A fluorescence nanosensor for lipase activity: enzyme-regulated quantum dots growth in situ†
Abstract
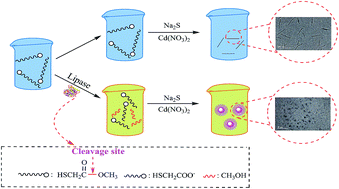
Lipase is a neglected enzyme in the field of quantum dots-based enzyme assays. We have developed a novel analytical assay to detect the lipase activity based on the enzyme-regulated quantum dots growth in situ. Catalytic hydrolytic cleavage of the ester bond in methyl thioglycolate (METG) will produce mercaptoacetate, and Na2S interacts with Cd(NO3)2 to give CdS nanocrystals stabilized by mercaptoacetate. The fluorescence intensity changes, which depends on the absence or presence of lipase, could be used to sense the lipase activity. The detection limit (3σ) as low as 1.2 × 10−2 mg mL−1 with a linear range from 0.05 to 1.6 mg mL−1 was achieved. A preliminary enzyme activity screening was carried out for four commercial purchased lipase samples, and the results are consistent with ones obtained from titration assay. Moreover, we applied this method to detecting lipase activity in fermentation broth of Bacillus subtilis without any pretreatment. The reported methodology shows potential applications for the development of a simple and inexpensive assay for the hydrolysis enzymes.

 Please wait while we load your content...
Please wait while we load your content...