ZnZn-L1/nitrocefin sample quenched at 10 ms.18 Our stopped-flow kinetic studies suggest that the sample quenched at 10 ms contains predominantly enzyme-intermediate complex (Fig. 2). These data strongly suggest that there was no change in the coordination number of either metal ion in the sample quenched at 10 ms.Fitting of the Zn K-edge of the EXAFS spectrum of ZnCo-L1/nitrocefin quenched at 50 ms suggests a first shell with 4 N/O ligands at 2.01 Å and a second shell coordinated with 3 His. Inclusion of a metal–metal interaction at 3.70 Å to the model improved the fit residual by 40% (Fig. S5 and Table S5†). Fitting of the Co K-edge of the EXAFS spectrum of ZnCo-L1/chromacef quenched at 50 ms indicates a first shell coordinated by 5 N/O ligands at 2.10 Å and a second shell coordinated with 2 His. Inclusion of a metal–metal distance of 3.68 Å improved the fit residual by 33%, and inclusion of 1 C interaction at 2.40 Å improved the fit residual by 20% (Fig. S6 and Table S6†). Inclusion of a Co–C interaction in the second shell resulted in an improvement in the fit residual, as observed in the analyses of the EXAFS data on the resting ZnCo-L1 sample. The metal–metal distance observed in the fits for the sample quenched at 50 ms was 0.12 Å shorter than that determined for the sample quenched at 10 ms. Stopped-flow kinetic studies suggest that this sample contains predominantly enzyme-intermediate complex; however, there are substantial amounts of enzyme-product (or product) complex in this sample (Fig. 2).
Fitting of the Zn K-edge of the EXAFS spectrum of ZnCo-L1/chromacef quenched at 100 ms resulted in a first shell coordinated by 4 N/O at 2.01 Å and a second shell coordinated by 3 His. Inclusion of a metal–metal interaction at 3.59 Å improved the fit residual by 18% (Fig. S7 and Table S7†). Best fits of the Co K-edge data for the ZnCo-L1/chromacef sample quenched at 100 ms showed 5 N/O scatterers at 2.07 Å; however, an improvement in the fit residual of 19% was observed when a model with 4 N/O ligands at 2.10 Å and 1 S ligand at 2.25 Å was used to fit the first shell data. The best fit of the data revealed a second shell with 2 His, and inclusion of a metal–metal interaction at 3.58 Å improved the fit residual by 10%. As with fits of the resting enzyme and the sample quenched at 50 ms, inclusion of 1 C at 2.40 Å in the second shell improved the fit residual by 37% (Fig. S8 and Table S8†). Five N/O scatterers in the first shell and 2 His, 1 C, and 1 Zn scatterers in second shell were used in the fit model to check the importance of Co–S contribution in the fit; however, fit results showed a 32% poorer fit as compared with the fit containing 1 S scatterer in the first shell along with 4 N/O scatterers (fit S-6, Table S8†). The observed first shell Co–S interaction in the spectrum of ZnCo-L1/chromacef quenched at 100 ms is a significant difference as compared with the fits of the resting sample and the samples freeze-quenched at 10 ms and 50 ms. The metal–metal distance observed in the sample quenched at 100 ms is 0.11 Å shorter than the metal–metal distance in the sample quenched at 50 ms. Stopped-flow kinetic studies suggest that this sample contains roughly equal amounts of enzyme-intermediate and enzyme-product (or product) (Fig. 2).
Best fits of the Zn K-edge data of the ZnCo-L1/chromacef product complex suggested a first shell with 4 N/O scatterers at 2.01 Å and a second shell coordinated by 3 His. Inclusion of a metal–metal interaction at 3.59 Å improved the fit residual by 53% (Fig. S9 and Table S9†). Best fit results of the Co K-edge showed a first shell coordinated by 4 N/O at 2.08 Å and 1 S at 2.30 Å, and the inclusion of the sulfur scatterer improved the fit residual by 40%. Multiple scattering analysis is consistent with the presence of 2 His scatterers, and inclusion of a metal–metal interaction at 3.57 Å improved the fit residual by 37%. Inclusion of 1 C interaction at 2.47 Å to the model improved the fit residual by 70% (Fig. S10 and Table S10†). The metal–metal distance observed in the ZnCo-L1/chromacef product complex is similar to the metal–metal distance in the sample quenched at 100 ms.
