A knot polymer mediated non-viral gene transfection for skin cells†
Lara
Cutlar‡
a,
Yongsheng
Gao‡
a,
Ahmed
Aied‡
a,
Udo
Greiser
a,
Eva Maria
Murauer
b,
Dezhong
Zhou
*a and
Wenxin
Wang
*a
aCharles Institute of Dermatology, University College Dublin, Dublin 4, Ireland. E-mail: dezhong.zhou@ucd.ie; wenxin.wang@ucd.ie
bDivision of Molecular Dermatology and EB House Austria, Department of Dermatology, Paracelsus Medical University, Salzburg, Austria
First published on 15th September 2015
Abstract
A knot polymer, poly[bis(2-acryloyl)oxyethyl disulphide-co-2-(dimethylamino) ethyl methacrylate] (DSP), was synthesized, optimized and evaluated as a non-viral vector for gene transfection for skin cells, keratinocytes. With recessive dystrophic epidermolysis bullosa keratinocytes (RDEBK-TA4), the DSP exhibited high transfection efficacy with both Gaussia luciferase marker DNA and the full length COL7A1 transcript encoding the therapeutic type VII collagen protein (C7). The effective restoration of C7 in C7 null-RDEB skin cells indicates that DSP is promising for non-viral gene therapy of recessive dystrophic epidermolysis bullosa (RDEB).
A wide range and variety of mutations in COL7A1 lead to the painful blistering skin condition RDEB.1COL7A1 codes for the protein type VII collagen (C7) and if this protein is either absent or expressed as a truncated product, any pressure or friction causes the epithelium sheet to detach beneath the lamina densa.2 While there are multiple avenues of research being pursued to treat this condition using cell and protein therapies, gene therapies hold the greatest promise for long-term correction of this severe condition.3 The subunits of the type VII pro-collagen are produced in both keratinocytes and fibroblasts, however keratinocytes along the stratified squamous epithelia are the preferred target for gene therapy4 because they are the main contributors to the expression of these precursors of the therapeutic gene. Chen et al. developed a retroviral system for packaging of a plasmid containing a truncated human C7 minigene and in vitro expression of a truncated mini C7 was detected for 8 months in RDEBKs.5 In an alternative approach by Mecklenbeck et al., expression of full length collagen type VII was observed in RDEBK transfected by a P1-derived artificial chromosome DNA vector carrying the COL7A1 cassette.6
Despite the progress with the successful viral vectors, however, the major safety issues, such as the possibility of the virus eliciting a severe immune reaction, greatly limit the application of viral vectors.7 A promising alternative is non-viral gene vectors.8 Over the past several decades, with the great progress in polymer chemistry, structurally diverse polymeric non-viral vectors have been developed.8,9 Among them, especially cyclized cationic polymers have attracted broad attention. Previously, our group has developed a novel type of cyclized polymer, termed knot, by in situ deactivation enhanced atom transfer radical polymerization (DE-ATRP). Due to the unique three-dimensional (3D) architecture and multiple terminal groups, the knot polymers demonstrated strong interaction with DNA and can mediate high gene transfection while inducing only a very low cytotoxicity, even in the primary cells and astrocytes, compared to the leading commercial gene transfection reagents Lipofectamine 2000 and SuperFect.10 Following this work, we then investigated the structure–function activity of the knot polymers in gene transfection and developed multi-knot polymers as highly efficacious vectors for gene delivery to a range of cell types.11 Here, we synthesize a knotted cationic polymer, termed DSP, and evaluate it as a non-viral gene vector for gene transfection of keratinocytes, especially with COL7A1 DNA to demonstrate the utility of DSP in RDEB gene therapy.
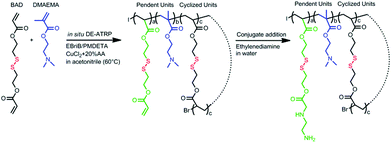
The DSP was synthesized from bis(2-acryloyl)oxyethyl disulphide (BAD) and 2-(dimethylamino) ethyl methacrylate (DMAEMA) via in situ DE-ATRP according to our previous work but employing ethylenediamine instead of 1,3-diaminopropane for end-capping given the relatively higher reactivity of the former.11 As outlined in Scheme 1, an acrylate terminated cyclized base polymer was first synthesized and then end-capped with ethylenediamine. The final product DSP has a Mw of 32.3 kDa with a PDI of 1.7. The chemical structure of the DSP was verified by 1H NMR.
