A green route for the synthesis of a bitter-taste dipeptide combining biocatalysis, heterogeneous metal catalysis and magnetic nanoparticles
Abstract
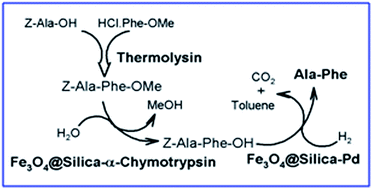
There is increasing demand for green technologies to produce high-solubility and low-toxicity compounds with potential application in the food industry. This study aimed to establish a clean, synthetic route for preparing the bitter-taste dipeptide Ala-Phe, a potential substitute for caffeine as a food additive. Synthesis of Z-Ala-Phe-OMe starting from Z-Ala-OH and HCl·Phe-OMe was catalysed by thermolysin at 50 °C in buffer (step 1). Z-Ala-Phe-OMe ester hydrolysis to give Z-Ala-Phe-OH at 37 °C in 30% acetonitrile/buffer was catalysed by α-bovine chymotrypsin (αCT), protease with esterase activity (step 2). Hydrogenation of Z-Ala-Phe to give the desired Ala-Phe was catalysed by C/Pd in methanol (step 3). Steps 2 and 3 were optimized by using the magnetically recoverable recycling enzyme Fe3O4@silica–αCT and the magnetically recoverable metal nanocatalyst Fe3O4@silica–Pd, respectively. This inspiring combination of technologies and the original results demonstrate the suitability of using enzymes, metal catalyst and magnetic nanoparticles for easy, economical, stereoselective, clean production of an important target compound. Besides, they add to the development of peptide chemistry and catalysis.

 Please wait while we load your content...
Please wait while we load your content...