2.4. Revised reaction mechanism
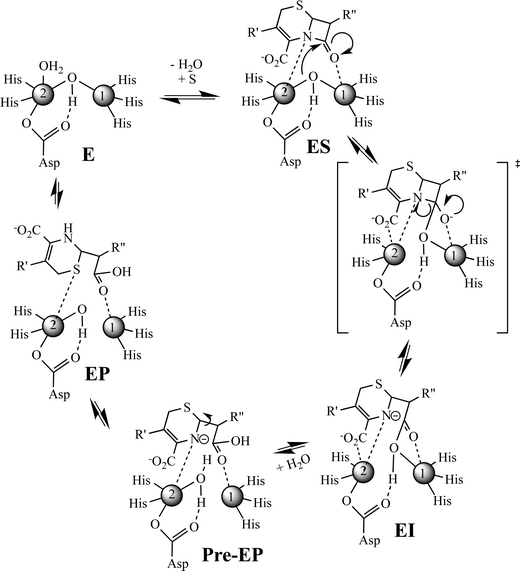
Previously, Hu et al. proposed a reaction mechanism for the hydrolysis of nitrocefin by L1.20 In this mechanism, the β-lactam carbonyl oxygen was proposed to interact with the metal ion in the Zn1 site, while the nitrogen lone pair on the nitrogen of the β-lactam interacted with the metal ion in the Zn2 site. Substrate binding was proposed to result in the loss of the bond between the metal ion in the Zn2 site and the bridging hydroxide, thereby generating a four-coordinate metal ion in the Zn2 site and a five-coordinate metal ion in the Zn1 site. The proposed mechanism showed that the Zn1-bound hydroxide is the reactive nucleophile, which was directed for attack on the β-lactam carbonyl by Asp120. The resulting, short-lived tetrahedral species was converted to a ring-opened, anionic intermediate (EI) after the loss of the β-lactam bond. The proposed mechanism showed that protonation of the anionic nitrogen occurs during the formation of a new bridging hydroxide/water, and it was assumed that this water came from bulk solvent. The resulting EP complex was in equilibrium with the resting enzyme, governed by the thermodynamic dissociation constant. The metal ion in the Zn2 site was proposed to be five-coordinate in the EP complex and in the resting state. The metal ion in the Zn1 site was proposed to be 4-coordinate in the resting enzyme and in the EI complex and 5-coordinate in the ES and EP complexes.EXAFS results on the ZnCo-L1/chromacef sample freeze quenched at 10 ms (EI is the predominant species, Fig. 2) showed a coordination number of 4 for Zn(II) and a coordination number of 5 for Co(II), which are identical to the coordination numbers of the resting enzyme. This result is not consistent with the reaction mechanism proposed earlier. Best fits of the EXAFS spectra of this sample demonstrate a large increase in the average metal–metal distance (3.81 vs. 3.49 Å), suggesting the loss of the bridging hydroxyl group in this sample, which is consistent the proposed structure of EI in the Hu mechanism.20 A similar increase in metal–metal distance was previously reported when the ZnZn analog of L1 was used in RFQ-EXAFS studies.18 There are several possible structures for EI that could explain a 4-coordinate metal ion the Zn1 site and a 5-coordinate metal ion in the Zn2 site. We favor a structure of EI that contains the Zn2 site with 3 enzyme–metal bonds and 2 substrate–metal bonds and the Zn1 site with 3 enzyme–metal bonds and 1 substrate–metal bond (Fig. 4). This structure is supported by several previous crystallographic studies on enzyme-product complexes.16,28
EXAFS results on the ZnCo-L1/chromacef sample freeze quenched at 50 ms also suggested a coordination number of 4 for Zn(II) and a coordination number of 5 for Co(II), which are identical to those of the resting enzyme and of the sample quenched at 10 ms. The metal–metal distance of this sample was found to be 3.69 Å, which is 0.12 Å smaller than the metal–metal distance of the sample quenched at 10 ms. We offer two possibilities to explain the decreased metal–metal distance in this sample compared to that of the sample quenched at 10 ms. First, the sample freeze quenched at 50 ms could be a mixture of EI (with a metal–metal distance of 3.82 Å) and of EP (with a metal–metal distance of 3.58 Å); however, EXAFS results indicated a uniform species at 50 ms rather than a mixture of EI and EP. Our stopped-flow studies also (Fig. 2) do not support a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 ratio of EI and EP at 50 ms; instead, there is roughly 25 μM EI and 15 μM EP. In addition, inclusion of a Co–S interaction in the first shell fit model of Co K-edge EXAFS data along with the 4 N/O scatterers did not improve the fit residual of the sample quenched at 50 ms, which argues against a sample containing a 50
