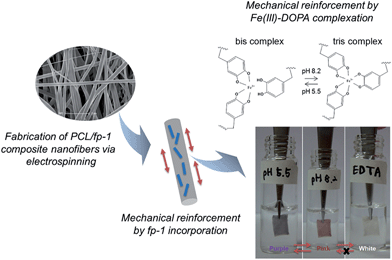
Mussel-inspired adhesive protein-based electrospun nanofibers reinforced by Fe(III)–DOPA complexation†
Bum Jin
Kim‡
ad,
Sangsik
Kim‡
bc,
Dongyeop X.
Oh
b,
Admir
Masic
f,
Hyung Joon
Cha
*ad and
Dong Soo
Hwang
*abce
aSchool of Interdisciplinary Bioscience and Bioengineering, Pohang University of Science and Technology, Pohang 790-784, Korea
bOcean Science and Technology Institute, Pohang University of Science and Technology, Pohang 790-784, Korea
cSchool of Environmental Science and Engineering, Pohang University of Science and Technology, Pohang 790-784, Korea
dDepartment of Chemical Engineering, Pohang University of Science and Technology, Pohang 790-784, Korea. E-mail: hjcha@postech.ac.kr
eIntegrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang 790-784, Korea. E-mail: dshwang@postech.ac.kr
fDepartment of Biomaterials, Max Planck Institute for Colloids and Interfaces, Potsdam 14424, Germany
First published on 8th October 2014
Abstract
Marine mussels utilize multiple bidentate complexes formed by Fe(III) and DOPA in a mussel adhesive protein (fp-1) to reinforce tough and elastic byssal fibers as a specialized underwater adhesive aid. In this study, mussel-inspired electrospun nanofibers were fabricated using a recombinant mussel adhesive protein (rfp-1), Fe(III)–DOPA complexes, and polycaprolactone. The mechanical properties of the fabricated nanofibers were reinforced by the Fe(III)–DOPA complex found in fp-1, which is a key component of the naturally occurring high-performance mussel fiber coating. Experimental results show that the stoichiometry of Fe(III)–DOPA complexes in the nanofibers could be controlled by buffer pH conditions and the stiffness of the nanofiber mat increased linearly with the concentration of the Fe(III)–DOPA complexes, as monitored by resonance Raman spectroscopy. This suggests the potential of Fe(III)–DOPA complexation as an effective strategy for modulating the mechanical properties of nanofibrous biomedical materials by using pH variations.
Introduction
Nanofiber technology has been widely utilized for fabricating non-woven fibrous matrices in diverse fields such as fabrics, filters, and biomedical engineering (e.g., drug delivery systems, sensors, and tissue engineering scaffolds).1 Particularly nanofiber technology is a versatile one owing to various advantages that make it suitable for numerous applications, such as fiber diameter ranging from tens to thousands of nanometers, microporous structure, and high surface-to-volume ratio. Among the various fabrication techniques for producing nanofibers, electrospinning is considered a relatively simple and an efficient method. Various types of polymeric materials have been successfully used to fabricate electrospun nanofibers.2–4 While fabrication methodology, basic physical and chemical characterization, and biological approach are the mainstream research topics related to nanofiber technology, studies on enhancing the mechanical properties of electrospun nanofibers should not be overlooked. Thus, the development of high-strength nanofibers via reinforcement strategies seems both necessary and challenging. Several studies on alignment techniques, micro/nanomesh composites, blending strategies using additives, and crosslinking with physical and chemical agents have been reported so far.5–9For improving the strength of fibrous structures, we drew inspiration from strong protein-based coatings found in marine mussels. Mussels secrete a bundle of threads called byssus for tethering themselves to marine substrata.10–12 Individual byssal threads are covered by a thin proteinaceous coating layer to protect the thread from tough mechanical stress in marine environments. The protective coating on the byssal threads of mussels (Mytilus species) has gained considerable interest in biomedical research fields owing to its extraordinary stiffness (comparable to that of epoxy resin) and excellent elongation properties (ε > ∼70%).13,14 Thus far, the only known biomacromolecule in the protective coating on mussel byssal threads is a mussel adhesive protein (MAP) named type-1 (fp-1). fp-1 from Mytilus edulis consists of about 80 tandem repetitions of decapeptides [AKPSY*OO*TY*K], in which O, O*, and Y* denote trans-4-hydroxyproline (HyP), trans-2,3,cis-3,4-dihydroxyproline (diHyP), and 3,4-dihydroxyphenylalanine (DOPA), respectively.15 It has been shown previously that the catechol moieties of DOPA in fp-1 form strong reversible bis- and tris-catecholate complexes with Fe(III) ions having a log stability constant greater than 40.16 Many recent studies have proved that the multiple bidentate complexes formed by Fe(III) and DOPA in fp-1 are mainly responsible for the mechanical co-existence of high extensibility and hardness in the byssal cuticle of Mytilus species.17–19
Previously, we reported the successful fabrication of electrospun nanofibers by blending the recombinant hybrid MAP fp-151 (rfp-151) with various types of synthetic polymers.20 In particular, mixing rfp-151 with polycaprolactone (PCL) as a representative polymer model, which is an FDA-approved biodegradable polymer with proven biocompatibility, and has been used previously to fabricate electrospun 3-D nanoporous scaffolds employed in tissue engineering,21 provided a 4-fold enhancement in the strength and stiffness compared to sole PCL nanofibers. In the present study, we fabricated electrospun composite nanofibers by using blends of PCL and a recombinant fp-1 (rfp-1), which was composed of 12 repeats of Mytilus fp-1 consensus decapeptides (AKPSYPPTYK),18 and investigated whether the mechanical properties of the nanofibers are influenced by the presence or absence of Fe(III) and DOPA complexes in DOPA-containing modified rfp-1 (mrfp-1) (Fig. 1).
