Tri-peptide cationic lipids for gene delivery†
Yinan
Zhao
ab,
Shubiao
Zhang
*b,
Yuan
Zhang
c,
Shaohui
Cui
b,
Huiying
Chen
b,
Defu
Zhi
b,
Yuhong
Zhen
d,
Shufen
Zhang
*a and
Leaf
Huang
e
aState Key Laboratory of Fine Chemicals, Dalian University of Technology, Dalian 116021, Liaoning, China. E-mail: zhangshf@dlut.edu.cn; Tel: +86-411-84986265
bSEAC-ME Key Laboratory of Biotechnology and Bio-resources Utilization, Dalian Nationalities University, Dalian 116600, Liaoning, China. E-mail: zsb@dlnu.edu.cn; Fax: +86-411-87656141; Tel: +86-411-87656141
cDepartment of Materials Science and Engineering, Department of Biological Engineering, The David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
dCollege of Pharmacy, Dalian Medical University, Dalian 116044, Liaoning, China
eDivision of Molecular Pharmaceutics, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA
First published on 30th September 2014
Abstract
Several novel tri-peptide cationic lipids were designed and synthesized for delivering DNA and siRNA. They have tri-lysine and tri-ornithine as headgroups, a carbamate group as a linker and 12 and 14 carbon atom alkyl groups as tails. These tri-peptide cationic lipids were prepared into cationic liposomes for the study of the physicochemical properties and gene delivery. Their particle size, zeta potential and DNA-binding were characterized to show that they were suitable for gene transfection. Further results indicate that these lipids can transfer DNA and siRNA very efficiently into NCI-H460 and Hep-2 tumor cells. The selected lipid, CDO14, was able to deliver combined siRNAs against c-Myc and VEGF for silencing distinct oncogenic pathways in lung tumors of mice, with little in vitro and in vivo toxicity.
1 Introduction
Viral vectors and non-viral vectors are widely used to modulate gene expression in target tissues to correct genetic defects for gene therapeutic applications. Viral vectors, such as adenovirus vectors, adeno-associated virus vectors, or retrovirus vectors, have been applied in clinical trials.1–3 However, some side effects including immunoresponse and infection have been reported.4,5 Non-viral gene delivery systems have, on one hand, been demonstrated as standard tools for in vitro transfection using many commercially available kits.6–8 On the other hand, they are regarded as a promising approach for the treatment of genetic diseases and cancers.9–12Among these non-viral vectors, peptide cationic lipids have drawn great attention,13–18 as peptides are more biocompatible. Peptides and their derivatives have been used for restraining cancer cell migration, curing anti-thrombosis, treating acute renal failure, reducing anti-inflammation, promoting skin regeneration, etc. But the isolated small peptides are easily degraded by enzymes in vivo, and they could be cleared very quickly.19,20 People found that the introduction of peptide groups into lipids can prolong their half-life in vivo and enhance targeting thereof.21
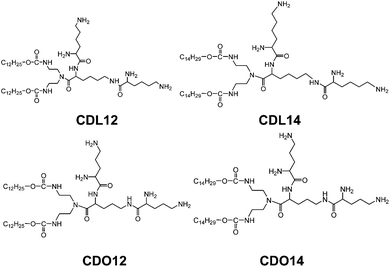
Therefore, peptide based cationic lipids have shown more superiorities over other cationic lipids, such as good biodegradability, excellent biocompatibility and targeting ability to cells, and potential applications in improving the delivery of gene therapeutics.22–24 However, the current peptide based cationic lipids still hold some limitations.25,26 Therefore, we designed and synthesized a series of novel tri-peptide cationic lipids CDL12, CDL14, CDO12 and CDO14 (Fig. 1) for facilitating the use of peptide based lipids for gene delivery.