50 ratio of EI and EP at 50 ms; instead, there is roughly 25 μM EI and 15 μM EP. In addition, inclusion of a Co–S interaction in the first shell fit model of Co K-edge EXAFS data along with the 4 N/O scatterers did not improve the fit residual of the sample quenched at 50 ms, which argues against a sample containing a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 ratio of EI and EP at 50 ms (see below). A second possibility is that a water binds to the Zn2 site, resulting in the loss of one metal–substrate bond (possibly the bond between Zn2 and oxygen of the carboxylate at the 2 position in substrate/intermediate) in EI. This “pre-EP” complex would be expected to have a metal–metal distance shorter than a species with three substrate–metal bonds (EI) because of the “strain” imposed by the substrate/intermediate. However, this “pre-EP” complex would be expected to have a metal–metal distance longer than a species with a bridging hydroxide. The newly-bound water could be oriented by Asp120 to protonate intermediate, and this hypothesis was offered several years ago based on mutagenesis studies.29 It is not clear from EXAFS data alone the exact structure of the species quenched at 50 ms.
50 ratio of EI and EP at 50 ms (see below). A second possibility is that a water binds to the Zn2 site, resulting in the loss of one metal–substrate bond (possibly the bond between Zn2 and oxygen of the carboxylate at the 2 position in substrate/intermediate) in EI. This “pre-EP” complex would be expected to have a metal–metal distance shorter than a species with three substrate–metal bonds (EI) because of the “strain” imposed by the substrate/intermediate. However, this “pre-EP” complex would be expected to have a metal–metal distance longer than a species with a bridging hydroxide. The newly-bound water could be oriented by Asp120 to protonate intermediate, and this hypothesis was offered several years ago based on mutagenesis studies.29 It is not clear from EXAFS data alone the exact structure of the species quenched at 50 ms.
EXAFS results on the ZnCo-L1/chromacef sample freeze quenched at 100 ms (EP) showed a coordination number of 4 for Zn(II) and a coordination number of 5 for Co(II), which is identical to the values in the resting sample, sample quenched at 10 ms, and the sample quenched at 50 ms. The metal–metal distance in this sample was 3.59 Å. Interestingly, the best fit of the EXAFS data for this sample required the inclusion of a sulfur scatterer. The EXAFS spectrum of the ZnCo-L1/product complex was very similar to that of the sample quenched at 100 ms, and our stopped-flow data show that, under the conditions used for these kinetic studies, the reaction contains roughly a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 mixture of EI and EP at 100 ms (Fig. 2).20 The only sulfur near the active site is the sulfur in substrate chromacef. Previously, RFQ-EXAFS data on ZnZn-L1 reacted with nitrocefin revealed the presence of a sulfur–Zn interaction in the spectrum of the enzyme-product complex, and this result was explained by a rotation of the C6–C7 bond in product.18,23 In this previous study, the authors could not ascertain whether the rotation of the bond occurred in the active site or whether the product released from the active site, there was the rotation, and the rotated product re-bound to the active site. With the data presented herein, it is most likely that the rotation of the bond occurs in the active site and that this species is kinetically-competent. These data also demonstrate that that sulfur coordinates to the metal ion in the Zn2 site, indicating that substrate binds with the 6-membered dihydrothiazine ring over the Zn2 site. This result is not consistent with either reaction mechanism presented earlier nor is it consistent with any of the published structures of MBL bound to reaction products.20,30 This result suggests that the β-lactam carbonyl is positioned over the Zn1 metal ion; however as discussed above, more studies are required to determine if the β-lactam carbonyl binds to Zn1.