Experimental
Preparation of DOPA-containing MAP
rfp-1 composed of 12 tandemly repeated decapeptides (AKPSYPPTYK) of fp-1 was produced using the Escherichia coli system, as previously reported.18 The molecular mass of rfp-1 was about 13.6 kDa from the matrix assisted laser desorption ionization-time of flight mass spectroscopy (MALDI-TOF MS) analysis. Prior to electrospinning, tyrosine residues of rfp-1 were modified into DOPA by using mushroom tyrosinase (Sigma, St. Louis, MO, USA); 1.5 mg mL−1 of rfp-1 solution in modification buffer (100 mM sodium phosphate dibasic, 20 mM boric acid, and 25 mM ascorbic acid; pH 6.8) was reacted with tyrosinase (100 μg mL−1) for 1 h and dialyzed with 1% acetic acid. The DOPA content in DOPA-containing modified rfp-1 (mrfp-1) was measured using a ninhydrin-based amino acid analyzer (S4300, SYKAM, Germany) after hydrolysis of mrfp-1 in 6 N HCl with 5% water-saturated phenol for 1 h at 156 °C.Electrospinning of PCL/MAP nanofibers
A polymer solution was prepared by dissolving PCL (Sigma; Mn = ∼80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and MAPs (rfp-1 and mrfp-1) at a concentration of 6 wt% in 1,1,1,3,3,3-hexafluoroisopropanol (HFIP; Sigma). The PCL solution was blended with the rfp-1 solution to prepare a series of PCL/rfp-1 solutions with varying PCL
000) and MAPs (rfp-1 and mrfp-1) at a concentration of 6 wt% in 1,1,1,3,3,3-hexafluoroisopropanol (HFIP; Sigma). The PCL solution was blended with the rfp-1 solution to prepare a series of PCL/rfp-1 solutions with varying PCL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) rfp-1 ratios of 100
rfp-1 ratios of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 90
0, 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, and 70
10, and 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30. The solutions were electrospun using a 5 mL syringe with a needle diameter of 0.4 mm; the mass flow rate of the solutions from the syringe was 1 mL h−1. A high voltage (13–15 kV) was applied to the tip of the needle attached to the syringe when the fluid jet was ejected. Random nanofibers were collected on a flat aluminum foil at a distance of 15 cm from the needle tip. All the electrospun nanofibers were vacuum-dried for at least 1 day to allow evaporation of any remaining solvents prior to use. For the fabrication of nanofibers containing Fe(III)–DOPA complexes, FeCl3 was added to a mrfp-1 solution at a molar ratio of DOPA
30. The solutions were electrospun using a 5 mL syringe with a needle diameter of 0.4 mm; the mass flow rate of the solutions from the syringe was 1 mL h−1. A high voltage (13–15 kV) was applied to the tip of the needle attached to the syringe when the fluid jet was ejected. Random nanofibers were collected on a flat aluminum foil at a distance of 15 cm from the needle tip. All the electrospun nanofibers were vacuum-dried for at least 1 day to allow evaporation of any remaining solvents prior to use. For the fabrication of nanofibers containing Fe(III)–DOPA complexes, FeCl3 was added to a mrfp-1 solution at a molar ratio of DOPA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fe(III) = 3
Fe(III) = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. This solution was mixed with a PCL solution, and the mixing ratio of PCL
1. This solution was mixed with a PCL solution, and the mixing ratio of PCL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mrfp-1 was adjusted to 70
mrfp-1 was adjusted to 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 before electrospinning.
30 before electrospinning.
Characterization of nanofibers
The morphology of the electrospun nanofibers was analyzed by scanning electron microscopy (SEM; JEOL JSM-7401F, Japan) at an accelerating voltage of 5 kV. The nanofibers were coated with gold via sputtering before the observations. The diameter of each nanofiber was measured from the SEM images by using image analysis software (Image J, National Institutes of Health, USA). The contact angle of the electrospun nanofibers was measured using a drop test method for determining the wettability of the nanofiber surface. A 5 μL water drop was applied to the nanofibers with a microsyringe. The contact angles were measured at intervals of 5 s by using a charge-coupled device (CCD) imaging system (Surface and Electro-Optics). The tensile properties of the nanofiber mats were analyzed using a universal testing machine (INSTRON) with a 2 kN load cell and a crosshead speed of 5 mm min−1 under ambient conditions. At least eight rectangular samples of each nanofiber mat were prepared with dimensions of 10 mm × 25 mm; the thickness of the nanofiber mat was measured using a micrometer prior to the tests. To keep the nanofiber mats wet, they were immediately loaded in the device after immersion in each of the buffers. The stress and strain values were monitored until the nanofiber mats were torn.Spectroscopy analysis
For Raman spectroscopy analysis, a continuous laser beam was focused on the samples through a confocal Raman microscope (α300; WITec) equipped with a piezoscanner (Physik Instrumente). Raman spectra were collected with a Nikon objective (20×, NA = 0.4) and a laser excitation wavelength of 532 nm. The Raman spectra of Fe(III)-incorporated PCL/mrfp-1 nanofibrous meshes immersed in each buffer (200 mM sodium acetate; pH 5.5, 200 mM tris–HCl; pH 8.2, and 50 mM EDTA in acetate buffer; pH 5.5) were acquired with a CCD camera (DV401-BV; Andor) behind a spectrometer (UHTS 300; WITec) with a spectral resolution of 3 cm−1. The samples were sensitive to burning by the laser beam; therefore, the laser power for all measurements ranged between 15 and 30 mW. ScanCtrlSpectroscopyPlus software (version 1.38, Witec) was used for the measurement setup whereas the acquired spectra were analyzed and processed with Witec Project Plus software (version 2.08). In order to obtain the spectra representative of the entire fiber, five spectra (2 s integration time each) were acquired at different positions of the fiber and averaged. Spectra were background subtracted (average background subtract function) and smoothed with a 9-point Savitzky–Golay filter (fourth order polynomial).The stoichiometry of DOPA–Fe(III) coordination was also monitored by using a UV-visible spectrophotometer (Mecasys, Korea) using a quartz cuvette with a path length of 1 cm. 0.1 mg mL−1 of mrfp-1 solution was used for the analysis.