2 Experimental procedures
Materials
9-Fluorenylmethyl N-succinimidyl carbonate (Fmoc-OSu), o-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU), lauryl alcohol, myristyl alcohol, N,N′-carbonyldiimidazole (CDI), diethylenetriamine, 3-amino-1,2-propanediol, lysine and ornithine were purchased from Shanghai Aladdin Industrial Inc. (China). Lipofectamine 2000 was purchased from Invitrogen Life Technologies (USA). 1,2-Dioleoyl-3-trimethylammonium-propane chloride salt (DOPE) and anti-luciferase siRNA were purchased from Sigma-Aldrich (USA). 1,2-Dioleoyl-3-trimethylammoniumpropane (DOTAP) was purchased from Roche Shanghai (China). Hep-2 and NCI-H460 cells were purchased from the Institute of Biochemistry and Cell Biology (China). A549 cells were from Leaf's lab of the University of North Carolina at Chapel Hill. DMEM, RPMI1640, fetal bovine serum (FBS), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Gibco (USA). Green fluorescent protein (pGFP-N2) and luciferase (pGL3) plasmid vectors were purchased from Sangon Biotech Co. Ltd. (China), and extracted in our lab. The Bright-Glo™ Luciferase assay system was purchased from Promega Biotech Co. Ltd. (Beijing, China). All other chemicals were of reagent grade.Synthesis of lipids
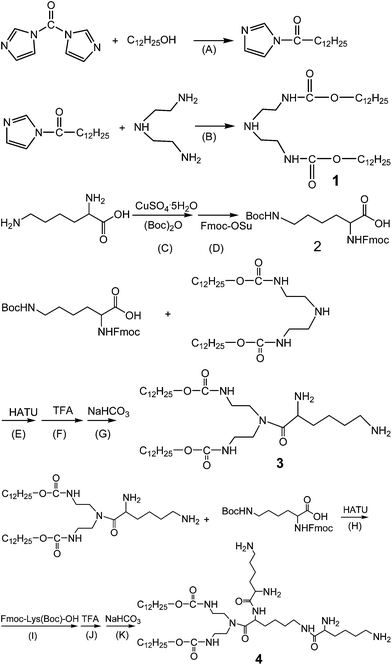
The synthesis of these lipids was performed as follows, using CDL12 as an example (Scheme 1): CDI was added to lauryl alcohol in toluene and kept at reflux for 3 h. Diethylenetriamine was added. After another 3 h, the solvent was evaporated. The crude product was recrystallised from the mixture solvent of ethanol–water to give intermediate 1. Boc2O in acetone was added to L-Lys aqueous solution. CuSO4·5H2O was added and kept stirring for 14 h at room temperature. Anhydrous sodium carbonate, 8-hydroxyquinoline and Fmoc-OSu were added. The solution was stirred at room temperature for 6–7 h before being filtered. The filtrate was cooled to 0 °C, and pH was adjusted to 2–3 with HCl solution. The crude product was recrystallised from the mixture solvent of acetonitrile and petroleum ether to give intermediate 2 as Fmoc-Lys (Boc)-OH.For the construction of CDL12, intermediate 2 was added to HATU in dichloromethane. After a reaction for 1 h, a dichloromethane solution of intermediate 1 was added. The Boc group was removed with CHCl3/TFA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the Fmoc group with dioxane/NaHCO3 to give intermediate 3 as H-Lys-OCD12. The reaction was repeated as the above steps to give the crude product with intermediates 2 and 3. The resulting sticky paste was recrystallised from the mixed solvent of acetonitrile and petroleum ether to give a white crystal as CDL12.
1) and the Fmoc group with dioxane/NaHCO3 to give intermediate 3 as H-Lys-OCD12. The reaction was repeated as the above steps to give the crude product with intermediates 2 and 3. The resulting sticky paste was recrystallised from the mixed solvent of acetonitrile and petroleum ether to give a white crystal as CDL12.
Chemical analysis of lipids
1H and 13C NMR spectra were recorded at 400 and 100 MHz, respectively, on a Varian MercuryPlus 400 spectrometer. High resolution electrospray ionization mass spectrometry was performed using a Q-Tof Micro (Micromass Inc., Manchester, UK). IR spectra were recorded using an IRPrestige-21 (Shimadzu, Japan). HPLC was performed on a LC-20AB (Shimadzu, Japan) with a C18 column, 1% TFA in the methanol–water mobile phase (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15), at a flow rate of 0.8 mL min−1.
15), at a flow rate of 0.8 mL min−1.
Preparation and characterisation of liposomes
The protocol for preparation of liposomes based on CDL12, CDL14, CDO12 and CDO14 lipids was optimised to obtain high-quality delivery systems. The lipids were formulated in combination with the neutral lipid DOPE in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. To prepare liposomes, suitable amounts of lipid and DOPE were dissolved in 1 mL of chloroform in a 5 mL glass vial. The solvent was removed under a stream of nitrogen gas, followed by high-vacuum desiccation. The dry lipid film was resuspended in 1 mL distilled water to give liposomes in a concentration of approximately 1 mg mL−1. The liposome/DNA complexes of varying weight ratios (N/P) were prepared by mixing liposomes with plasmid solutions 20 min before use.
1 molar ratio. To prepare liposomes, suitable amounts of lipid and DOPE were dissolved in 1 mL of chloroform in a 5 mL glass vial. The solvent was removed under a stream of nitrogen gas, followed by high-vacuum desiccation. The dry lipid film was resuspended in 1 mL distilled water to give liposomes in a concentration of approximately 1 mg mL−1. The liposome/DNA complexes of varying weight ratios (N/P) were prepared by mixing liposomes with plasmid solutions 20 min before use.