50 mixture of EI and EP at 100 ms (Fig. 2).20 The only sulfur near the active site is the sulfur in substrate chromacef. Previously, RFQ-EXAFS data on ZnZn-L1 reacted with nitrocefin revealed the presence of a sulfur–Zn interaction in the spectrum of the enzyme-product complex, and this result was explained by a rotation of the C6–C7 bond in product.18,23 In this previous study, the authors could not ascertain whether the rotation of the bond occurred in the active site or whether the product released from the active site, there was the rotation, and the rotated product re-bound to the active site. With the data presented herein, it is most likely that the rotation of the bond occurs in the active site and that this species is kinetically-competent. These data also demonstrate that that sulfur coordinates to the metal ion in the Zn2 site, indicating that substrate binds with the 6-membered dihydrothiazine ring over the Zn2 site. This result is not consistent with either reaction mechanism presented earlier nor is it consistent with any of the published structures of MBL bound to reaction products.20,30 This result suggests that the β-lactam carbonyl is positioned over the Zn1 metal ion; however as discussed above, more studies are required to determine if the β-lactam carbonyl binds to Zn1.
3. Conclusions
In summary, this study was designed to build on a previous RFQ-EXAFS study, in which several questions remained. The preparation of a ZnCo-heterometallic analog of L1 allowed for refinement of the reaction mechanism gleaned from this previous study (Fig. 4). In this mechanism, the free enzyme has a 4-coordinate metal ion in the Zn1 site and a 5-coordinate metal ion in the Zn2 site. Our current RFQ system and the kinetics of L1 prevented EXAFS characterization of the ES complex for this reaction (see structure in Fig. 4); therefore, we included one possible structure for ES that would lead to an EI that is supported by our present results. In our proposed mechanism, we show the formation of a transition state species that has a tetrahedral carbon at the former β-lactam carbonyl. The breakdown of this transition state species results in the rapid formation of an anionic intermediate, which can be detected with stopped-flow UV-vis and our RFQ-EXAFS data. The loss of the bridging hydroxide results in a lengthening of the metal–metal distance during the very early stages of the reaction (before 10 ms). The corresponding EI converts to a pre-EP complex, which favors the protonation of the anionic nitrogen. We propose a loss of the Zn2–oxygen (of the carboxylate of the 6-membered ring of the substrate) bond in the pre-EP complex, which is replaced by a terminally-bound water to the Zn2 site. This loss of one metal ion-substrate/intermediate bond results in a decrease in the metal–metal distance of the pre-EP complex. A rotation of the substrate occurred in the sample quenched at 100 ms and in the product complex, and it suggests substrate rearrangement occurs in the active site (Fig. 4). It is important to note that there is no direct evidence yet available for interaction between the Zn1 and β-lactam carbonyl in substrate during the reaction catalyzed by the MBLs. Given the lack of a conserved positively-charged amino acid in the vicinity of the β-lactam carbonyl, it would be surprising that the metal ion in the Zn1 site does not activate the β-lactam carbonyl during catalysis. Future studies are planned to address this issue. This study suggests that the involvement of both (Zn1 and Zn2) sites in the β-lactam hydrolysis; therefore, both metal sites should be targeted for inhibitors in future design efforts.4. Experimental section
4.1. Over-expression and purification of L1