Cytotoxicity validation of nanofibers
For cell culture experiments, the electrospun nanofibers were exposed to UV radiation for 2 h, and were washed thrice with phosphate-buffered saline (PBS) for 30 min each time before cell seeding. A mouse preosteoblast cell line, MC3T3-E1, was cultured in minimal essential medium-alpha (MEM-α; Hyclone) supplemented with 10% fetal bovine serum (FBS; Hyclone) and 1% penicillin/streptomycin (Hyclone) at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. The subconfluent cells were detached using 0.25% trypsin-EDTA (Hyclone), and the viable cells were counted by a trypan blue assay. The cells were seeded onto electrospun nanofibers that were placed in a 24-well plate, at a density of 5 × 104 cells per well, and cultured for further analysis. The morphology and viability of the cells were imaged using a live/dead cell staining kit (Invitrogen). After 3 days of cell seeding, live cells were stained by 2 μM calcein AM (green) and dead cells were stained by 4 μM ethidium homodimer-1 (red) for 30 min; the stained images were then observed using a fluorescence microscope (Leica). Further, to quantify cell viability on each nanofiber, after cell seeding for two different periods of 1 h and 3 days, media containing the samples were treated with CCK-8 (Dojindo) reagents and incubated for 2 h at 37 °C in a humidified atmosphere of 5% CO2. Aliquots of each sample medium were collected in a 96-well plate to measure the absorbance at 450 nm. All experiments were performed in triplicate to test for cell viability.Results and discussion
Fabrication of composite nanofibers using PCL and rfp-1
fp-1 is the only known biomacromolecule in the tough and extensible coating on mussel fibers.15 We used recombinant fp-1 (rfp-1) to enhance the mechanical properties of PCL nanofibers by mimicking the mussel coating. With the help of recombinant DNA technology employed for the high-yield production of MAPs in an E. coli system,18,22 we obtained a sufficient amount of rfp-1 proteins for electrospinning. Since rfp-1 is highly solubilized in HFIP, 6 wt% of an rfp-1 solution was easily prepared and electrospun nanofibers were successfully fabricated under typical electrospinning conditions by using 6 wt% PCL and rfp-1, mixed in various ratios of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 90
0, 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, and 70
10, and 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
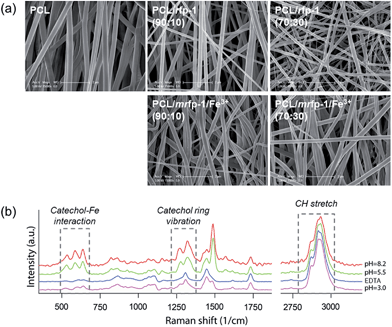
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30. As we had observed in a previous report,20 we could not find any dissolvation or release of MAPs as well as any morphological collapse of nanofibers after their exposure to aqueous solutions for 3 days. At each mixing ratio, the structure and morphology of the electrospun nanofibers were observed by SEM (upper panels in Fig. 2a). At all the three ratios, homogeneous fibrous morphologies with diameters of ∼200 nm (Table 1) and hardly any bead formations were observed. The fiber diameter was found to be constant regardless of the concentration of rfp-1 in PCL/rfp-1 nanofibers. Normally, ribbon-shape formation and thinness of electrospun fibers are the representative evidence of uneven blending component distribution.23 However, our PCL/rfp-1 nanofiber does not have ribbon-shaped but has more of round-shaped fibers. Additionally, we could not observe any wrinkles in the skin of our round-shaped PCL/rfp-1 nanofiber, which also indicates skin formation.23 This may be due to the low concentration (6 wt%) of the spinning solution, the similar solubility between PCL and rfp-1 in HFIP, and the relatively thin fiber diameter (200–300 nm). Therefore, it is more likely that the two blending solutes spread evenly rather than towards a surface direction. Moreover, the surface of the nanofibers was homogeneous, indicating that the rfp-1 and PCL were mixed well in the nanofibers. To determine whether incorporated rfp-1 would be well exposed on the surface of the composite nanofibers, water contact angles were measured (Fig. S1†). The results revealed that the contact angle gradually decreased according to the rfp-1 concentration, which might be attributed to the hydrophilic residues of surface-exposed rfp-1. Increasing the hydrophilicity will be beneficial for general biomedical applications of these nanofibers.24–26
30. As we had observed in a previous report,20 we could not find any dissolvation or release of MAPs as well as any morphological collapse of nanofibers after their exposure to aqueous solutions for 3 days. At each mixing ratio, the structure and morphology of the electrospun nanofibers were observed by SEM (upper panels in Fig. 2a). At all the three ratios, homogeneous fibrous morphologies with diameters of ∼200 nm (Table 1) and hardly any bead formations were observed. The fiber diameter was found to be constant regardless of the concentration of rfp-1 in PCL/rfp-1 nanofibers. Normally, ribbon-shape formation and thinness of electrospun fibers are the representative evidence of uneven blending component distribution.23 However, our PCL/rfp-1 nanofiber does not have ribbon-shaped but has more of round-shaped fibers. Additionally, we could not observe any wrinkles in the skin of our round-shaped PCL/rfp-1 nanofiber, which also indicates skin formation.23 This may be due to the low concentration (6 wt%) of the spinning solution, the similar solubility between PCL and rfp-1 in HFIP, and the relatively thin fiber diameter (200–300 nm). Therefore, it is more likely that the two blending solutes spread evenly rather than towards a surface direction. Moreover, the surface of the nanofibers was homogeneous, indicating that the rfp-1 and PCL were mixed well in the nanofibers. To determine whether incorporated rfp-1 would be well exposed on the surface of the composite nanofibers, water contact angles were measured (Fig. S1†). The results revealed that the contact angle gradually decreased according to the rfp-1 concentration, which might be attributed to the hydrophilic residues of surface-exposed rfp-1. Increasing the hydrophilicity will be beneficial for general biomedical applications of these nanofibers.24–26