Particle size and zeta potential measurements
The particle size and zeta potential were measured with a Malvern ZetaSizer Nano series (Westborough, MA) at room temperature. All the lipoplexes of varying N/P ratios ranging from 1 to 8 were prepared with pGL3 plasmid. For the measurement of particle size and zeta potential, 20 μL of the liposomes or lipoplexes were diluted to 1 mL with distilled water.Agarose gel retardation assay
The lipoplexes of varying N/P ratios ranging from 0 to 8 were prepared with pGL3 plasmid. The lipoplexes were electrophoresed in 1.2% agarose gel containing GelRed and Tris-acetate (TAE) running buffer at 90 V for 40 min. DNA bands were visualised in a gel documentation unit (Syngene, Britain).In vitro pDNA transfection
Hep-2 and NCI-H460 cells were seeded in 24-well plates (5.0 × 104 cells per well), and incubated at 37 °C under 5% CO2 until approximately 80% confluence was attained. The medium was removed and replaced with 100 μL serum-free DMEM per well. Cationic liposome/DNA lipoplexes were then added to the plates and incubated for 4 h at 37 °C. The medium was then replaced with DMEM containing 10% FBS and 1% antibiotics, and the cultures were maintained at 37 °C under 5% CO2 for 48 h.The expression of green fluorescent protein was measured using an inverted fluorescence microscope (Olympus IX71, Japan). Relative luciferase activity was assessed using a Bright-Glo™ Luciferase Assay System and a Synergy™ 2 Multi-Detection Microplate Reader. Briefly, the growth medium was removed from each well, and luciferase activity was measured after lysis buffer was added. The total protein concentration in the cell lysate was measured using a BCA protein assay kit and luciferase activity was expressed as relative light units (RLU) per mg protein.
In vitro siRNA transfection
A549 cells expressing firefly luciferase were seeded in 12-well plates (1.0 × 105 per well) approximately 24 h before experiments. Cells were treated with different lipid formulations at an anti-luciferase siRNA concentration of 40 nM in Opti-MEM at 37 °C for 4 h. Cells were washed with DPBS, followed by incubation with lysis buffer at room temperature for 20 min.Fluorescence intensity in cell lysate was determined using a Perkin-Elmer LS 50B luminescence spectrometer (Norwalk, CT) (λex, 494 nm; λem, 519 nm). Cell lysate (5 μg per well) was dissolved in BCA protein assay reagent, and total protein concentrations were determined at 570 nm. The silencing rate is expressed as relative light units (RLU) per microgram of total protein.
Cytotoxicity of the cationic liposomes
The assay was performed in 96-well plates by maintaining the N/P weight ratios as used in the transfection experiments. The cells were seeded in 96-well plates and incubated at 37 °C under 5% CO2 for 24 h to get a confluence of about 80%. After the replacement of the medium, the liposome/DNA complexes were added to the cells for further culture at 37 °C under 5% CO2 for 24 h, and then MTT (20 μL, 5 mg mL−1) was added and kept for 4.5 h. The absorbance at 570 nm was monitored by using the enzyme mark instrument (Sunrise Tecan, Australia).Animals
In vivo studies were carried out on BALB/c nude mice 4–6 weeks of age. All experiments performed on the animals were in accordance with and approved by the Institutional Animal Care and Use Committee at University of North Carolina.In vivo gene silencing study
Combined siRNAs against c-Myc and VEGF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were co-delivered by the CDO14 liposome. The tumor-bearing mice were injected 4 times every 3 days via tail vein with lipoplexes at a N/P ratio of 3
1) were co-delivered by the CDO14 liposome. The tumor-bearing mice were injected 4 times every 3 days via tail vein with lipoplexes at a N/P ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (with a dose of 0.45 mg total siRNA per kg). One day after the third injection, the mice were killed and the tumor-loaded lungs were collected for the preparation of paraffin embedded sections (4–5 μm thick). Expressions of c-Myc and VEGF in the sections were examined immunohistochemically using the antibodies from a kit in accordance with the product protocol.
1 (with a dose of 0.45 mg total siRNA per kg). One day after the third injection, the mice were killed and the tumor-loaded lungs were collected for the preparation of paraffin embedded sections (4–5 μm thick). Expressions of c-Myc and VEGF in the sections were examined immunohistochemically using the antibodies from a kit in accordance with the product protocol.
TUNEL assay
TUNEL staining was performed as recommended by the manufacturer's protocol (Promega, Madison, WI). Tumor-bearing mice were given i.v. injections of the siRNA formulated in the CDO14 on days 9 and 10. Twenty-four hours after the second injection, the mice were killed, and the tumor-loaded lungs were collected for the TUNEL staining. The nuclei were counterstained with hematoxylin, and the samples were imaged using an inverted fluorescence microscope (Olympus IX71, Japan).Toxicity of the cationic liposomes in vivo
The tumor-bearing mice were i.v. injected with siRNA formulated in the targeted liposome at 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 doses, and the serum cytokine level was determined with an Olympus AU400 Automatic Biochemistry Analyzer. The mouse body weight was also monitored.
1 doses, and the serum cytokine level was determined with an Olympus AU400 Automatic Biochemistry Analyzer. The mouse body weight was also monitored.
