Nanoparticles' interference in the evaluation of in vitro toxicity of silver nanoparticles†
Abstract
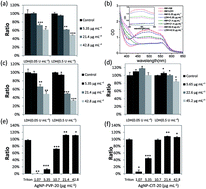
The biological safety of nanomaterials is a worldwide concern considering the extensive usage of nanoparticles in daily life as well as in medical care. However, toxicity evaluation of nanomaterials has met difficulties because of unique properties of nanomaterials. Here, we have investigated the interference of nanoparticles on the toxicity evaluations. Silver nanoparticles (AgNPs) with different sizes and surface coatings were used as model materials. Several widely used assays, such as lactate dehydrogenase (LDH) release, MTS assay and nitric oxide (NO) measurement, have been investigated in our study. Our results showed that 20 nm PVP-coated AgNPs with a concentration of 42.8 μg mL−1 affected LDH detection by about 50%, while citrate-coated 20 nm AgNPs with a concentration of 42.8 μg mL−1 affected LDH detection by about 70%. Moreover, 20 nm AgNPs with a concentration of 42.8 μg mL−1 disturbed MTS assay and NO measurement by less than 20% and 10%, respectively. Based on our results, nanoparticles with higher concentrations gave more interference. Therefore, for accurate toxicity evaluation of nanoparticles, it is very necessary to limit particle concentrations or choose other approaches free from the interference.

 Please wait while we load your content...
Please wait while we load your content...