| Ratio [PCL/rfp-1] | Fiber mean diameter [nm] | Tensile strength [MPa] | Extension [mm mm−1] | Young's modulus [MPa] |
|---|---|---|---|---|
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0 |
140.0 ± 46.7 | 5.6 ± 2.2 | 2.9 ± 1.2 | 34.4 ± 6.7 |
90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
169.7 ± 23.2 | 24.3 ± 3.5 | 0.6 ± 0.1 | 56.8 ± 9.9 |
70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 30 |
158.4 ± 21.8 | 7.8 ± 4.3 | 0.5 ± 0.2 | 15.3 ± 5.0 |
Fabrication and identification of composite nanofibers containing Fe(III)–DOPA complexes
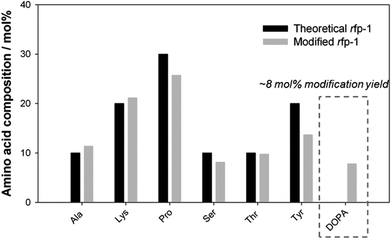
To mimic the mussel protective coating, crosslinks formed using Fe(III)–DOPA complexes were introduced to enhance the mechanical properties of the PCL/rfp-1 nanofibers. Prior to electrospinning with PCL, tyrosine residues of rfp-1 were converted into DOPA residues via an enzymatic reaction with mushroom tyrosinase in vitro,27 and it was determined that ∼8 mol% of the total amino acids (∼37% of total tyrosine residues) in rfp-1 were converted into DOPA molecules (Fig. 3). FeCl3 was directly added to a solution containing 6 wt% of mrfp-1 dissolved in HFIP with a DOPA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fe(III) molar ratio of 3
Fe(III) molar ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 before the electrospinning process. The color of the FeCl3-added solution turned dark purple without any precipitation, implying that the complexation of Fe(III) and DOPA had occurred.16–19,28 The absorbance around 560 nm (purple) of the FeCl3-added solution steeply increased after the addition of Fe(III) and reached a plateau within 10 min, indicating that the formation of Fe(III)–DOPA complexes saturated within 10 min.
1 before the electrospinning process. The color of the FeCl3-added solution turned dark purple without any precipitation, implying that the complexation of Fe(III) and DOPA had occurred.16–19,28 The absorbance around 560 nm (purple) of the FeCl3-added solution steeply increased after the addition of Fe(III) and reached a plateau within 10 min, indicating that the formation of Fe(III)–DOPA complexes saturated within 10 min.
Electrospinning was carried out for the successful fabrication of fine nanofibers by using FeCl3-added mrfp-1 with PCL at PCL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mrfp-1 weight ratios of 90
mrfp-1 weight ratios of 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 and 70
10 and 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 (Fig. 2a). The diameter and morphology of Fe(III)-incorporated mrfp-1 nanofibers were not notably different from those of rfp-1 nanofibers. Interestingly, we observed that the color of the nanofiber mat changed according to the pH of the immersing buffers (Fig. 1). Previously, it has been shown that the three binding states of Fe(III)–DOPA complexes, namely, mono-, bis-, or tris-, are controlled by the buffer pH and DOPA
30 (Fig. 2a). The diameter and morphology of Fe(III)-incorporated mrfp-1 nanofibers were not notably different from those of rfp-1 nanofibers. Interestingly, we observed that the color of the nanofiber mat changed according to the pH of the immersing buffers (Fig. 1). Previously, it has been shown that the three binding states of Fe(III)–DOPA complexes, namely, mono-, bis-, or tris-, are controlled by the buffer pH and DOPA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fe(III) molar ratio. Because the Fe(III) concentration was fixed to maintain a DOPA
Fe(III) molar ratio. Because the Fe(III) concentration was fixed to maintain a DOPA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fe(III) molar ratio of 3
Fe(III) molar ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in mrfp-1, the binding state of the Fe(III)–DOPA complexes was governed solely by the pH of the buffer in which the electrospun nanofiber mat was incubated.16–19,28 Further, the maximum absorbance of the respective complex, as determined using a spectrophotometer, was observed at around 500 nm (pink) and around 560 nm (purple), assigned to tris- and bis-, respectively (Fig. S2†).16 The Fe(III)-incorporated PCL/mrfp-1 (70
1 in mrfp-1, the binding state of the Fe(III)–DOPA complexes was governed solely by the pH of the buffer in which the electrospun nanofiber mat was incubated.16–19,28 Further, the maximum absorbance of the respective complex, as determined using a spectrophotometer, was observed at around 500 nm (pink) and around 560 nm (purple), assigned to tris- and bis-, respectively (Fig. S2†).16 The Fe(III)-incorporated PCL/mrfp-1 (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) nanofiber mat fabricated in this study immediately showed a purple color when immersed in a sodium acetate buffer (pH 5.5) and this color changed to pink in the tris buffer (pH 8.2). Further, the color changed reversibly when the opposite buffer pH change occurred. In addition, when the color-changed nanofiber mat was immersed in an EDTA solution (pH 5.5), the colors disappeared in a few seconds and could not be recovered by immersing the nanofibers in higher buffer (pH 8.2) (Fig. 1). Presumably, Fe(III) was removed from the DOPA molecules in the nanofibers by EDTA. It has been reported that EDTA in a pH 5.5 buffer could steal iron and calcium ions from a mussel byssal cuticle or fp-1 in the cuticle, thereby resulting in significant reduction in mechanical hardness.17,29 Thus, our results indicate that the unique color change might have been due to the Fe(III)–DOPA complexes that were stably incorporated into the nanofibers.
