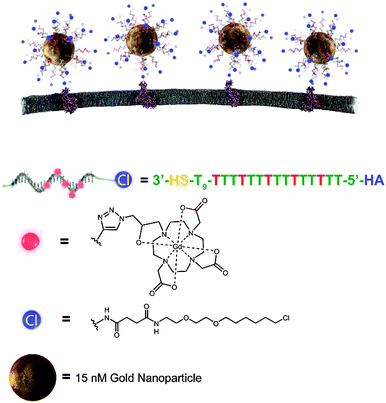
Targeted delivery of gold nanoparticle contrast agents for reporting gene detection by magnetic resonance imaging†
Luke F.
Vistain
 ,
Matthew W.
Rotz
,
Richa
Rathore
,
Adam T.
Preslar
and
Thomas J.
Meade
*
,
Matthew W.
Rotz
,
Richa
Rathore
,
Adam T.
Preslar
and
Thomas J.
Meade
*
Department of Chemistry, Molecular Biosciences, Neurobiology, Biomedical Engineering, and Radiology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113, USA. E-mail: tmeade@northwestern.edu
First published on 21st October 2015
Abstract
Detection of protein expression by MRI requires a high payload of Gd(III) per protein binding event. Presented here is a targeted AuDNA nanoparticle capable of delivering several hundred Gd(III) chelates to the HaloTag reporter protein. Incubating this particle with HaloTag-expressing cells produced a 9.4 contrast-to-noise ratio compared to non-expressing cells.
The field of molecular imaging is motivated by the need for techniques that enable in vivo visualization of biochemical processes, biomarkers, and gene expression.1–3 Magnetic resonance imaging (MRI) is an appealing modality for molecular imaging because it provides excellent spatial resolution (<100 μm), detailed anatomical information, and does not require exposing the subject to potentially harmful ionizing radiation.4 Where native MR contrast is insufficient, contrast agents (CAs), such as those based on paramagnetic gadolinium, are used to shorten water proton relaxation times, increasing image contrast. However, the low sensitivity of Gd(III) CAs has limited their utility in molecular imaging due to the high concentrations required to produce contrast (10–100 μM).5 Crucially, many biomolecules are present at concentrations (0.1–1 μM) that are below the detection limit of Gd(III) CAs.6 To date, molecular imaging using Gd(III) has been limited to a small number of biomarkers present at high concentrations in vivo.7–13
The low sensitivity of Gd(III) CAs has made it challenging to develop MR reporter genes. Many of these genes are from endogenous proteins, produce negative contrast (bright-to-dark), and generate only modest contrast overall.14–19 Furthermore, none of the genes in these systems have been integrated into existing reporter gene platforms. As such, their utility is limited because they require a unique genetic element dedicated solely to MR detection.
An ideal reporter platform for MR monitoring of gene expression presents extracellularly, integrates into an existing reporter gene platform, provides irreversible binding of molecular probes, and contains the necessary signal amplification to overcome the low sensitivity of Gd(III) probes. The HaloTag reporter gene system addresses these challenges.20 HaloTag is an engineered haloalkane dehalogenase that can be expressed on the outer surface of the plasma membrane.21 The enzyme active site has been modified to catalyze covalent bond formation with terminal haloalkanes, promoting superior probe retention.20 Because haloalkanes are virtually absent from eukaryotic systems, HaloTag and its targeting group create an orthogonal binding pair. Furthermore, HaloTag can readily form functional fusions with a variety of proteins.22 The specificity and versatility of the HaloTag system make it attractive as an MR reporter gene. In addition, it operates as a variable-output reporter gene, whereby the researcher can select the nature of the output by choosing the appropriate HaloTag-targeted agent. For this reason, a variety of imaging agents, including fluorophores, PET agents, MR agents, and quantum dots have been successfully targeted to HaloTag.21,23–25 However, coupling HaloTag expression to the production of T1 contrast demands significant signal amplification.
Spherical nucleic acids (SNAs) have the potential to address this design requirement.26 Extensive work on over 100 cell types showed that SNAs exhibit high biocompatibility and low toxicity in vitro and in vivo.27–29 Furthermore, previous work with SNAs developed a multiplexing strategy to deliver a high payload of Gd(III) chelates.30 In this case, the SNAs were not targeted and their cellular uptake was a result of SNAs binding to scavenger receptors on the cell surface.31 Although SNAs can be targeted using antibodies or aptamers, there is no precedent for SNA targeting using small molecule ligands.32,33 We demonstrate that HaloTag-dependent MR contrast enhancement can be achieved by using a HT-targeted AuDNA–Gd(III) nanoparticle.