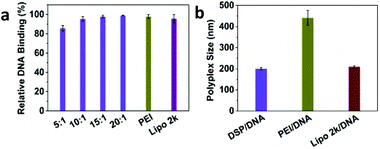
The DNA binding affinity of the cyclized DSP to DNA was first examined using a PicoGreen assay (Fig. 1a). Over the four w/w ratios, the DSP showed high DNA binding affinity, even at the lowest w/w ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the relative DNA binding was still over 80%. The high DNA binding affinity of DSP led the translation to formulate small polyplexes with high zeta potential. At the w/w ratio of 10
1, the relative DNA binding was still over 80%. The high DNA binding affinity of DSP led the translation to formulate small polyplexes with high zeta potential. At the w/w ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the DSP/DNA polyplexes have an average size of 195 nm with a zeta potential of +50 mV (Fig. 1b and Fig. S1†).
1, the DSP/DNA polyplexes have an average size of 195 nm with a zeta potential of +50 mV (Fig. 1b and Fig. S1†).
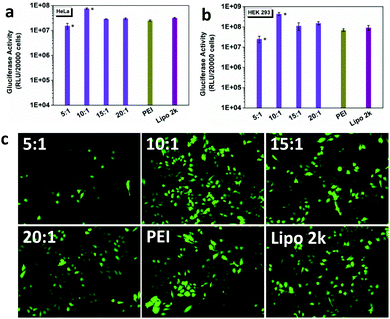
The gene transfection efficiency of DSP was first evaluated with Gaussia luciferase in HeLa and HEK 293 cells (Fig. 2a and b), and compared with that of the commercial gene transfection reagents PEI and Lipofectamine 2000. In both cell types, the DSP showed high gene transfection efficiency, especially at w/w = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, The gluciferase activity of the cells after treatment was 5–10-fold higher than that of PEI and Lipofectamine 2000. The high transfection efficiency of DSP was further supported by the high GFP expression of cells after transfection (Fig. 2c and Fig. S2†). In addition, the DSP can preserve much higher cell viability compared with PEI and Lipofectamine 2000 (Fig. S3†). Because of the high transfection efficacy and cell viability observed, w/w = 10
1, The gluciferase activity of the cells after treatment was 5–10-fold higher than that of PEI and Lipofectamine 2000. The high transfection efficiency of DSP was further supported by the high GFP expression of cells after transfection (Fig. 2c and Fig. S2†). In addition, the DSP can preserve much higher cell viability compared with PEI and Lipofectamine 2000 (Fig. S3†). Because of the high transfection efficacy and cell viability observed, w/w = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was chosen for all of the following studies.
1 was chosen for all of the following studies.
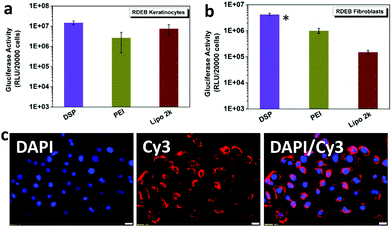
The levels of expression of the Gaussia luciferase were next compared with the cell metabolic activity for RDEB-TA4 keratinocytes (RDEBKs) and fibroblasts (RDEBFs). At w/w = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2–4-fold higher Gluciferase activity in RDEBK and 2–28-fold higher activity in RDEBF were seen after transfection with DSP compared with cells transfected with PEI and Lipofectamine2000 (Fig. 3a and b). It should be pointed out that here PEI was chosen as a comparative reagent as it is the gold standard with a relatively high non-viral transfection capability. Lipofectamine 2000 was used as a lipid based control since lipid based systems have shown high transfection efficiency in keratinocytes. The fact that DSP showed much better transfection efficiency in comparison with PEI and Lipofectamine 2000 demonstrates that DSP is highly potent in transfecting not only the easy-to-transfect HeLa and HEK 293 cells but also the more challenging human RDEBK and RDEBF. Interestingly, the overall gene expression is ∼10-fold lower in RDEBF than it is in RDEBK, indicating that this cell type is comparatively harder to transfect. The high transfection of DSP in RDEBK can also be illustrated by the high cellular uptake efficiency of DSP/DNA polyplexes. It can be seen that after 4 hours of incubation many polyplexes were taken up by the RDEBK and distributed around the nucleus (Fig. 3c). In terms of cytotoxicity in RDEBK and RDEBF, it can be seen that PEI and Lipofectamine 2000 had a significant higher cytotoxicity in particular for RDEBK and in comparison with DSP3, which maintained above 90% of cell viability (Fig. S4†). This is also reflected in the morphology of the RDEBK which better retained a uniform polygonal basal morphology.