30) nanofiber mat fabricated in this study immediately showed a purple color when immersed in a sodium acetate buffer (pH 5.5) and this color changed to pink in the tris buffer (pH 8.2). Further, the color changed reversibly when the opposite buffer pH change occurred. In addition, when the color-changed nanofiber mat was immersed in an EDTA solution (pH 5.5), the colors disappeared in a few seconds and could not be recovered by immersing the nanofibers in higher buffer (pH 8.2) (Fig. 1). Presumably, Fe(III) was removed from the DOPA molecules in the nanofibers by EDTA. It has been reported that EDTA in a pH 5.5 buffer could steal iron and calcium ions from a mussel byssal cuticle or fp-1 in the cuticle, thereby resulting in significant reduction in mechanical hardness.17,29 Thus, our results indicate that the unique color change might have been due to the Fe(III)–DOPA complexes that were stably incorporated into the nanofibers.
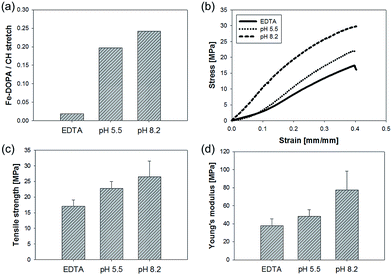
Resonance Raman spectroscopic analysis was employed to gather further chemical evidence for the existence of Fe(III)–DOPA complexes in the nanofibers (Fig. 2b). The absorption by Fe(III)–DOPA complexes in the visible spectral range can be utilized to produce resonance Raman spectra by using the laser line that falls in the range of the characteristic absorption band. In this study, a green laser (532 nm) was used and Raman spectra characteristic of typical Fe(III)–DOPA coordination bands, namely, bidentate Fe-catechol (550, 587, and 636 cm−1) and catechol ring modes (1270, 1322, 1423, and 1476 cm−1), as well as CH stretching (2850–3010 cm−1), were obtained.17,19,30 At a high pH value (pH 8.2), a significant increase in the resonance signal intensity was observed, suggesting more tris Fe(III)–DOPA coordination.31 Notably, the intensity of the Raman spectra dramatically decreased for Fe(III)–DOPA complexes in EDTA-treated nanofibers. We calculated the amount of Fe(III)–DOPA complexes in a single nanofiber molecule by calculating the peak area corresponding to Fe(III)–DOPA complexation and dividing it by the area corresponding to CH stretching (Fig. 4a). Interestingly, the relative amount of Fe(III)–DOPA complexes in the nanofiber increased at higher buffer pH values, implying that the formation of DOPA involved in the complex increased at higher pH values. Collectively, it was clearly demonstrated that the incorporation of Fe(III)–DOPA complexes into the mrfp-1-based electrospun nanofibers fabricated in this study could be successfully achieved, and that the stoichiometry of Fe(III)–DOPA complexes could be controlled by buffer pH conditions. Interestingly, the pH of the tris Fe(III)–DOPA complex, toward which the pH shifted in this study, is almost similar to the actual pH of marine environments (pH ∼ 8.2). Therefore, the artificial fibrous structure containing Fe(III)–DOPA complexes fabricated in this study might be a good model for further research on marine mussel byssus.
Mechanical reinforcement of composite nanofibers mediated by Fe(III)–DOPA complexes
Next, the contribution of Fe(III)–DOPA complexes to the mechanical properties of the fabricated electrospun nanofibers was investigated. The tensile strength, elongation, and Young's modulus of the nanofibers were calculated from the obtained stress–strain curves for each nanofiber mat. As we expected from the previous results using electrospun nanofibers based on hybrid rfp-151,20 simple addition of unmodified rfp-1 into the PCL nanofiber changed the mechanical properties to be more rigid and stiff compared to the soft and flexible sole PCL nanofiber (Table 1). Its tensile strength and Young's modulus were the highest at a certain mixing ratio (PCL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) rfp-1 = 90
rfp-1 = 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10), while elongation dramatically decreased as the rfp-1 concentration increased. Particularly, the composite nanofiber of PCL/rfp-1 (90
10), while elongation dramatically decreased as the rfp-1 concentration increased. Particularly, the composite nanofiber of PCL/rfp-1 (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) showed a tensile strength value which is 5-times higher than that of the sole PCL nanofiber. The mechanical reinforcement via incorporation of rfp-1, which has been demonstrated by similar trends reported in other studies using gelatin, collagen, and MAP (hybrid rfp-151), might be attributed to increased crystallinity and tight interaction between the molecules in composite nanofibers.8,20,32,33 In addition, further mechanical reinforcement of the PCL/rfp-1 composite nanofibers was achieved using the Fe(III)–DOPA complexation strategy (Fig. 4b–d).