Statistical analysis
The data were presented as mean values ± SD. The statistical significance was determined using two-way analysis of variance. P values <0.05 were considered significant.3 Results and discussion
Design and synthesis of cationic lipids
The tri-peptide cationic lipids designed are composed of three biocompatible molecular moieties: tri-lysine peptides or tri-ornithine peptides (the cationic headgroup), carbamate (the linker), and two hydrocarbon chains (hydrophobic tails). These tri-peptide cationic lipids were prepared by adapting a straightforward procedure (Scheme 1). Intermediate 1 was obtained from carbonyldiimidazole, dodecyl alcohol and triethylenetetraamine. The synthesis of intermediate 2 started with Fmoc and Boc group protected lysine. After the condensation under HATU activation and the deprotection by using TFA and NaHCO3, intermediate 3 could be obtained. Following the same procedure by using another 2 mol Boc and Fmoc protected lysine; we got tri-peptide cationic lipids for further chemical analysis and biological study.The structures and purities of tri-peptide cationic lipids were characterised by electrospray ionisation-mass spectrometry (ESI-MS), proton nuclear magnetic resonance spectroscopy (1H NMR, 13C NMR), infrared (IR) spectroscopy, and high pressure liquid chromatography (HPLC) (Fig. S1–S4, ESI†). The analytical data of these compounds are listed under the figures in the ESI† to show that tri-peptide cationic lipids were successfully obtained.
Particle size, and zeta-potential of tri-peptide cationic liposomes and lipoplexes
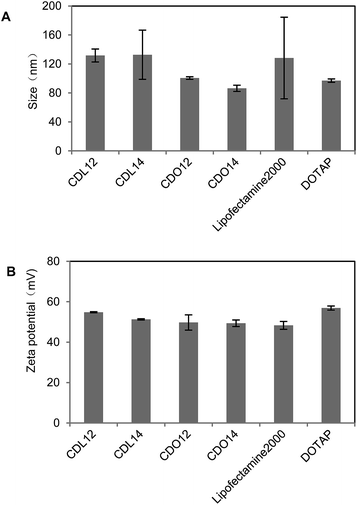
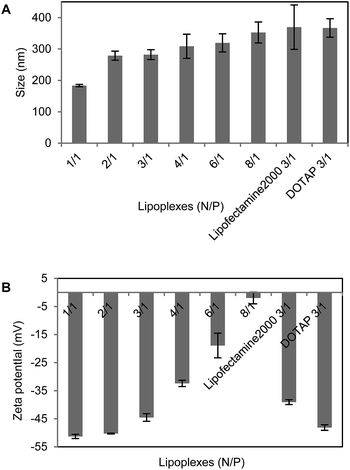
Our previous study showed that liposomes had higher transfection efficiency with an equivalent molar ratio of DOPE, so the liposomes were prepared by using the tri-peptides with DOPE as a co-lipid at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The dynamic light scattering showed that the particle sizes of these liposomes were from 70 nm to 150 nm, those of the lipoplexes of CDO14/DNA were from 200 nm to 400 nm in the N/P ratio range of 1
1. The dynamic light scattering showed that the particle sizes of these liposomes were from 70 nm to 150 nm, those of the lipoplexes of CDO14/DNA were from 200 nm to 400 nm in the N/P ratio range of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 8
1 to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 2 and 3). Zeta potentials of liposomes were between 40 and 60 mV, which ensured the stability of the liposomes for at least three months. Zeta potentials of the lipoplexes of CDO14/DNA were found to increase from −50 mV to nearly 0 mV with the increase of the N/P ratios from 1
1 (Fig. 2 and 3). Zeta potentials of liposomes were between 40 and 60 mV, which ensured the stability of the liposomes for at least three months. Zeta potentials of the lipoplexes of CDO14/DNA were found to increase from −50 mV to nearly 0 mV with the increase of the N/P ratios from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 8
1 to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 2 and 3). The sizes and zeta potentials were in the ranges suitable for gene transfection.27 The morphological characteristics were directly visualised by transmission electron microscopy (TEM) (Fig. S5, ESI†). Images obtained from the CDO14 liposome and CDO14 liposome/DNA lipoplex at a N/P ratio of 3
1 (Fig. 2 and 3). The sizes and zeta potentials were in the ranges suitable for gene transfection.27 The morphological characteristics were directly visualised by transmission electron microscopy (TEM) (Fig. S5, ESI†). Images obtained from the CDO14 liposome and CDO14 liposome/DNA lipoplex at a N/P ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 revealed well-defined spherical structures.
1 revealed well-defined spherical structures.
DNA-binding assay
DNA-binding with liposomes is a key factor for gene transfection.28 Therefore, the electrostatic binding interactions between plasmid DNA and the tri-peptide cationic liposomes were measured by the gel retardation assay across the N/P ratios of 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–8
1–8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Findings in the gel retardation assay revealed all the liposomes could start to retard the plasmid DNA at a N/P ratio of 0.5
1. Findings in the gel retardation assay revealed all the liposomes could start to retard the plasmid DNA at a N/P ratio of 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. They could nearly completely retard pDNA at higher N/P ratios of 6
1. They could nearly completely retard pDNA at higher N/P ratios of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 6
1, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 6
1, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 3
1 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for CDL12, CDL14, CDO12 and CDO14, respectively. Though all the above liposomes could bind pDNA at higher N/P ratios, the lower N/P ratio of CDO14 for retardation compared with other liposomes may reduce its dose required to obtain maximal transfection efficiency. The above results demonstrate that cationic liposomes can form DNA/liposomes complexes at different N/P ratios. This ensured the entry of pDNA into cells and the protection of pDNA from degradation by DNase. (Fig. S6, ESI†).