Heterobimetallic (ZnCo) L1 was prepared as previously described.20 Briefly, four liters of minimal medium were inoculated with 50 ml of an overnight culture, and the cultures were shaken at 37 °C until the OD600 reached 0.6–0.8. Cells were cooled to 18 °C for 15 min, and protein production was induced by making the cultures 0.5 mM in IPTG. After the cultures were shaken overnight (16 h), cells were harvested by centrifugation at 7000 RPM for 10 min in a GS3 rotor. The bacterial pellet was resuspended in 30 ml of Chelex100-treated 50 mM HEPES, pH 6.8, containing 500 mM NaCl. Cells were ruptured by passing the suspension two times through a French press at 1000 psi. The lysed cells were centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RPM for 25 min to clear the mixture, which was dialyzed overnight against 2 L of Chelex100-treated 50 mM HEPES, pH 6.8, to remove the NaCl. After centrifugation the resulting supernatant was loaded on a (1.5 × 20 with a 28 ml bed volume) SP-Sepharose column, which was equilibrated with 50 mM HEPES, pH 6.8, and protein was eluted with a 0–500 mM NaCl linear gradient in the same buffer. Protein purity was confirmed by SDS-PAGE, and protein concentration was determined by using a Nanodrop.31
000 RPM for 25 min to clear the mixture, which was dialyzed overnight against 2 L of Chelex100-treated 50 mM HEPES, pH 6.8, to remove the NaCl. After centrifugation the resulting supernatant was loaded on a (1.5 × 20 with a 28 ml bed volume) SP-Sepharose column, which was equilibrated with 50 mM HEPES, pH 6.8, and protein was eluted with a 0–500 mM NaCl linear gradient in the same buffer. Protein purity was confirmed by SDS-PAGE, and protein concentration was determined by using a Nanodrop.31
4.2 Metal analyses
A Perkin-Elmer Optima 7300 DV Inductively Coupled Plasma spectrometer with optical emission spectroscopy detection (ICP-OES) was used to determine the metal content of the L1 samples. Protein samples were diluted to 1 μM by using 50 mM Tris, pH 7.6. Standard calibration curves were generated with correlation coefficients greater than 0.999 by using Zn and Co metal standard solutions.224.3 Steady state kinetics
All steady state kinetic studies were conducted on an Agilent 8453 UV-vis diode array spectrophotometer at 25 °C. Michaelis constants (Km) and turnover numbers (kcat) were determined by monitoring product formation at 442 nm using chromacef as substrate in 50 mM cacodylate, pH 7.0. Rate data were converted to concentration data using the extinction coefficient of hydrolyzed chromacef (ε442nm = 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 M−1 cm−1).22,26 Chromacef was obtained from Dr. Larry Sutton from Sopharmia, Inc.321H NMR spectra of chromacef dissolved in 100% DMSO-d6 were similar to those spectra previously published,32 except we observed peaks corresponding to the cis-analog (double bond in the styrylbenzene substituent) that accounted for 20–30% of the total sample. Stocks of chromacef were prepared by dissolving the powder directly in 50 mM cacodylate, pH 7.0. Concentration versus time data were then fitted to the Michaelis–Menton equation, as previously reported.22
600 M−1 cm−1).22,26 Chromacef was obtained from Dr. Larry Sutton from Sopharmia, Inc.321H NMR spectra of chromacef dissolved in 100% DMSO-d6 were similar to those spectra previously published,32 except we observed peaks corresponding to the cis-analog (double bond in the styrylbenzene substituent) that accounted for 20–30% of the total sample. Stocks of chromacef were prepared by dissolving the powder directly in 50 mM cacodylate, pH 7.0. Concentration versus time data were then fitted to the Michaelis–Menton equation, as previously reported.22
4.4 Stopped-flow studies
Stopped-flow UV-vis studies were conducted using an Applied Photophysics SX 20 stopped-flow spectrophotometer, equipped with a photodiode array detector. Reactions of 40 μM L1 and 40 μM chromacef were thermostated at 10 °C, and the buffer was 30 mM Tris, pH 7.6. Reaction progress curves were generated by converting the absorbance data to concentration data using the following extinction coefficients: substrate ε378 = 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1, product ε442 = 18
000 M−1 cm−1, product ε442 = 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 M−1 cm−1, and intermediate ε575 = 22
600 M−1 cm−1, and intermediate ε575 = 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1.22,26
000 M−1 cm−1.22,26
4.5 EXAFS sample preparation