10) showed a tensile strength value which is 5-times higher than that of the sole PCL nanofiber. The mechanical reinforcement via incorporation of rfp-1, which has been demonstrated by similar trends reported in other studies using gelatin, collagen, and MAP (hybrid rfp-151), might be attributed to increased crystallinity and tight interaction between the molecules in composite nanofibers.8,20,32,33 In addition, further mechanical reinforcement of the PCL/rfp-1 composite nanofibers was achieved using the Fe(III)–DOPA complexation strategy (Fig. 4b–d).
In order to evaluate the effect of Fe(III)–DOPA complex stoichiometry on the mechanical behavior of the fabricated nanofibers, tensile tests were carried out immediately after immersing the nanofibers in each of the buffers with different pH values. The nanofibers immersed in higher-pH buffers showed higher tensile strength and Young's modulus. Moreover, the Fe–DOPA peak area values divided by the CH stretching area values obtained from the resonance Raman spectroscopic analysis, in which the normalized concentration of the Fe(III)–DOPA complexes in the nanofibers was used, closely correlated with the tensile strength and Young's modulus of the nanofibers (Fig. 4). EDTA treatment of the nanofibers significantly degraded their tensile properties and reduced Raman signals corresponding to the Fe(III)–DOPA complexes in the nanofibers (Fig. 2b). These results indicate that the extent of augmented crosslinking between Fe(III) and DOPA might be a major reason for enhancement in the hardness of the nanofibers. Meanwhile, Fe(III)–DOPA complexes in the nanofibers did not affect their stretchability (Fig. 4b).
In addition, to check the reversibility of Fe(III)–DOPA stoichiometry depending on pH changes which can affect the mechanical properties of the nanofibers, we sequentially immersed the nanofiber mats in buffer pH 5.5 and pH 8.2, from pH 8.2 to pH 5.5, and then reversely from pH 5.5 to pH 8.2. As expected, the tensile strength and modulus of the nanofiber mats immersed in pH 8.2 decreased when placed in pH 5.5 buffer. Moreover, the decreased strength and modulus recovered again after changing back to pH 8.2 buffer. The observed changes in mechanical strength due to pH variation add support to Fe(III)–DOPA complexation taking the main role in mechanical reinforcement (Fig. S3†).
Recently, it has been reported that metal-infiltration improves the mechanical properties of load bearing biomacromolecules, suggesting that simple addition of Fe(III) could improve the mechanical properties of fibrous materials.34,35 In addition, DOPA alone could enhance the stiffness of the fibrous materials by undergoing a variety of crosslinking reactions with nearby available functional groups in neutral and basic pH.36,37 To identify whether Fe(III) or DOPA each independently affects the mechanical properties or not, control experiments were conducted using rfp-1 nanofibers with and without Fe(III), and mrfp-1 nanofibers without Fe(III). Indeed, Fe(III) or DOPA each enhanced the Young's modulus and tensile strength but the improvement in the mechanical properties due to Fe(III) or DOPA alone was much less than the improvement due to Fe(III)–DOPA complexes (Table 2). In basic pH (>8.2), the PCL/mrfp-1 nanofiber mat containing DOPA without Fe(III) had similar Young's modulus to the PCL/mrfp-1 nanofiber mat containing Fe(III)–DOPA complexes but the DOPA containing nanofibers without Fe(III) showed lower tensile strength than that of the nanofibers with Fe(III) (Table S1†).
| Nanofibers | Tensile strength [MPa] | Extension [mm mm−1] | Young's modulus [MPa] |
|---|---|---|---|
PCL/rfp-1 (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) 30) |
7.8 ± 4.3 | 0.5 ± 0.2 | 15.3 ± 2.2 |
PCL/rfp-1 (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) w/Fe(III) 30) w/Fe(III) |
10.7 ± 6.8 | 0.5 ± 0.2 | 19.2 ± 6.4 |
PCL/mrfp-1 (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) 30) |
10.7 ± 7.2 | 0.5 ± 0.2 | 23.4 ± 1.3 |
PCL/mrfp-1 (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) w/Fe(III) 30) w/Fe(III) |
19.4 ± 5.2 | 0.5 ± 0.1 | 46.0 ± 5.7 |
It has been reported that the byssal cuticle of mussels (Mytilus species) has high strength and flexibility.13 These mechanical properties seem to be due to diverse chemical compositions such as DOPA–DOPA covalent coupling, DOPA–thiol bonding, cation–π interaction, and metal–DOPA chelation, as well as structural aspects such as the secondary structure of mussel cuticle proteins in the cuticle and a granule-embedded cuticle structure with a high density of metal–DOPA complexes.13,14,17,38,39 Of these, DOPA can modulate the mechanical properties of the cuticle via two pathways: (1) covalent and (2) coordinative crosslinking. Covalent crosslinking by DOPA oxidation in basic pH is quick and robust, but uncontrollable after the crosslinking reaction. In contrast, coordinative crosslinking by DOPA–metal ion complexation could be controlled by surrounding conditions (e.g., pH, type and concentration of metal ions). In addition, coordinative crosslinking by DOPA–metal ion complexation is reversible, self-healable, and has bonding energy which approaches half of that of covalent bonds.10,16,18,28,40,44 Therefore, the use of covalent bonds alone cannot maximize the benefit of DOPA-mediated adhesion; reinforcement with DOPA–metal ion complexation which provide reversibility is necessary for a functional biomaterial. Although the mechanical effect of simple crosslinks between Fe(III) and DOPA was the focus of this work, the newly developed rfp-1-based nanofibers containing Fe(III)–DOPA complexes and partial amino acid sequences of natural MAP fp-1 have potential to be investigated for further physical and structural mussel-mimetic studies.