1 for CDL12, CDL14, CDO12 and CDO14, respectively. Though all the above liposomes could bind pDNA at higher N/P ratios, the lower N/P ratio of CDO14 for retardation compared with other liposomes may reduce its dose required to obtain maximal transfection efficiency. The above results demonstrate that cationic liposomes can form DNA/liposomes complexes at different N/P ratios. This ensured the entry of pDNA into cells and the protection of pDNA from degradation by DNase. (Fig. S6, ESI†).
In vitro pDNA and siRNA transfection
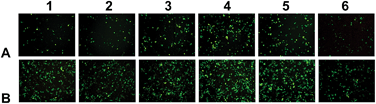
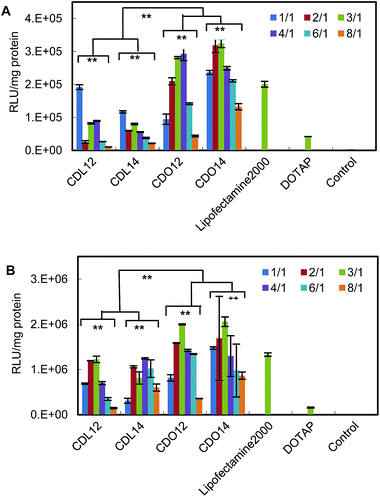
The in vitro transfection of these liposomes was evaluated using pGFP-N2 plasmid DNA and pGL3 plasmid DNA against Hep-2 and NCI-H460 cells. The expression of pGFP-N2 and pGL3 is illustrated in Fig. 4 (for more details see Fig. S7 and S8, ESI†) and Fig. 5, respectively. The results indicate that all the lipids were comparable to lipofectamine 2000 and DOTAP in terms of GFP and luciferase expression. They had higher transfection against Hep-2 cells than against NCI-H460 cells. The liposomes with the ornithine headgroup also showed higher transfection than those with the lysine headgroup, perhaps because they have smaller heads.Through the above experiments, the transfection in cells will undergo the following process. First, the plasmid DNA binds to the tri-peptide head-groups of the cationic lipids. Then, the lipid/DNA lipoplexes will enter the Hep-2 cells and NCI-H460 cells (e.g., via endocytosis), and trypsin in the cell will hydrolyze the carbamate bond in the lipids. The hydrolization assists DNA to escape from the endosome for the subsequent transcription. The whole process is readily performed and thus enhances DNA transfection efficiency.
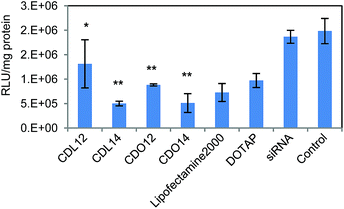
Tri-peptide lipids were screened for the ability to deliver siRNA to A549 lung carcinoma cells which can stably express firefly luciferase (Fig. 6). The efficacy of siRNA delivery by these tri-peptide lipids was determined by treating cells with siRNA-complexes, prepared using a firefly luciferase targeting siRNA (siLuc).29,30 Overall, all four lipids showed gene silencing compared with the control. Three of them were better than DOTAP, and CDO14 and CDL14 were slightly better than lipofectamine 2000, silencing nearly 75% of luciferase in A549 cells. Much work has been carried out on modifying cationic lipids for use in gene transfection, to determine if there is a ‘best’ chain length.31 However, solid conclusions are rarely obtained, for the conclusions of these studies are frequently contradictory. In this study, CDL14 and CDO14 showed superior siRNA delivery. It may maintain an appropriate balance between fluidity and rigidity for entering into cells, and have the ability to release siRNA in cells due to the carbamate linker.