HaloTag-targeted AuDNA–Gd(III) nanoparticles were synthesized according to Scheme 1. A 24 mer polydeoxythymidine (dT) oligonucleotide bearing a protected 3′ thiol and a 5′ terminal haloalkane (HA) moiety for HaloTag binding was synthesized (Schemes S1 and S2, ESI†). The oligonucleotide included modified dT bases bearing terminal alkyne functionality at five positions internal to each strand. Using a Gd(III) chelate bearing an azide functionality, a Cu(I)-catalyzed 1,3 dipolar cycloaddition was conducted to produce the complete HaloTag-targeted Gd(III) DNA (Scheme S3, ESI†). The purified oligonucleotide was deprotected to expose the 3′ thiol and conjugated to gold nanoparticles using a salt aging procedure.34
The density of oligonucleotide loading on the particle surface was determined by calculation of the Gd/Au ratio using Inductively Coupled Plasma Mass Spectrometry (ICP-MS).30 Results indicate that the average loading of DNA was 100 ± 10 strands per particle, yielding a Gd(III)-chelate payload of 500 ± 60 per particle. The T1 relaxivity (r1) was measured to be 16 ± 3 mM−1 s−1 per Gd(III) at 37 °C and 1.41 T, and the T2 relaxivity (r2) was measured to be 28 ± 3 mM−1 s−1 per Gd(III) (Fig. S3 and S4, ESI†). We hypothesized that this degree of signal amplification would enable visualization of surface receptors that would be below the detection limit of individual Gd(III) chelates.
The U-2 OS HT-ECS (HT+) cell line constitutively expresses extracellular HaloTag. Flow cytometry was used to quantify the number of HaloTag proteins expressed on the outer surface of the plasma membrane by using cell-impermeable HaloTag-targeted AlexaFluor488 dye. Unlike antibody-based cell surface stains, each HaloTag protein binds irreversibly to only one molecule of AlexaFluor488.20 Therefore, the number of HaloTag proteins present on the surface of these cells could be quantified by fluorescence. The HT+ cell line was observed to express 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800
800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 500
000 ± 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies of HaloTag on its surface (Fig. S5, ESI†). Using the common volume approximation of 2 pL per cell, this yields a concentration of 1.6 ± 0.4 μM HaloTag that is accessible to the cell surface.35 Though this concentration of HaloTag corresponds to the top decile of protein expression in the mammalian cell, a Gd(III) agent bound to HaloTag in one-to-one stoichiometry would still fail to achieve a detectable concentration (Fig. S9, ESI†).36
000 copies of HaloTag on its surface (Fig. S5, ESI†). Using the common volume approximation of 2 pL per cell, this yields a concentration of 1.6 ± 0.4 μM HaloTag that is accessible to the cell surface.35 Though this concentration of HaloTag corresponds to the top decile of protein expression in the mammalian cell, a Gd(III) agent bound to HaloTag in one-to-one stoichiometry would still fail to achieve a detectable concentration (Fig. S9, ESI†).36
To directly observe AuDNA–Gd(III)–HA binding to HaloTag, transmission electron microscopy (TEM) was used to identify membrane binding. A change in the subcellular localization of AuDNA–Gd(III)–HA was observed when comparing HT+ cells to otherwise identical cells that do not express HaloTag (U-2 OS (HT−)) (Fig. 1a, b and Fig. S10, ESI†). Both cell lines showed intracellular clusters of nanoparticles, which are likely endosomes or lysosomes (Fig. 1b). This observation is consistent with the previously proposed mechanism of scavenger receptor uptake.31 However, only HT+ cells showed large numbers of particles adjacent to the plasma membrane (Fig. 1a).
Flow cytometry was used to measure binding of AuDNA–Gd(III)–HA to HaloTag on the plasma membrane. HT+ cells were first incubated with AuDNA–Gd(III)–HA, followed by labeling with HaloTag-targeted AlexaFluor488. When AuDNA–Gd(III)–HA binds to HaloTag on the cell surface, fewer sites remain for AlexaFluor488 binding. Therefore, AuDNA–Gd(III)–HA binding to HaloTag was monitored by the loss of AlexaFluor488 fluorescence in both a time and dose dependent manner (Fig. 1c and d).