Cytotoxicity of composite nanofibers containing Fe(III)–DOPA complexes
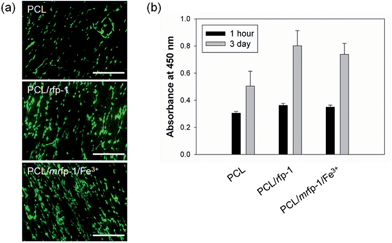
Electrospun nanofibers can be utilized as scaffolds for providing suitable cell adhesion substrata and tissue regeneration cues in tissue engineering applications.23–25 Moreover, the mechanical stiffness of scaffolds in which cell–matrix interactions occur has been known to significantly affect the self-renewal and differentiation behavior of stem cells.41,42 In addition, free Fe(III) has been known to be able to derive severe cytotoxicity through generating free radical-mediated mitochondrial damage.43 It was observed that rfp-1 addition to the nanofibers enhanced cell attachment and proliferation; further, Fe(III)–DOPA complexes in the nanofibers did not show any detectable cytotoxic and inhibitory effects on in vitro cell culture experiments (Fig. 5). Therefore, the controllable stiffness of the Fe(III)-incorporated nanofibers might be useful for stem cell research, particularly in investigating the effect of substrate elasticity on stem cell differentiation. In addition, the high strength and stiffness of the nanofibers might be adaptable for tissue engineering scaffolds because our Fe(III)–DOPA complex-based reinforcement can occur immediately under physiological conditions (pH 7.4)44 and provide sufficient mechanical endurance.Conclusions
In this study, we developed mussel-inspired electrospun nanofibers based on recombinant MAP rfp-1 and its Fe(III)–DOPA complexation chemistry. Stepwise mechanical reinforcement occurred via the incorporation of rfp-1 into PCL nanofibers and tris Fe(III)–DOPA coordinative crosslinking induced by the pH of basic buffers. The mechanical behavior of the nanofibers due to rfp-1 and its Fe(III)–DOPA complexation may be potentially exploited for basic research on fibrous mussel byssal threads and in application fields related to tissue engineering scaffolds.Acknowledgements
Financial support for this research was provided by the Marine Biotechnology Program funded by the Ministry of Oceans and Fisheries, Korea (to D. S. Hwang & H. J. Cha) and the Rising Star Program funded by POSTECH (to H. J. Cha). D. X. Oh acknowledges the National Research Foundation of Korea Grant funded by the Korean Government (MEST) (NRF-C1ABA001-2011-0029960 & NRF-2013-Fostering Core Leaders of the Future Basic Science Program).Notes and references
- C. P. Barnes, S. A. Sell, E. D. Boland, D. G. Simpson and G. L. Bowlin, Adv. Drug Deliver. Rev., 2007, 59, 1413–1433 CrossRef CAS PubMed.
- W. He, Z. Ma, T. Yong, W. E. Teo and S. Ramakrishna, Biomaterials, 2005, 26, 7606–7615 CrossRef CAS PubMed.
- J. Stitzel, J. Liu, S. J. Lee, M. Komura, J. Berry, S. Soker, G. Lim, M. Van Dyke, R. Czerw, J. J. Yoo and A. Atala, Biomaterials, 2006, 27, 1088–1094 CrossRef CAS PubMed.
- J. R. Paletta, S. Bockelmann, A. Walz, C. Theisen, J. H. Wendorff, A. Greiner, S. Fuchs-Winkelmann and M. D. Schofer, J. Mater. Sci.: Mater. Med., 2010, 21, 1363–1369 CrossRef CAS PubMed.
- X. Wang, Y. Si, J. Yang, B. Ding, L. Chen, Z. Hu and J. Yu, Nanoscale, 2013, 5, 886–889 RSC.
- X. Wang, Y. Yan, M. J. Yost, S. A. Fann, S. Dong and X. Li, J. Biomed. Mater. Res., Part A, 2007, 83, 130–135 CrossRef PubMed.
- J. O. Zoppe, M. S. Peresin, Y. Habibi, R. A. Venditti and O. J. Rojas, ACS Appl. Mater. Inter., 2009, 1, 1996–2004 CrossRef CAS PubMed.
- J. Lee, G. Tae, Y. H. Kim, I. S. Park and S. H. Kim, Biomaterials, 2008, 29, 1872–1879 CrossRef CAS PubMed.
- M. S. Kim, I. Jun, Y. M. Shin, W. Jang, S. I. Kim and H. Shin, Macromol. Biosci., 2010, 10, 91–100 CrossRef CAS PubMed.
- B. P. Lee, P. B. Messersmith, J. N. Israelachvili and J. H. Waite, Annu. Rev. Mater. Res., 2011, 41, 99–132 CrossRef CAS PubMed.
- H. J. Cha, D. S. Hwang and S. Lim, Biotechnol. J., 2008, 3, 631–638 CrossRef CAS PubMed.
- H. G. Silverman and F. F. Roberto, Mar. Biotechnol., 2007, 9, 661–681 CrossRef CAS PubMed.
- E. Vaccaro and J. H. Waite, Biomacromolecules, 2001, 2, 906–911 CrossRef CAS.
- N. Holten-Andersen, G. E. Fantner, S. Hohlbauch, J. H. Waite and F. W. Zok, Nat. Mater., 2007, 6, 669–672 CrossRef CAS PubMed.
- J. H. Waite, J. Biol. Chem., 1983, 258, 2911–2915 CAS.