1, 2–4-fold higher Gluciferase activity in RDEBK and 2–28-fold higher activity in RDEBF were seen after transfection with DSP compared with cells transfected with PEI and Lipofectamine2000 (Fig. 3a and b). It should be pointed out that here PEI was chosen as a comparative reagent as it is the gold standard with a relatively high non-viral transfection capability. Lipofectamine 2000 was used as a lipid based control since lipid based systems have shown high transfection efficiency in keratinocytes. The fact that DSP showed much better transfection efficiency in comparison with PEI and Lipofectamine 2000 demonstrates that DSP is highly potent in transfecting not only the easy-to-transfect HeLa and HEK 293 cells but also the more challenging human RDEBK and RDEBF. Interestingly, the overall gene expression is ∼10-fold lower in RDEBF than it is in RDEBK, indicating that this cell type is comparatively harder to transfect. The high transfection of DSP in RDEBK can also be illustrated by the high cellular uptake efficiency of DSP/DNA polyplexes. It can be seen that after 4 hours of incubation many polyplexes were taken up by the RDEBK and distributed around the nucleus (Fig. 3c). In terms of cytotoxicity in RDEBK and RDEBF, it can be seen that PEI and Lipofectamine 2000 had a significant higher cytotoxicity in particular for RDEBK and in comparison with DSP3, which maintained above 90% of cell viability (Fig. S4†). This is also reflected in the morphology of the RDEBK which better retained a uniform polygonal basal morphology.
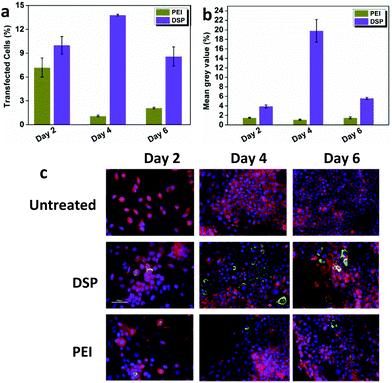
In gene transfection, one of the most significant challenges is that the high efficiency marker gene expression mediated by the vectors usually does not correlate very well with that of the therapeutic gene.12 Therefore, the effectiveness of DSP mediating the expression of COL7A1 was further assessed. Immunofluorescence staining was carried out on RDEBK transfected with polyplexes carrying the COL7A1 sequence after 2, 4 and 6 days. Expression of the C7 in transfected cells was visualized using an affinity-purified antibody to the non-collagenous domain of the type VII collagen. Large amounts of the protein were seen in the cells as it binds to the intracellular pro-collagen chains, with some low level fluorescence seen in the extracellular matrix detecting the secreted protein. A quantitative evaluation of the number of RDEBK cells transfected at each time point was calculated using Image J software (Fig. 4a and b). The results indicate that compared to PEI, both the number of transfected cells and the mean fluorescence intensity mediated by the DSP were much higher, which correlates very well with the expression of the Gaussia luciferase. This indicates that DSP3 could also mediate high efficiency therapeutic gene expression. Also, a significantly higher percentage of transfected cells was found on day 4 compared to days 2 and 6. This indicates that a reduction in C7 production occurred over time with the highest accumulation seen on day 4 (Fig. 4c). These results have been expected for a temporary expression cassette in a relatively quickly replicating cell line such as RDEBK. The strong expression of C7 mediated by DSP was also evident by the western blot assay (Fig. S5†).
While other studies have successfully transformed RDEB cells with viral vectors, we believe this is the first successful transfection of RDEBK with COL7A1 by a non-viral approach. The current focus on gene therapy for RDEB uses SIN lentiviral or retroviral vectors. However, the highly controllable nature and the safety profile of the potential gene therapy make the use of non-viral vectors an intriguing option for further research. High recombinant protein expression levels were seen as expected and similar ectopic expression has so far demonstrated to have no negative effects. The most notable DSP showed significantly higher cell metabolic activity over conventional branched polymer and liposome based transfection agents. This is a key feature as it allows for repeated application of the polymer to the cells. This promising approach holds great potential to restore the structural integrity needed to reverse the RDEB phenotype in vivo.