After a 24 hour incubation period, HaloTag binding was observed to plateau at an incubation concentration of 40 nM nanoparticles, with greater than 60% binding as low as 10 nM (Fig. 1e). In addition, HaloTag was saturated after 8 hours of incubation at 40 nM (Fig. 1f and Fig. S11, ESI†). The binding kinetics of AuDNA–Gd(III)–HA are slower than HaloTag-targeted small molecules.20 This is likely the result of a complex protein corona that forms around nanoparticles when exposed to serum proteins and reduces access to targeting groups.37,38 Importantly, these data suggest that saturated cells will have an average of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 730
730![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nanoparticles associated with the cell as a result of HaloTag binding. If each particle contributes 500 Gd(III) chelates, we predict that HaloTag saturation will result in 1.5 fmol Gd(III) per cell. This concentration is an order of magnitude above the most conservative estimates for the detection limit.6
000 nanoparticles associated with the cell as a result of HaloTag binding. If each particle contributes 500 Gd(III) chelates, we predict that HaloTag saturation will result in 1.5 fmol Gd(III) per cell. This concentration is an order of magnitude above the most conservative estimates for the detection limit.6
To determine the accuracy of these uptake approximations, HT− and HT+ cells were incubated with AuDNA–Gd(III)–HA and Gd(III) uptake was measured using ICP-MS. These data can be used to determine the signal contributions that depend on the expression of HaloTag and untargeted uptake of AuDNA–Gd(III)–HA. It is likely that untargeted uptake of AuDNA–Gd(III)–HA is due to AuDNA nanoparticles binding scavenger receptors, as previously reported.31 HT− cells display saturation for both incubation concentration and time, which is the expected trend as available scavenger receptors are depleted (Fig. 2a and b). The Gd(III) uptake values for HT+ cells continue to increase beyond the measured values for HT− cells at several time points and concentrations (Fig. 2a and b). After 8 hours of incubation with 40 nM AuDNA–Gd(III)–HA, HT+ cells accumulated threefold more Gd(III) than HT− cells (Fig. 2b). HaloTag expression resulted in an additional accumulation of 1.16 ± 0.08 fmol Gd(III) per cell over HT− cells. This value is very close to the predicted value of 1.5 fmol Gd(III) per cell calculated from the expression level of HaloTag (Fig. 1). A likely explanation for the reduced uptake is slow degradation of the nanoparticles over the course of the incubation. While AuDNA nanoparticles are resistant to the activity of DNase, the reaction still proceeds at a measurable rate.39
Cell pellet MR images were taken to verify that the additional uptake conferred by HaloTag expression would effectively translate to contrast in an MR image acquired at 7 T. As expected, both cells lines showed an increase in signal intensity over unlabeled cells after incubation with AuDNA–Gd(III)–HA (Fig. 2c). Significantly, HaloTag-expressing cells are clearly distinguishable from cells that do not express HaloTag. This contrast enhancement was apparent in both the greyscale T1 weighted image (Fig. S7, ESI†) and the corresponding intensity map (Fig. 2c). The observed T1 shortening was significant both for the image slice shown and for the average of 4 slices (Fig. 2c and Fig. S5, ESI†). From this image the contrast-to-noise ratio (CNR) between HT+ and HT− cells was calculated to be 9.4. Based on the clinical standard for MR imaging, CNR values above 5 are considered to be visually “obvious.”5
We have shown that a Gd(III)-conjugated AuDNA nanoparticle can be targeted the HaloTag protein and produce detectable MR contrast. The signal amplification afforded by nanoparticle targeting enabled delivery of Gd(III) at millimolar cellular concentrations for a 2 pL cell. As a result, HaloTag's reporter gene functionality has been expanded to include an MR output. Beyond HaloTag, this technology has shown that in principle, cell surface receptors can be detected using a T1 contrast agent. The straightforward chemistry and broad applicability of SNA's suggests that these nanoparticles can be easily adapted to other surface receptors that bind to a small molecule ligands.
This work was supported by the National Institutes of Health (NIH grants R0I EB005866 and P01 HL108795) and by a National Cancer Institute Ruth L. Kirschstein National Research Service Award (F31 CA174281). The U-2 OS HT-ECS cell line was a generous gift from Promega.