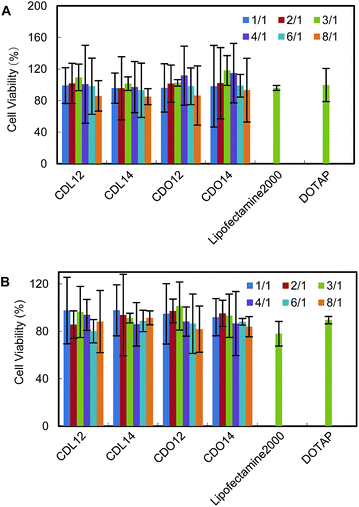
Cytotoxicity of the cationic liposomes
The cytotoxicity of gene delivery materials could be measured by the MTT method. Cell viabilities after exposure to lipids, CDL12, CDL14, CDO12 and CDO14, were evaluated in NCI-H460 cells and Hep-2 cells across the entire range of N/P weight ratios used in the transfection experiments. Cells could tolerate all the lipids at relatively low N/P ratios (under 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), better than the commercial liposomes, lipofectamine 2000 and DOTAP (Fig. 7). The lower cytotoxicity of these lipids should be attributed to the nature of the peptide headgroups, as they are more biocompatible over quaternary ammonium with cells, thus increasing the tolerance of cells to them.19,24
1), better than the commercial liposomes, lipofectamine 2000 and DOTAP (Fig. 7). The lower cytotoxicity of these lipids should be attributed to the nature of the peptide headgroups, as they are more biocompatible over quaternary ammonium with cells, thus increasing the tolerance of cells to them.19,24
In vivo gene silencing study
Since cationic lipid CDO14 showed optimal in vitro pDNA and siRNA transfection, its in vivo delivery was studied. Combined siRNAs against c-Myc and VEGF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were delivered by the CDO14 liposome. The A549 lung tumor bearing mice were injected 3 times every 3 days via tail vein with lipoplexes at a N/P ratio of 3
1) were delivered by the CDO14 liposome. The A549 lung tumor bearing mice were injected 3 times every 3 days via tail vein with lipoplexes at a N/P ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (with a dose of 0.45 mg total siRNA per kg). As immunohistochemistry could give clear conclusion of RNA interference,17,31,32 the gene silencing activity was measured by using this method. As shown in Fig. 8, the protein expression of c-Myc and VEGF in the lung tumor was suppressed by the combined siRNAs delivered with the CDO14 liposome. The results indicate that the combined siRNAs formulated in the CDO14 liposome were able to simultaneously silence the expressions of the target oncogenes in the lung tumors. The successful interference of combined siRNAs to different genes opens a door by silencing multi-oncogenic pathways by using tri-peptide cationic lipids. This increases the possibility of CDO14 as a potential candidate for further in vivo studies and clinical trials.
1 (with a dose of 0.45 mg total siRNA per kg). As immunohistochemistry could give clear conclusion of RNA interference,17,31,32 the gene silencing activity was measured by using this method. As shown in Fig. 8, the protein expression of c-Myc and VEGF in the lung tumor was suppressed by the combined siRNAs delivered with the CDO14 liposome. The results indicate that the combined siRNAs formulated in the CDO14 liposome were able to simultaneously silence the expressions of the target oncogenes in the lung tumors. The successful interference of combined siRNAs to different genes opens a door by silencing multi-oncogenic pathways by using tri-peptide cationic lipids. This increases the possibility of CDO14 as a potential candidate for further in vivo studies and clinical trials.
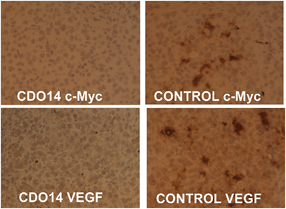 | ||
| Fig. 8 Immunohistochemical analysis of the lung tumor. SiRNAs against c-Myc and VEGF were delivered with (left) or without (right) the CDO14 liposome. | ||
TUNEL assay
Apoptotic induction plays an essential role in the suppression of tumor cell proliferation.33 We examined the apoptosis induced by siRNAs using the TdT dUTP nick end-labeling (TUNEL) staining (Fig. 9). As shown in the figure, the number of TUNEL-positive cells increased after intravenous injections of siRNA formulated in the CDO14 liposome. Intravenous injections of siRNA in the control group showed only a slight increase in TUNEL-positive cells. No significant apoptotic induction was observed in other treatment groups. These results suggest that this formulation triggered cell death and might play a critical role in regulating the survival of A549 lung cancer cells in vivo.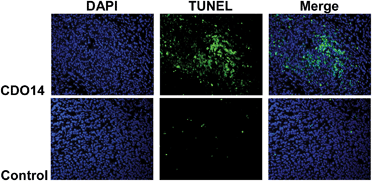 | ||
| Fig. 9 TUNEL staining of A549 lung tumors on day 9 after 3 consecutive i.v. injections on days 0, 3, and 6 of combined siRNAs. | ||
Toxicity of the cationic liposomes in vivo
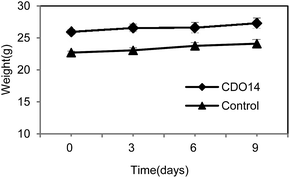
In addition to therapeutic effects, toxicity is a crucial parameter of a therapeutic agent for clinical use. At the therapeutic dose (0.45 mg kg−1), the formulation did not induce significant elevation of renal indicators and liver enzymes, including blood urea nitrogen (BUN), creatinine, aspartate aminotransferase (AST) and alanine aminotransferase (ALT) (Table 1). Additionally, the body weights of mice were stably increased after post-injection of lipoplexes at the therapeutic dose (Fig. 10). The results indicate that the formulation containing CDO14 did not induce obvious toxicity to mice. The toxicity to cells and animals is a major obstacle for the clinical trials of cationic lipids, as the cationic lipids with a quaternary ammonium headgroup can become cytotoxic by interacting with critical enzymes such as PKC.34| Kidney | Liver | |||
|---|---|---|---|---|
| BUN mg dL−1 | Creatinine mg dL−1 | AST U L−1 | ALT U L−1 | |
| CDO14 lipoplex | 18 | 0.1 | 87 | 51 |
| 20 | 0.2 | 102 | 59 | |
| 24 | 0.2 | 82 | 59 | |
| Control | 20 | 0.1 | 112 | 97 |
| 19 | 0.1 | 157 | 71 | |
| Normal | 8–33 | 0.1–0.2 | 54–298 | 17–77 |
Therefore, the toxicity of cationic lipids is mainly determined by their cationic nature. The replacement of quaternary ammonium with the tri-peptide increased the affinity and biocompatibility, thereby, increasing their transfection and lowering their toxicity.