EXAFS samples (approximately 1 mM in protein) were prepared with 20% (v/v) glycerol as a glassing agent. Product samples were prepared by incubating an equal amount of 1 mM enzyme and 5 mM chromacef for one hour on ice. All EXAFS samples of resting L1 or the L1-product complex were loaded in Lucite cuvettes with 6 μm polypropylene windows, frozen rapidly, and stored in liquid nitrogen.Freeze-quenched EXAFS samples were generated using a modified Update Instruments (Madison, WI) rapid-freeze-quench (RFQ) system. All enzyme and substrate starting concentrations were 1 mM and 5 mM, respectively, prepared in metal-free (Chelex100, Bio-Rad) 50 mM HEPES, pH 7.0, containing 20% v/v glycerol. A model 715 Update Instruments ram controller was used to drive a PMI-Kollmorgen stepping motor (model 00D12F-02001-1) connected to a ram that in turn drove the Update Instrument syringes. The syringes, mixer, and tubing were all contained in a watertight bath that was maintained at 2 °C.25,33 Immediately prior to sample collection, the nozzle (and, for the shortest reaction times, the attached mixer) was removed from the bath and held 5 mm above the surface of 2-methylbutane contained in a collecting funnel and maintained at −130 °C by a surrounding bath (Update Instruments) of liquid nitrogen cooled 2-methylbutane. Samples were packed into home-designed EXAFS sample holders at −130 °C; excess 2-methylbutane was decanted, and samples were stored in liquid nitrogen until data collection. The RFQ system was calibrated by comparing the development of a low-spin Fe(III) EPR signal and the disappearance of a high-spin Fe(III) EPR signal with the associated optical changes at 636 nm using stopped-flow spectrophotometry, upon mixing myoglobin with an excess of sodium azide. The shortest, total effective reaction time that could be achieved with the RFQ system was 10 ms.21,33
4.6 EXAFS spectroscopy
X-ray absorption spectra were measured at the National Synchrotron Light Source (NSLS), beamline X3B, using a Si (111) double-crystal monochromator; harmonic rejection was accomplished with a Ni mirror. Fluorescence excitation spectra for all samples were measured with a 31-element solid-state Ge detector array. Samples were held at approximately 15 K in a Displex cryostat. EXAFS data collection and reduction were performed according to published procedures.34 Data were measured in duplicate, six scans each on two independently-prepared samples. Fits to the two data sets were equivalent. The experimental spectra presented here are the averaged data sets (12 scans per sample). The data were converted from energy to k-space using E0 = 9680 eV and 7730 eV for Zn and Co, respectively.Fourier-filtered EXAFS data were fitted using the nonlinear least-squares engine of IFEFFIT, which is distributed with SixPack (SixPack is available free of charge from http://www-ssrl.slac.stanford.edu/exafspak.html; IFEFFIT is open source software available from http://cars9.uchicago.edu/ifeffit/Ifeffit). Theoretical amplitude and phase functions were calculated with FEFF v. 8.00.35 Zinc–nitrogen single-scattering and zinc–imidazole multiple-scattering was calibrated to the experimental EXAFS of zinc tetrakis-1-methylimidazole Zn(II) perchlorate, [Zn(MeIm)4][ClO4]2. Zinc–sulfur scattering was calibrated to the experimental EXAFS of tetrabutylammonium zinc tetramesitylthiolate, [Bu4N]2[Zn(Smes)4]. Optimum scale factors (Sc) and ΔE0 were derived from fits to the model data (Sc = 0.78 (Zn–N) or 0.91 (Zn–S); ΔE0 = −21 eV), and they were held fixed at these values for fits to metalloprotein data. The models used for the calibration of cobalt–nitrogen (and cobalt–imidazole) and cobalt–sulfur scattering were, respectively, hexakis-imidazole cobalt(II) perchlorate, [Co(Im)6][ClO4]2 and tetrabutylammonium cobalt(II) tetramesitylthiolate, [Bu4N]2[Co(Smes)4], respectively. The resulting Sc and ΔE0 (Sc = 0.79 (Co–N) or 0.85 (Co–S); ΔE0 = −21 eV) were held fixed at these calibrated values in subsequent fits to metalloprotein data. First shell fits were then obtained for all reasonable coordination numbers, including mixed nitrogen/oxygen/sulfur ligation, while allowing the absorber-scattered distance, Ras, and the Debye–Waller factor, σas2, to vary. Multiple scattering contributions from histidine ligands were fitted according to published procedures.36 Metal–metal (zinc–cobalt and cobalt–cobalt) scattering were modeled with reference to the experimental EXAFS of Zn2 (salpn)2 and Co2 (salpn)2.