Notes and references
- M. L. James and S. S. Gambhir, Physiol. Rev., 2012, 92, 897–965 CrossRef CAS PubMed.
- A. S. Harney and T. J. Meade, Future Med. Chem., 2010, 2, 503–519 CrossRef CAS PubMed.
- J. L. Major and T. J. Meade, Acc. Chem. Res., 2009, 42, 893–903 CrossRef CAS PubMed.
- M. Rudin and R. Weissleder, Nat. Rev. Drug Discovery, 2003, 2, 123–131 CrossRef CAS PubMed.
- E. T. Ahrens, U. Rothbächer, R. E. Jacobs and S. E. Fraser, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 8443–8448 CrossRef CAS.
- K. Hanaoka, A. J. Lubag, A. Castillo-Muzquiz, T. Kodadek and A. D. Sherry, Magn. Reson. Imaging, 2008, 26, 608–617 CrossRef CAS PubMed.
- J. Vymazal, E. Spuentrup, G. Cardenas-Molina, A. J. Wiethoff, M. G. Hartmann, P. Caravan and E. C. J. Parsons, Invest. Radiol., 2009, 44, 697–704 CrossRef CAS PubMed.
- L. M. De León-Rodríguez, A. Lubag, D. G. Udugamasooriya, B. Proneth, R. A. Brekken, X. Sun, T. Kodadek and A. Dean Sherry, J. Am. Chem. Soc., 2010, 132, 12829–12831 CrossRef PubMed.
- S. Geninatti Crich, D. Alberti, I. Szabo, S. Aime and K. Djanashvili, Angew. Chem., 2013, 52, 1161–1164 CrossRef CAS PubMed.
- T. L. Kalber, N. Kamaly, P. W. So, J. A. Pugh, J. Bunch, C. W. McLeod, M. R. Jorgensen, A. D. Miller and J. D. Bell, Mol. Imag. Biol., 2011, 13, 653–662 CrossRef PubMed.
- W.-h. Li, G. Parigi, M. Fragai, C. Luchinat and T. J. Meade, Inorg. Chem., 2002, 41, 4018–4024 CrossRef CAS PubMed.
- M. C. Heffern, L. M. Matosziuk and T. J. Meade, Chem. Rev., 2014, 114, 4496–4539 CrossRef CAS PubMed.
- S. R. Banerjee, E. J. Ngen, M. W. Rotz, S. Kakkad, A. Lisok, R. Pracitto, M. Pullambhatla, Z. Chen, T. Shah, D. Artemov, T. J. Meade, Z. M. Bhujwalla and M. G. Pomper, Angew. Chem., Int. Ed., 2015, 54, 10778–10782 CrossRef CAS PubMed.
- B. Cohen, H. Dafni, G. Meir, A. Harmelin and M. Neeman, Neoplasia, 2005, 7, 109–117 CrossRef CAS PubMed.
- O. Zurkiya, A. W. Chan and X. Hu, Magn. Reson. Med., 2008, 59, 1225–1231 CrossRef CAS PubMed.
- B. B. Bartelle, K. U. Szulc, G. A. Suero-Abreu, J. J. Rodriguez and D. H. Turnbull, Magn. Reson. Med., 2013, 70, 842–850 CrossRef PubMed.
- A. Y. Louie, M. M. Huber, E. T. Ahrens, U. Rothbacher, R. Moats, R. E. Jacobs, S. E. Fraser and T. J. Meade, Nat. Biotechnol., 2000, 18, 321–325 CrossRef CAS PubMed.
- P. S. Patrick, J. Hammersley, L. Loizou, M. I. Kettunen, T. B. Rodrigues, D.-E. Hu, S.-S. Tee, R. Hesketh, S. K. Lyons, D. Soloviev, D. Y. Lewis, S. Aime, S. M. Fulton and K. M. Brindle, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 415–420 CrossRef CAS PubMed.
- R. A. Moats, S. E. Fraser and T. J. Meade, Angew. Chem., 1997, 36, 726–728 CAS.
- G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina, C. Zimprich, M. G. Wood, R. Learish, R. F. Ohana, M. Urh, D. Simpson, J. Mendez, K. Zimmerman, P. Otto, G. Vidugiris, J. Zhu, A. Darzins, D. H. Klaubert, R. F. Bulleit and K. V. Wood, ACS Chem. Biol., 2008, 3, 373–382 CrossRef CAS PubMed.