4 Conclusion
In summary, we selected the tri-peptides Lys–Lys–Lys and Orn–Orn–Orn as the headgroups for the gene delivery study, because both lysine and ornithine are found in a number of structurally characterized DNA binding domains of proteins. These cationic lipids are relatively simple to synthesize by using standard peptide chemistry, and their chemical structures can readily be tailored for effective interaction with DNA. The in vitro delivery results of pDNA and siRNA by these tri-peptide lipids against NCI-H460 and Hep-2 tumor cells show that they could lead to superior transfection compared to conventional non-viral transfection reagents. They also showed lower cytotoxicity against the two tumor cells. The lipid CDO14 with ornithine as the headgroup and 14 carbon atom alkyl groups as tails demonstrated better transfection over other lipids. It was selected to be used for in vivo evaluation. It showed not only enhanced transfection activity, but also very low toxicity in vivo. CDO14 could be able to deliver combined siRNAs against c-Myc and VEGF for silencing distinct oncogenic pathways in lung tumors of mice. The results of this study strengthen the validity of the peptide based cationic lipids as drug delivery vehicles for cancer therapy.Acknowledgements
This research is financially supported by the National High Technology Research and Development Program (863 Program) of China (2014AA020707), the National Natural Science Foundation of China (20876027 and 21176046), the Fundamental Research Funds for the Central Universities (DC12010104) and NIH grants (CA129835, CA129421, CA149363).Notes and references
- R. G. Crystal, Science, 1995, 270, 148–151 Search PubMed.
- V. Chandrashekhar, M. Srujan, R. Prabhakar, R. C. Reddy, B. Sreedhar, K. K. R. Rentam, S. Kanjilal and A. Chaudhuri, Bioconjugate Chem., 2011, 3, 497–509 CrossRef PubMed.
- K. M. Xiu, N. N. Zhao, W. T. Yang and F. Xu, Acta Biomater., 2013, 9, 7439–7448 CrossRef CAS PubMed.
- R. E. Donahue, S. W. Kessler and D. Bodine, J. Exp. Med., 1992, 176, 1125–1135 CrossRef CAS.
- J. Y. Sun, V. Anand-Jawa and S. Chatterjee, Gene Ther., 2003, 10, 964–976 CrossRef CAS PubMed.
- O. Nakagawa, X. Ming, L. Huang and R. L. Juliano, J. Am. Chem. Soc., 2010, 132, 8848–8849 CrossRef CAS PubMed.
- D. F. Zhi, S. B. Zhang, F. Qureshi, Y. N. Zhao, S. H. Cui, B. Wang, H. Y. Chen, B. L. Yang and D. F. Zhao, Colloids Surf., B, 2013, 1, 537–541 CrossRef PubMed.
- C. J. Bishop, T.-M. Ketola, S. Y. Tzeng, J. C. Sunshine, A. Urtti, H. Lemmetyinen, E. Vuorimaa-Laukkanen, M. Yliperttula and J. J. Green, J. Am. Chem. Soc., 2013, 18, 6951–6957 CrossRef PubMed.
- S. Altieri, M. Balzi, S. Bortolussi, P. Bruschi, L. Ciani, A. M. Clerici, P. Faraoni, C. Ferrari, M. A. Gadan, L. Panza, D. Pietrangeli, G. Ricciardi and S. Ristori, J. Med. Chem., 2009, 52, 7829–7835 CrossRef CAS PubMed.
- S. B. Zhang, Y. N. Zhao, B. D. Zhao and B. Wang, Bioconjugate Chem., 2010, 21, 1003–1009 CrossRef CAS PubMed.
- D. F. Zhi, S. B. Zhang, S. H. Cui, Y. N. Zhao, Y. H. Wang and D. F. Zhao, Bioconjugate Chem., 2013, 24, 487–519 CrossRef CAS PubMed.
- S. B. Zhang, Y. N. Zhao, D. F. Zhi and S. F. Zhang, Bioorg. Chem., 2012, 40, 10–18 CrossRef PubMed.