Acknowledgements
This work was supported by the National Institutes of Health (GM093987 to MWC and DLT; P30-EB-009998 to the Center for Synchrotron Biosciences from the NIBIB, which supports beamline X3B at the NSLS), and the National Science Foundation (CHE-1151658 to MWC and DLT).References
- J. R. Knowles, Enzyme Inhibitors, Weinheim, Verlag Chemie, 1980, pp. 163–167 Search PubMed.
- J. R. Knowles, Acc. Chem. Res., 1985, 18, 97–104 CrossRef.
- K. Bush, J. Infect. Chemother., 2013, 19, 549–559 CrossRef CAS PubMed.
- R. P. Ambler, Philos. Trans. R. Soc., B, 1980, 289, 321–331 CrossRef CAS.
- B. Jaurin and T. Grundstrom, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 4897–4901 CrossRef CAS.
- A. Mederios, Br. Med. Bull., 1984, 40, 18–27 Search PubMed.
- S. M. Drawz and R. A. Bonomo, Clin. Microbiol. Rev., 2010, 23, 160–201 CrossRef CAS PubMed.
- M. W. Crowder, J. Spencer and A. J. Vila, Acc. Chem. Res., 2006, 39, 721–728 CrossRef PubMed.
- A. Carfi, S. Pares, E. Duée, M. Galleni, C. Duez, J. M. Frère and O. Dideberg, EMBO J., 1995, 14, 4914–4921 CAS.
- Z. Wang, W. Fast and S. J. Benkovic, Biochemistry, 1999, 38, 10013–10023 CrossRef CAS PubMed.
- I. Garcia-Saez, J. Hopkins, C. Papamicael, N. Franceschini, G. Amicosante, G. M. Rossolini, M. Galleni, J. M. Frère and O. Dideberg, J. Biol. Chem., 2003, 278, 23868–23873 CrossRef CAS PubMed.
- J. H. Ullah, T. R. Walsh, I. A. Taylor, D. C. Emery, C. S. Verma, S. J. Gamblin and J. Spencer, J. Mol. Biol., 1998, 284, 125–136 CrossRef CAS PubMed.
- J. H. Toney, G. G. Hammond, P. M. Fitzgerald, N. Sharma, J. M. Balkovec, G. P. Rouen, S. H. Olson, M. L. Hammond, M. L. Greenlee and Y. D. Gao, J. Biol. Chem., 2001, 276, 31913–31918 CrossRef CAS PubMed.
- N. Franceschini, B. Caravelli, J. D. Docquier, M. Galleni, J. M. Frère, G. Amicosante and G. M. Rossolini, Antimicrob. Agents Chemother., 2000, 44, 3003–3007 CrossRef CAS PubMed.
- D. Yong, M. A. Toleman, C. G. Giske, H. S. Cho, K. Sundman, K. Lee and T. R. Walsh, Antimicrob. Agents Chemother., 2009, 53, 5046–5054 CrossRef CAS PubMed.
- J. Spencer, J. Read, R. B. Sessions, S. Howell, G. M. Blackburn and S. J. Gamblin, J. Am. Chem. Soc., 2005, 127, 14439–14444 CrossRef CAS PubMed.
- M. Aitha, L. Moritz, I. D. Sahu, O. Sanyurah, Z. Roche, R. McCarrick, G. A. Lorigan, B. Bennett and M. W. Crowder, J. Biol. Inorg. Chem., 2015, 20, 585–594 CrossRef CAS PubMed.
- R. M. Breece, Z. Hu, B. Bennett, M. W. Crowder and D. L. Tierney, J. Am. Chem. Soc., 2009, 131, 11642–11643 CrossRef CAS PubMed.