Conclusions
Knot polymer DSP was successfully synthesized by DE-ATRP and used as a non-viral gene delivery vector for transfection of keratinocytes. DSP can effectively mediate both marker gene and therapeutic gene expression. The high potency of DSP in mediating C7 expression makes it a highly promising non-viral gene delivery vector for the treatment of RDEB.Acknowledgements
We would like to acknowledge Dr Andrew South (Division of Cancer Research, University of Dundee, Ninewells, Dundee, UK) for kindly providing the COL7A1 plasmid. We also acknowledge Dr Fernando Larcher for kindly providing the RDEBK-TA4 and RDEBF cell lines. This work was funded by Science Foundation Ireland (SFI), grant number 10/IN.1/B2981(T) (SFI Principal Investigator Program), grant number 14/TIDA/2367 (Technology Innovation Development Award), Health Research Board of Ireland (HRA-POR-2013-412) and University College Dublin.Notes and references
- J. S. Kern, J. Kohlhase, L. Bruckner-Tuderman and C. Has, J. Invest. Dermatol., 2006, 126, 1006–1012 CrossRef CAS PubMed; P. C. van den Akker, A. J. van Essen, M. M. Kraak, R. Meijer, M. Nijenhuis, G. Meijer, R. M. Hofstra, H. H. Pas, H. Scheffer and M. F. Jonkman, J. Dermatol. Sci., 2009, 56, 9–18 CrossRef PubMed.
- K. Wertheim-Tysarowska, A. Sobczynska-Tomaszewska, C. Kowalewski, M. Skronski, G. Swieckowski, A. Kutkowska-Kazmierczak, K. Wozniak and J. Bal, Hum. mutat., 2012, 33, 327–331 CrossRef CAS PubMed; A. M. Christiano, S. Amano, L. F. Eichenfield, R. E. Burgeson and J. Uitto, J. Invest. Dermatol., 1997, 109, 390–394 CrossRef PubMed.
- L. De Rosa, S. Carulli, F. Cocchiarella, D. Quaglino, E. Enzo, E. Franchini, A. Giannetti, G. De Santis, A. Recchia, G. Pellegrini and M. De Luca, Stem Cell Rep., 2014, 2, 1–8 CrossRef CAS PubMed; J. Remington, X. Wang, Y. Hou, H. Zhou, J. Burnett, T. Muirhead, J. Uitto, D. R. Keene, D. T. Woodley and M. Chen, Mol. Ther., 2009, 17, 26–33 CrossRef PubMed.
- R. E. Burgeson, J. Invest. Dermatol., 1993, 101, 252–255 CrossRef CAS PubMed.
- M. Chen, E. A. O'Toole, M. Muellenhoff, E. Medina, N. Kasahara and D. T. Woodley, J. Biol. Chem., 2000, 275, 24429–24435 CrossRef CAS PubMed.
- S. Mecklenbeck, S. H. Compton, J. E. Mejia, R. Cervini, A. Hovnanian, L. Bruckner-Tuderman and Y. Barrandon, Hum. Gene Ther., 2002, 13, 1655–1662 CrossRef CAS PubMed.
- D. T. Woodley, D. R. Keene, T. Atha, Y. Huang, R. Ram, N. Kasahara and M. Chen, Mol. Ther., 2004, 10, 318–326 CrossRef CAS PubMed.
- X. G. a. L. Huang, Acc. Chem. Res., 2012, 45, 9 Search PubMed.
- D. W. Pack, A. S. Hoffman, S. Pun and P. S. Stayton, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS PubMed.
- B. Newland, Y. Zheng, Y. Jin, M. Abu-Rub, H. Cao, W. Wang and A. Pandit, J. Am. Chem. Soc., 2012, 134, 4782–4789 CrossRef CAS PubMed.
- A. Aied, Y. Zheng, B. Newland and W. Wang, Biomacromolecules, 2014, 15, 4520–4527 CrossRef CAS PubMed.
- M. A. M. a. E. E. Simanek, Chem. Rev., 2009, 109, 259–302 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1 and S2 and experimental procedure. See DOI: 10.1039/c5bm00216h |
| ‡ Equal contributions. |
| This journal is © The Royal Society of Chemistry 2016 |