- J. C. Rea, R. F. Gibly and A. E. Barron, Acta Biomater., 2009, 5, 903–912 CrossRef CAS PubMed.
- D. J. Coles, A. Esposito and H. T. Chuah, Tetrahedron, 2010, 66, 5435–5441 CrossRef CAS PubMed.
- D. Zhou, C. Li, Y. L. Hu, H. Zhou, J. T. Chen, Z. P. Zhang and T. Y. Guo, Chem. Commun., 2012, 48, 4594–4596 RSC.
- M. Jiang, L. Gan, C. L. Zhu, Y. Dong, J. P. Liu and Y. Gan, Biomaterials, 2012, 30, 7621–7630 CrossRef PubMed.
- C. Gehin, J. Montenegro, E.-K. Bang, A. Cajaraville, S. Takayama, H. Hirose, S. Futaki, S. Matile and H. Riezman, J. Am. Chem. Soc., 2013, 135, 9295–9298 CrossRef CAS PubMed.
- A. Saha, G. Mondal, A. Biswas, I. Chakraborty, B. Jana and S. Ghosh, Chem. Commun., 2013, 49, 6119–6122 RSC.
- Y. N. Zhao, S. B. Zhang, S. H. Cui, B. Wang and S. F. Zhang, Expert Opin. Drug Delivery, 2012, 1, 127–139 CrossRef PubMed.
- K. W. Wang, X. H. Yan, Y. Cui, Q. He and J. B. Li, Bioconjugate Chem., 2007, 18, 1735–1738 CrossRef CAS PubMed.
- A. Weiss, P. Neuberg, S. Philippot, P. Erbacher and C. O. Weill, Biotechnol. Bioeng., 2011, 108, 2477–2487 CrossRef CAS PubMed.
- C. E. Ashley, E. C. Carnes, K. E. Epler, D. P. Padilla, G. K. Phillips, R. E. Castillo, D. C. Wilkinson, B. S. Wilkinson, C. A. Burgard, R. M. Kalinich, J. L. Townson, B. Chackerian, C. L. Willman, D. S. Peabody, W. Wharton and C. Jeffrey Brinker, ACS Nano, 2012, 3, 2174–2188 CrossRef PubMed.
- M. Rajesh, J. Sen, M. Srujan, K. Mukherjee, B. Sreedhar and A. Chaudhuri, J. Am. Chem. Soc., 2007, 129, 11408–11420 CrossRef CAS PubMed.
- C. M. Lamanna, H. Lusic, M. Camplo, T. J. Mclntosh, P. Barthelemy and M. W. Grinstaff, Acc. Chem. Res., 2012, 7, 1026–1038 CrossRef PubMed.
- T. Takeuchi, J. Montenegro, A. Hennig and S. Matile, Chem. Sci., 2011, 2, 303–307 RSC.
- M. E. Davis, J. E. Zuckerman, C. H. J. Choi, D. Seligson, A. Tolcher, C. A. Alabi, Y. Yen, J. D. Heidel and A. Ribas, Nature, 2010, 464, 1067–1070 CrossRef CAS PubMed.
- S. R. Sarker, Y. Aoshima, R. Hokama, T. Inoue, K. Sou and S. Takeoka, Int. J. Nanomed., 2013, 8, 1361–1375 CrossRef PubMed.
- Y. Obata, S. Saito, N. Takeda and S. Takeoka, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 1148–1158 CrossRef CAS PubMed.
- W. Qu, S. Y. Qin, Y. Kuang, R. X. Zhuo and X. Z. Zhang, J. Mater. Chem. B, 2013, 1, 2147–2154 RSC.
- H. X. Zeng, H. C. Little, T. N. Tiambeng, G. A. Williams and Z. B. Guan, J. Am. Chem. Soc., 2013, 135, 4962–4965 CrossRef CAS PubMed.
- J. Willibald, J. Harder, K. Sparrer, K. K. Conzelmann and T. Carell, J. Am. Chem. Soc., 2012, 134, 12330–12333 CrossRef CAS PubMed.
- H. Hatakeyama, H. Akita, E. Ito, Y. Hayashi, M. Oishi, Y. Nagasaki, R. Danev, K. Nagayama, N. Kaji, H. Kikuchi, Y. Baba and H. Harashima, Biomaterials, 2011, 32, 4306–4316 CrossRef CAS PubMed.
- H. Li, H. Zhu and C. J. Xu, Cell, 2008, 94, 491–501 CrossRef.
- H. T. Lv, S. B. Zhang, B. Wang, S. H. Cui and J. Yan, J. Controlled Release, 2006, 114, 100–109 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: IR, NMR, HR-ESI-MS and HPLC characterisation of all the lipids synthesized; TEM images of the CDO14 liposome and CDO14 liposome/DNA lipoplex; gel electrophoresis of liposome/pDNA complexes; in vitro transfection images against NCI-H460 and Hep-2 cells. See DOI: 10.1039/c4tb01312c |
| This journal is © The Royal Society of Chemistry 2015 |