- R. M. Breece, L. I. Llarrull, M. F. Tioni, A. J. Vila and D. L. Tierney, J. Inorg. Biochem., 2012, 111, 182–186 CrossRef CAS PubMed.
- Z. Hu, G. Periyannan, B. Bennett and M. W. Crowder, J. Am. Chem. Soc., 2008, 130, 14207–14216 CrossRef CAS PubMed.
- N. P. Sharma, C. Hajdin, S. Chandrasekar, B. Bennett, K. W. Yang and M. W. Crowder, Biochemistry, 2006, 45, 10729–10738 CrossRef CAS PubMed.
- H. Yang, M. Aitha, A. M. Hetrick, T. K. Richmond, D. L. Tierney and M. W. Crowder, Biochemistry, 2012, 51, 3839–3847 CrossRef CAS.
- A. Costello, G. Periyannan, K. W. Yang, M. W. Crowder and D. L. Tierney, J. Biol. Inorg. Chem., 2006, 11, 351–358 CrossRef CAS PubMed.
- Z. Hu, G. R. Periyannan and M. W. Crowder, Anal. Biochem., 2008, 378, 177–183 CrossRef CAS PubMed.
- M. Aitha, T. K. Richmond, Z. Hu, A. Hetrick, R. Reese, A. Gunther, R. McCarrick, B. Bennett and M. W. Crowder, J. Inorg. Biochem., 2014, 136, 40–46 CrossRef CAS PubMed.
- M. Aitha, A. Marts, A. Bergstrom, A. Moller, L. Moritz, L. Turner, J. Nix, R. Bonomo, R. Page, D. L. Tierney and M. W. Crowder, Biochemistry, 2014, 53, 7321–7331 CrossRef CAS PubMed.
- H. Yang, M. Aitha, A. R. Marts, A. Hetrick, B. Bennett, M. W. Crowder and D. L. Tierney, J. Am. Chem. Soc., 2014, 136, 7273–7285 CrossRef CAS PubMed.
- H. Feng, J. Ding, D. Zhu, X. Liu, X. Xu, Y. Zhang, S. Zang, D.-C. Wang and W. Liu, J. Am. Chem. Soc., 2014, 136, 14694–14697 CrossRef CAS PubMed.
- J. D. Garrity, A. L. Carenbauer, L. R. Herron and M. W. Crowder, J. Biol. Chem., 2004, 279, 920–927 CrossRef CAS PubMed.
- M. F. Tioni, L. I. Llarrull, A. A. Poeylaut-Palena, M. A. Marti, M. Saggu, G. R. Periyannan, E. G. Mata, B. Bennett, D. H. Murgida and A. J. Vila, J. Am. Chem. Soc., 2008, 130, 15852–15863 CrossRef CAS PubMed.
- M. W. Crowder, T. R. Walsh, L. Banovic, M. Pettit and J. Spencer, Antimicrob. Agents Chemother., 1998, 42, 921–926 CAS.
- S. Yu, A. Vosbeek, K. Corbella, J. Severson, J. Schesser and L. D. Sutton, Anal. Biochem., 2012, 428, 96–98 CrossRef CAS PubMed.
- J. D. Garrity, B. Bennett and M. W. Crowder, Biochemistry, 2005, 44, 1078–1087 CrossRef CAS PubMed.
- D. L. Tierney and G. Schenk, Biophys. J., 2014, 107, 1263–1272 CrossRef CAS PubMed.
- A. L. Ankudinov, B. Ravel, J. J. Rehr and S. D. Conradson, Phys. Rev. B: Condens. Matter Mater. Phys., 1998, 58, 7565–7576 CrossRef CAS.
- P. W. Thomas, E. M. Stone, A. Costello, D. L. Tierney and W. Fast, Biochemistry, 2005, 44, 7559–7569 CrossRef CAS PubMed.
Footnotes |
| † The authors declare no competing interests. |
| ‡ Electronic supplementary information (ESI) available. See DOI: 10.1039/c5md00358j |
| This journal is © The Royal Society of Chemistry 2016 |