- N. Kosaka, M. Ogawa, P. L. Choyke, N. Karassina, C. Corona, M. McDougall, D. T. Lynch, C. C. Hoyt, R. M. Levenson, G. V. Los and H. Kobayashi, Bioconjugate Chem., 2009, 20, 1367–1374 CrossRef CAS PubMed.
- R. F. Ohana, R. Hurst, J. Vidugiriene, M. R. Slater, K. V. Wood and M. Urh, Protein Expression Purif., 2011, 76, 154–164 CrossRef CAS PubMed.
- Y. Zhang, M. K. So, A. M. Loening, H. Yao, S. S. Gambhir and J. Rao, Angew. Chem., 2006, 45, 4936–4940 CrossRef CAS PubMed.
- R. C. Strauch, D. J. Mastarone, P. A. Sukerkar, Y. Song, J. J. Ipsaro and T. J. Meade, J. Am. Chem. Soc., 2011, 133, 16346–16349 CrossRef CAS PubMed.
- H. Hong, H. A. Benink, Y. Zhang, Y. Yang, H. T. Uyeda, J. W. Engle, G. W. Severin, M. G. McDougall, T. E. Barnhart, D. H. Klaubert, R. J. Nickles, F. Fan and W. Cai, Am. J. Transl. Res., 2011, 3, 392–403 CAS.
- J. I. Cutler, E. Auyeung and C. A. Mirkin, J. Am. Chem. Soc., 2012, 134, 1376–1391 CrossRef CAS PubMed.
- D. A. Giljohann, D. S. Seferos, W. L. Daniel, M. D. Massich, P. C. Patel and C. A. Mirkin, Angew. Chem., 2010, 49, 3280–3294 CrossRef CAS PubMed.
- S. A. Jensen, E. S. Day, C. H. Ko, L. A. Hurley, J. P. Luciano, F. M. Kouri, T. J. Merkel, A. J. Luthi, P. C. Patel, J. I. Cutler, W. L. Daniel, A. W. Scott, M. W. Rotz, T. J. Meade, D. A. Giljohann, C. A. Mirkin and A. H. Stegh, Sci. Transl. Med., 2013, 5, 209ra152 Search PubMed.
- M. D. Massich, D. A. Giljohann, A. L. Schmucker, P. C. Patel and C. A. Mirkin, ACS Nano, 2010, 4, 5641–5646 CrossRef CAS PubMed.
- Y. Song, X. Xu, K. W. MacRenaris, X. Q. Zhang, C. A. Mirkin and T. J. Meade, Angew. Chem., 2009, 48, 9143–9147 CrossRef CAS PubMed.
- C. H. J. Choi, L. Hao, S. P. Narayan, E. Auyeung and C. A. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7625–7630 CrossRef CAS PubMed.
- K. Zhang, L. Hao, S. J. Hurst and C. A. Mirkin, J. Am. Chem. Soc., 2012, 134, 16488–16491 CrossRef CAS PubMed.
- D. Zheng, D. S. Seferos, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Nano Lett., 2009, 9, 3258–3261 CrossRef CAS PubMed.
- S. J. Hurst, A. K. R. Lytton-Jean and C. A. Mirkin, Anal. Chem., 2006, 78, 8313–8318 CrossRef CAS PubMed.
- O. Padovan-Merhar, G. P. Nair, A. G. Biaesch, A. Mayer, S. Scarfone, S. W. Foley, A. R. Wu, L. S. Churchman, A. Singh and A. Raj, Mol. Cell, 2015, 58, 339–352 CrossRef CAS PubMed.
- B. Schwanhausser, D. Busse, N. Li, G. Dittmar, J. Schuchhardt, J. Wolf, W. Chen and M. Selbach, Nature, 2011, 473, 337–342 CrossRef PubMed.
- Q. Dai, C. Walkey and W. C. W. Chan, Angew. Chem., Int. Ed., 2014, 53, 5093–5096 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS PubMed.
- D. S. Seferos, A. E. Prigodich, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Nano Lett., 2009, 9, 308–311 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, nanoparticle characterization, relaxivity, HaloTag quantification, cell viability, MR images, and additional acknowledgements. See DOI: 10.1039/c5cc06565h |
| This journal is © The Royal Society of Chemistry 2016 |