- S. W. Taylor, D. B. Chase, M. H. Emptage, M. J. Nelson and J. H. Waite, Inorg. Chem., 1996, 35, 7572–7577 CrossRef CAS.
- M. J. Harrington, A. Masic, N. Holten-Andersen, J. H. Waite and P. Fratzl, Science, 2010, 328, 216–220 CrossRef CAS PubMed.
- H. Zeng, D. S. Hwang, J. N. Israelachvili and J. H. Waite, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 12850–12853 CrossRef CAS PubMed.
- D. S. Hwang, A. Masic, E. Prajatelistia, M. Iordachescu and J. H. Waite, Acta Biomater., 2013, 9, 8110–8117 CrossRef CAS PubMed.
- B. J. Kim, Y. S. Choi and H. J. Cha, Angew. Chem., Int. Ed., 2012, 51, 675–678 CrossRef CAS PubMed.
- Y. Z. Zhang, J. Venugopal, Z. M. Huang, C. T. Lim and S. Ramakrishna, Biomacromolecules, 2005, 6, 2583–2589 CrossRef CAS PubMed.
- D. S. Hwang, Y. Gim, H. J. Yoo and H. J. Cha, Biomaterials, 2007, 28, 3560–3568 CrossRef CAS PubMed.
- S. Koombhongse, W. Liu and D. H. Reneker, J. Polym. Sci., Part B: Polym. Phys., 2001, 39, 2598–2606 CrossRef CAS.
- D. Liang, B. S. Hsiao and B. Chu, Adv. Drug Deliver. Rev., 2007, 59, 1392–1412 CrossRef CAS PubMed.
- D. R. Nisbet, J. S. Forsythe, W. Shen, D. I. Finkelstein and M. K. Horne, J. Biomater. Appl., 2009, 24, 7–29 CrossRef CAS PubMed.
- B. M. Baker, A. M. Handorf, L. C. Ionescu, W. J. Li and R. L. Mauck, Expert Rev. Med. Devices, 2009, 6, 515–532 CrossRef CAS PubMed.
- S. W. Taylor, Anal. Biochem., 2002, 302, 70–74 CrossRef CAS PubMed.
- N. Holten-Andersen, M. J. Harrington, H. Birkedal, B. P. Lee, P. B. Messersmith, K. Y. Lee and J. H. Waite, Proc. Natl. Acad. Sci. U. S. A., 2010, 108, 2651–2655 CrossRef PubMed.
- N. Holten-Andersen, T. E. Mates, M. S. Toprak, G. D. Stucky, F. W. Zok and J. H. Waite, Langmuir, 2009, 25, 3323–3326 CrossRef CAS PubMed.
- K. K. Andersson, D. D. Cox, L. Que Jr, T. Flatmark and J. Haavik, J. Biol. Chem., 1988, 263, 18621–18626 CAS.
- M. J. Sever, J. T. Weisser, J. Monahan, S. Srinivasan and J. J. Wilker, Angew. Chem., Int. Ed., 2004, 43, 448–450 CrossRef CAS PubMed.
- Y. Zhang, H. Ouyang, C. T. Lim, S. Ramakrishna and Z. M. Huang, J. Biomed. Mater. Res., Part B., 2005, 72, 156–165 CrossRef PubMed.
- I. K. Kwon and T. Matsuda, Biomacromolecules, 2005, 6, 2096–2105 CrossRef CAS PubMed.
- S. M. Lee, E. Pippel, U. Gçsele, C. Dresbach, Y. Qin, C. V. Chandran, T. Brauniger, G. Hause and M. Knez, Science, 2009, 324, 488–492 CrossRef CAS PubMed.
- S. M. Lee, G. Grass, G. M. Kim, C. Dresbach, L. Zhang, U. Gçsele and M. Knez, Phys. Chem. Chem. Phys., 2009, 11, 3608–3614 RSC.
- M. Yu, J. Hwang and T. J. Deming, J. Am. Chem. Soc., 1999, 121, 5825–5826 CrossRef CAS.
- J. H. Waite, Integr. Comp. Biol., 2002, 42, 1172–1180 CrossRef CAS PubMed.
- Q. Lu, D. S. Hwang, L. Yan and H. Zeng, Biomaterials, 2012, 33, 1903–1911 CrossRef CAS PubMed.
- D. S. Hwang, H. Zeng, Q. Lu, J. N. Israelachvili and J. H. Waite, Soft Matter, 2012, 8, 5640–5648 RSC.
- H. Lee, N. F. Scherer and P. B. Messersmith, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12999–13003 CrossRef CAS PubMed.
- D. Li, J. Zhou, F. Chowdhury, J. Cheng, N. Wang and F. Wang, Regener. Med., 2011, 6, 229–240 CrossRef PubMed.
- M. M. Nava, M. T. Raimondi and R. Pietrabissa, J. Biomed. Biotechnol., 2012, 2012, 797410 Search PubMed.
- M. Valko, H. Morris and M. T. Cronin, Curr. Med. Chem., 2005, 12, 1161–1208 CrossRef CAS.
- B. J. Kim, D. X. Oh, S. Kim, J. H. Seo, D. S. Hwang, A. Masic, D. K. Han and H. J. Cha, Biomacromolecules, 2014, 15, 1579–1585 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Contact angles of PCL/rfp-1 composite nanofibers with various mixing ratios, UV-visible spectra of PCL/rfp-1 solutions, pH dependent changes in the tensile properties of PCL/mrfp-1 nanofibers with Fe(III), pH dependent changes in the tensile properties of PCL/mrfp-1 nanofibers without Fe(III). See DOI: 10.1039/c4tb01496k |
| ‡ B. J. Kim and S. Kim contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |