Protein adsorption and cellular uptake of AuNPs capped with alkyl acids of different length†
Jun Deng,
Honghao Zheng,
Sai Wu,
Pan Zhang and
Changyou Gao*
MOE Key Laboratory of Macromolecular Synthesis and Functionalization, Department of Polymer Science and Engineering, Zhejiang University, Hangzhou 310027, China. E-mail: cygao@mail.hz.zj.cn; Fax: +86-571-87951108
First published on 19th February 2015
Abstract
The chain length of ligands plays an important role in many biomedical applications. Yet the interaction of nanoparticles with surface-capped ligands of different length (especial the hydrophobic chains) and cells is neglected, and thereby the underlying mechanism influencing cellular uptake is unclear. Herein the role of alkyl length on AuNPs in mediating serum protein adsorption and the subsequent cellular uptake by A549 cells was studied. The AuNPs were modified with carboxylic alkyl thiols of different chain length (3MA, 11MA and 16MA), resulting in MA-AuNPs with a size of 5 nm in dry state and several tens of nm in media. The MA-AuNPs possessed a negatively charged surface, whose zeta potential was around −20 mV in serum-containing medium regardless of the length of ligands. Whereas the total amount of adsorbed serum proteins had no significant difference, the relative amounts of adhesion mediators especially vitronectin (Vn) depended on the alkyl length significantly. The 16MA-AuNPs adsorbed the largest amount of Vn, and were internalized by A549 cells with the largest quantity and good distribution in cytoplasm. Taking account all the results, the protein adsorption and its role in linking alkyl length on AuNPs to cellular uptake are figured out.
1. Introduction
Cellular delivery of drugs and biomolecules such as DNAs and proteins is a central issue in biomedicine.1 Promising delivery vehicles based on nanoparticles (NPs) have been extensively investigated.2 In particular, gold nanoparticles (AuNPs) are promising candidates for cellular delivery due to their low toxicity, biocompatibility and tunable surface functionalities.3–5 Verma et al. have reported that cationic tetraalkyl ammonium-functionalized AuNPs recognize the surface of an anionic protein through complementary electrostatic interaction and inhibit its activity.6 The activity is recovered due to the release of free protein by treating the protein–particle complex with glutathione (GSH), showing the potential of AuNPs as protein transporters.6 Many efforts have been paid to understand the cellular uptake of AuNPs, revealing that the physicochemical properties of AuNPs such as size,7 shape,7,8 and surface chemistry9 influence the uptake behaviors. Bartneck et al. reported that the uptake of nanorods by macrophages is more efficient than that of nanospheres.10 Sykes et al. showed that tumor accumulation of actively targeted NPs is 5 times faster and approximately 2-fold higher relative to their passive counterparts within the 60 nm diameter range.11Physiological environments such as blood, interstitial fluid, and cellular cytoplasm contain a complex spectrum of proteins. When NPs come into contact with the physiological environment, proteins rapidly adsorb onto their surface and form what is known as the protein “corona”.12 The interaction between proteins and NPs can change the protein conformation, and alter the composition of the nanomaterials along with their aggregation state, giving it a “biological identity” that is distinct from its “synthetic identity”-the surface chemistry, size, and shape of the nanomaterials right after synthesis.13 The biological identity is considered as the form of a NP “seen” by cells, and is responsible for the kinetics, transport, and reactivity of a NP in a physiological system.14 Consequently, many efforts have been paid to study protein adsorption on various NPs. It is known that many factors such as surface chemistry,9 size15 and shape16 of the NPs influence the protein adsorption. Lundqvist et al. identified the types of serum proteins that adsorbed onto NPs using mass spectrometry, and clarified their relationship with the particle size and surface hydrophobicity.17 Recently, Calatayud et al. showed that the proteins adsorbed on Fe3O4 NPs influence their subsequent cellular uptake rather than their original charging property on the surface.18 Kah et al. showed how amphiphilic ligands (ALs) of four different types (polyoxyethylene, cetyl ether, oligofectamine, and phosphatidylserine) affect the formation of protein corona on gold nanorods (NRs) and their impact on cellular response.19 The cellular behavior toward amphiphilic ligands-modified nanorods (NR-AL) is mediated by not only the type of ALs and the protein corona but also the resulting colloidal stability and interaction with cell culture supplements.19 Walkey et al. showed that poly(ethylene glycol) (PEG) density and NPs size together determine the mechanism and efficiency of subsequent macrophage uptake, by controlling either the identity or accessibility of adsorbed serum proteins.20 However, extensive efforts should still be paid to investigate how serum protein adsorption on NPs surfaces influences the cellular uptake, since most previous studies focus mainly on the amount of adsorbed serum proteins or a single type of model proteins such as albumin.
The chain length of ligands plays an important role at many fields. For example, Ayala et al. reported that the hydrophobicity (endowed by, for example, the different length of alkyls) of the extracellular matrix plays a considerable role in dictating cellular behaviors such as adhesion, migration and differentiation.21 Previous study showed that long-chain polyunsaturated fatty acids (LCPU-FAs) affect receptor activator of nuclear factor κβ (RANK), a receptor found on osteoclasts responsible for bone resorption, which controls osteoclast formation.22 Rai et al. showed that medium chain length polyhydroxyalkanoates (mcl-PHAs (C6–C14 carbon atoms)) are suitable for a range of biomedical applications where flexible biomaterials are required, such as heart valves and other cardiovascular devices as well as matrices for controlled drug delivery.23 Moreover, some other studies also report that the length of chains grafted on NPs influence cellular uptake as well. For example, Daou et al. reported that quantum dots grafted with PEG chains of longer length slow down reticulo-endothelial system uptake.24 In general, there are still very few studies elucidating the effect of length of ligands grafted onto NPs on cellular uptake.
Moreover, most previous studies are concentrated on the hydrophilic molecules such as PEG, whereas those hydrophobic molecules are neglected in terms of grafting on NPs, although the length of the hydrophobic ligands such as in the case of lipids may have special influence on cellular uptake and protein adsorption. In the present study, the hydrophobic alkyls with different chain length (3, 11, and 16 carbon atoms) are modified onto gold particles, since they are varied in a long enough scale and may have different influence on protein adsorption and cellular uptake. The lung cell model (A549) is used in the in vitro cell culture since inhalation is one of the most important exposure pathways for NPs to mammalian animals. Lung is one of the major organs that nanoparticles would accumulate when they enter into the body. Therefore, lung cells, i.e. A549 cells are widely used as a model to study cell–nanoparticle interactions. Our main finding is that the length of hydrophobic alkyls is a critical factor for controlling the binding of adhesion mediators (especially Vn), which in turn enhances the cellular uptake of AuNPs. By understanding the influence of these key design parameters, this study establishes principles for the rational design of alkylated NPs with controlled protein adsorption and effective phagocyte evasion. To the best of our knowledge, this is the first attempt to correlate the length of alkyls grafted on NPs with the serum protein adsorption and the cellular uptake.
2. Results and discussion
2.1 Synthesis and characterization of MA-AuNPs
Nanomaterials, which are of similar size as typical biomacromolecular assemblies, are often seen to utilize the endocytosis machinery for intruding cells, where they can give rise to permanent cell damage.25,26 In recent years, a wide variety of different NPs with different surface chemical properties have been synthesized ranging from bare inorganic surfaces over organic coatings to intricate polymeric structures. However, the chain length of ligands especially the alkyls grafted on NPs, which influences significantly the biological performances of NPs, is neglected. In this study, three small molecules with same chemical terminal group but different chain length were grafted onto AuNPs to explore their biological impacts. The AuNPs with a diameter about 5 nm was synthesized by citrate and NaBH4 reduction.15 Following the synthesis and ligand exchange protocols, the AuNPs with desired size and ligands of different length (3MA, 11MA, 16MA) were successfully obtained. TEM (Fig. 1a–c) and UV-vis spectroscopy (Fig. 1d) reveal that the MA-modified AuNPs (MA-AuNPs) had a very uniform size and were well dispersed in mediums. Statistical analysis of the particles' sizes from the TEM images shows their narrow distribution (Fig. 1a1–c1) with values about 5.1 ± 0.6, 4.4 ± 0.3, and 4.5 ± 0.4 nm for the 3MA, 11MA and 16MA modified AuNPs (3MA-AuNPs, 11MA-AuNPs and 16MA-AuNPs), respectively (Table 1). Slight red shift (from 512 nm to 526 nm) was observed in the surface plasmon resonance (SPR) peaks of the MA-functionalized AuNPs along with the increase of ligand length from 3MA to 16MA, without significant peak broadening which is indicative of particle aggregation (Fig. 1d). The hydrodynamic diameters determined by dynamic light scattering (DLS, Table 1) are several to tens folds larger (depending on the mediums) than those determined by TEM. This phenomenon is well understood as the effect of hydrophilic corona layers on the particle surfaces. They increased along with the increase of ligand length regardless of the types of mediums, which should be attributed to the increase of ligand length. For the same MA-AuNPs, the particle size increased along with the sequence of mediums PB, BSA/PB, and 10% FBS/DMEM. However, the overall variation of sizes of the MA-AuNPs in the same type of mediums such as 10% FBS/DMEM is within a small range (i.e. ∼10%). All the AuNPs possessed a zeta potential of around −40 mV in PB regardless of their ligand's length due to the same terminal carboxyl acid group. In the proteins-containing mediums such as BSA/PB and 10% FBS/DMEM, the zeta potentials of the MA-AuNPs became less negative and possessed values around −30 mV and −20 mV, respectively. This alteration conveys the adsorption of protein molecules on the AuNPs, leading to the difference in particles-aggregation degree and surface charge.| Nanoparticles | Diameter (nm, TEM) | Diameter in medium (nm, DLS) | Z-potential in mediums (mV) | ||||
|---|---|---|---|---|---|---|---|
| PB | BSA/PB | 10% FBS/DMEM | PB | BSA/PB | 10% FBS/DMEM | ||
| 3MA-AuNPs | 5.1 ± 0.6 | 32.9 ± 0.2 | 34.3 ± 1.7 | 70.5 ± 0.8 | −35.7 ± 0.8 | −31.1 ± 0.5 | −16.7 ± 2.1 |
| 11MA-AuNPs | 4.4 ± 0.3 | 38.8 ± 1.8 | 43.1 ± 1.6 | 73.4 ± 3.8 | −40.1 ± 4.1 | −28.8 ± 2.2 | −18.3 ± 0.9 |
| 16MA-AuNPs | 4.5 ± 0.4 | 39.1 ± 1.6 | 49.1 ± 1.8 | 80.2 ± 4.7 | −37.4 ± 0.7 | −23.5 ± 2.6 | −18.8 ± 0.4 |
2.2 Protein adsorption on MA-AuNPs
It is known that NPs become coated with proteins and other biomolecules to form a “protein corona” when exposed to a biological fluid. The overall NPs–protein corona formation is a multifactorial process that depends on the characteristics of the NPs surface (hydrophobicity, functional groups, and size etc.) as well as on the interacting proteins and mediums,9,27 which in turn influences the NPs properties such as size and surface charge etc.16,18 This process in general is still very fast28,29 compared to the longer time for cellular uptake.20,30,31 Therefore, in this study, we chose 24 h as the time point to study protein adsorption, since at this moment the protein corona should be completely formed and became stable enough.The shape of UV-vis spectra of the MA-AuNPs in PB, 4 mg mL−1 BSA/PB solution (10 mM, pH 7.4), and 10% FBS/DMEM (Fig. 2) was similar with each other, without significant peak broadening. Compared to those in the PB, the SPR peaks of the 3MA-AuNPs (Fig. 2a) and 11MA-AuNPs (Fig. 2b) were red shifted for 9 and 9 nm in the BSA/PB, and 28 and 19 nm in the 10% FBS/DMEM, respectively. No obvious change in the peak position was found for the 16MA-AuNPs in all types of the mediums (Fig. 2c). However, in 4 mg mL−1 BSA/DMEM or DMEM solutions, all the MA-NPs solutions changed from oil red to purple, and precipitates were observed after a period of time. The SPR peaks were largely red-shifted, accompanying with the large reduction of peak intensity (data not shown). Therefore, the MA-AuNPs are not stable in mediums of high-ionic strength like DMEM even in the presence of albumin. This phenomenon was verified in NaCl solutions of different concentrations (Table S1†), in which all the MA-AuNPs were aggregated when the NaCl concentration reached 50 mM. By contrast, although their sizes were enlarged to 70–80 nm the MA-AuNPs were stable in 10% FBS/DMEM (pH 7), and even showed pretty good colloidal stability when the medium pH was adjusted to 5. Therefore, one can conclude that protein adsorption can stabilize the MA-AuNPs in high ion concentration solution, and besides albumin other types of proteins should be adsorbed on the MA-AuNPs as well.
 | ||
| Fig. 2 UV-vis absorbance spectra of (a) 3MA-AuNPs, (b) 11MA-AuNPs, and (c) 16MA-AuNPs measured in water, 4 mg mL−1 BSA, and 10% FBS/DMEM, respectively. | ||
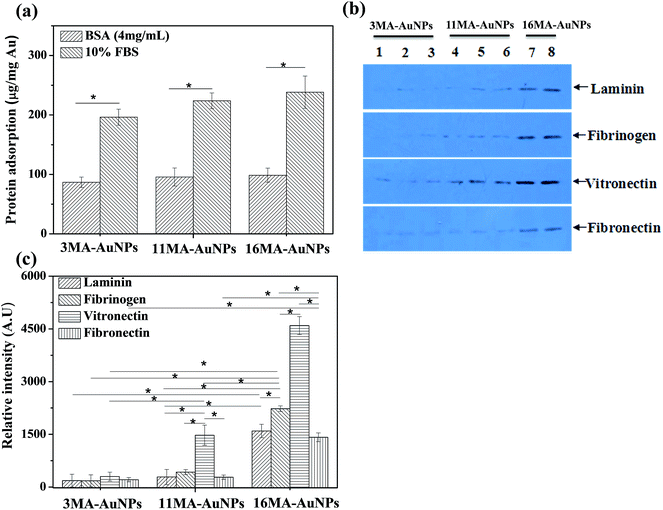
To quantitatively measure the adsorption amount of proteins by the μBCA, the MA-AuNPs were incubated in protein solutions (4 mg mL−1 BSA/PB which is same as the BSA concentration in 10% FBS, and 10% FBS/DMEM) for 24 h. Fig. 3a shows that the BSA molecules adsorbed on all the MA-AuNPs at a density of nearly 90 μg protein per mg Au, and the absolute value increased slightly along with the increase of ligand length. The amount of adsorbed serum proteins had the same trend with the adsorbed BSA molecules. However, the absolute value on each type of the MA-AuNPs was about 2 times of that of the adsorbed BSA, revealing that other types of proteins should be adsorbed besides BSA.
Besides the adsorption amount of proteins, the types of adsorbed proteins also influence the biological performance of NPs. The serum protein corona is very complex, consisting of various of proteins including apolipoproteins, adhesion mediators, signaling and transport proteins, complement components, and coagulation factors.13 Many of these proteins can mediate the cellular uptake either in their native state32 or after undergoing a conformational change.33 It was known that the adhesion mediators such as Fn, Vn, Ln and Fgn can interact with receptors of cells through their RGD domains, which in turn modulate the cellular uptake.34 Therefore, the western blot method was used to investigate the adsorption of the above adhesion mediators (Fn, Vn, Ln and Fgn) from 10% FBS solution (Fig. 3b and c). The image of these proteins adsorbed from the same amount of serum proteins on the AuNPs was shown in Fig. 3b. The data of Fig. 3c is obtained by multiplying the grey value in Fig. 3b with the amount of proteins adsorbed on the AuNPs (Fig. 3a). Generally, all the checked adhesion mediators could adsorb onto the MA-AuNPs. However, their amounts increased along with the increase of ligand length on the MA-AuNPs, in particular for the Vn. The amount of adsorbed Vn on the 11MA-AuNPs and 16MA-AuNPs was basically doubled compared with those of the Fn, Fgn and Ln, respectively, but was not significantly different on the 3MA-AuNPs. It is worth indicating that the amounts of Ln, Fn and Fgn adsorbed on the 16MA-AuNPs were significantly larger than those on the 3MA-AuNPs and 11MA-AuNPs, whereas no significant difference was found between the 3MA-AuNPs and 11MA-AuNPs.
2.3 Cellular uptake
Cellular uptake usually takes longer time than the protein adsorption. According to literatures and our experiences, the cells usually achieve the highest cellular loading of NPs at 24 h.35,36 Since the cells were co-cultured with the NPs during all the time, the results reflect the net accumulated amount of NPs regardless of the exocytosis. Previous studies demonstrate that the surface charge and grafting density of PEG on the AuNPs decide the intracellular location and distribution.18,20 To elucidate the intracellular fate of the MA-AuNPs after being co-cultured with A549 cells, ultrathin cell section TEM analysis was carried out. All the MA-AuNPs were located within cytoplasm but not inside the cell nucleus regardless of the ligand length (Fig. 4a–c and 5). However, the distribution state was different. The 3MA-AuNPs were present in densely aggregated clusters inside cytoplasm (Fig. 4a1 and a2), whereas the 11MA-AuNPs and 16MA-AuNPs were well dispersed in cytoplasm (Fig. 4b1, b2, c1 and c2) likely due to the better stability of the alkyl ligands with longer length. This hypothesis is partially supported by the DLS results (Table S1†). In 10% FBS/DMEM (pH 5) the diameter of 3MA-AuNPs (170.4 ± 1.2 nm) was nearly 2 times larger than those of the 11MA-AuNPs (88.4 ± 0.8) and 16MA-AuNPs (94.5 ± 0.9). Hence, the grafting of longer alkyls is beneficial to enhance the stability and dispersity of AuNPs in lysosomes-like environment.In order to verify whether the black dots in the nucleus and cytoplasm are AuNPs, the EDS (GENESIS XM2) combined with TEM (H7650, Hitachi) was used. The elemental analysis results (Fig. S1†) show that the black dots or aggregates in the cytoplasm contained Au. However, no Au was found in the black dots or aggregates in the nucleus. This result indicates that the MA-AuNPs are only located in cytoplasm regardless of the ligand's length.
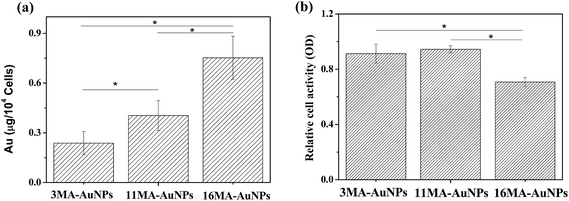
The different AuNPs were incubated with cells in same Au concentration (50 μg mL−1), and the cellular uptake amount (described as Au content in the cells) was quantified by ICP-MS with high detection sensitivity. It has to mention that here the cells-associated amount include both internalized ones into cells and adhered ones on cell surface since the ICP-MS is not able to differentiate. However, the in vitro cell culture was long enough (24 h) to allow the sufficient cellular uptake of NPs, and the contribution from the surface-associated ones would be neglectable as evidenced by the TEM observation (Fig. 4 and 5). Therefore, the cellular uptake is still used to keep the consistence and clear presentation. As shown in Fig. 6a, the amount of internalized MA-AuNPs increased with the increase of ligand length, and the internalized amount of 16MA-AuNPs was nearly 4.5 times higher than that of the 3MA-AuNPs. These results were on the contrary to previous work, in which the amount of internalized M-NPs (M represents the ligand of same EG length but different end group such as methyl and phenyl) by HeLa cells decreased with the hydrophobic index increase in 10% FBS/DMEM.37 This inconsistency should be attributed to the difference in chemical structures of the ligands on NPs surface. Nonetheless, the results indicate that the length of ligand grafted onto the AuNPs plays a significant role on uptake of MA-AuNPs by A549 cells.
Taking into account the protein adsorption results, it is reasonably to conclude that the higher adsorption of adhesion mediators (especially Vn) from 10% FBS onto the 11MA-AuNPs and 16MA-AuNPs favors their uptake by A549 cells compared to the 3MA-AuNPs. One can assume that the interaction between the MA-AuNPs and the cells involves the whole AuNPs–protein complex and not the bare AuNPs. Therefore, the properties of this complex are the parameters to influence the uptake by the cells. Indeed, uptake of NPs is a two-step process. First, NPs are covered with protein corona, and adhere onto the cell membrane and interact with lipids and proteins of the membrane. This step is followed by the activation of some energy-dependent uptake mechanisms, which allow the NPs to be internalized by a cell. The proteins–NPs complex could mediate binding to a cell by two mechanisms, specific and non-specific. In the specific interactions, the adsorbed proteins on the NPs interact with the binding sites of receptor proteins on a cell surface. The non-specific interactions involve random binding between the proteins–NPs complex and cell surface. Since all the MA-AuNPs studied here display a similar size and surface charge in biological medium, our results support the existence of specific interactions instead of non-specific ones. Multiple serum proteins attached to the MA-AuNPs may allow entry through multiple receptor sites.
As discussed above, the adhesion mediators (Fn, Vn, Ln and Fgn) adsorbed on the AuNPs can interact with receptors of cells through their RGD domains, which can modulate cellular uptake either in their native state or after undergoing a conformational change. The relative amount of Vn adsorbed on the MA-AuNPs increased significantly with the MA length. Compared to the other three proteins (Fn, Ln and Fgn), the Vn was largely adsorbed on all the three MA-AuNPs. By repotting the cellular uptake amount of MA-AuNPs (Fig. 6a) against the adsorption amount of Vn (Fig. 3c), one can find that the internalized MA-AuNPs increased linearly with the Vn adsorption amount. Therefore, the adsorbed Vn should take a major role in mediating the cellular uptake of the MA-AuNPs.
The cytotoxicity of MA-AuNPs was evaluated by MTT assay (Fig. 6b), which reflects the cell metabolic activity based on the ability of the mitochondrial succinate/tetrazolium reductase system in living cells. The 16MA-AuNPs exhibited higher cell toxicity than the 3MA-AuNPs and 11MA-AuNPs, which is highly attributed to the significant larger amount of internalized AuNPs in the A549 cells. No significant difference between the 3MA-AuNPs and 11MA-AuNPs was found, suggesting the uptake amount of 11MA-AuNPs is not high enough for inducing apparent cytotoxicity.
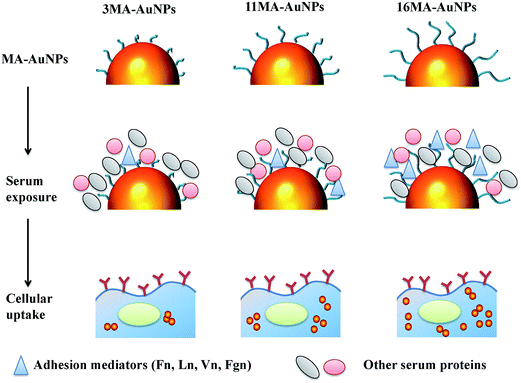
The increase of alkyl length on the AuNPs improves not only the hydrophobic index but also the ability to stabilize the AuNPs in complex cytoplasm. Many previous works reported that the alkyl length can influence the adsorption amount of serum proteins.38,39 Here the amount of serum proteins adsorbed on all the three MA-AuNPs has no significant difference though slightly increases with the longer alkyl length. However, the amount of adhesion mediators (Fn, Ln, Vn, Fgn) especially Vn on the MA-AuNP, which interact with the receptors on cell membrane, increases significantly with the increase of alkyl length (middle panel in Fig. 7). As there is no significant difference of MA-AuNPs' properties in terms of surface charge and size even after protein adsorption, it is safe to conclude that the difference in cellular uptake is caused by the interaction of protein–AuNPs complex rather than the AuNPs with the cells. The increased amount of adhesion mediators especially Vn on the AuNPs of longer alkyl ligands enhances the cellular uptake (lower panel in Fig. 7). The 3MA-AuNPs are present in densely aggregated clusters inside cytoplasm, whereas the 11MA-AuNPs and 16MA-AuNPs are better dispersed in cytoplasm (lower panel in Fig. 7). Therefore, the MA-AuNPs with longer alkyl ligands adsorb more mediator proteins especially Vn, enhance cellular uptake, and maintain better dispersion in cytoplasm. It has been found that the cellular uptake of NPs is influenced by the adsorption amount of FBS31,40 and the properties of NPs.20,41 Zhu et al. reported that the surface hydrophobicity is a critical factor for controlling serum albumin binding, which in turn decreases the cellular uptake of AuNPs.37 In this study, the adsorption amount of adhesion mediators (especial Vn) rather the BSA or FBS was governed by the alkyl's length. It is the protein (especially Vn)–NPs complexes that influence the subsequent cellular uptake and cytoplasm distribution. Therefore, the current study provides important but usually overlooked information that not only the properties of hydrophilic coatings of NPs influence the protein adsorption and subsequent cells–NPs interactions, but also the length of hydrophobic part of surface coating of NPs can dominate their interactions between proteins and cells.
3. Conclusion
Three types of AuNPs modified with alkyl acids of different chain length were successfully prepared. These particles had a diameter about 5 nm and narrow size distribution regardless of the ligand length in a dry state. However, their hydrodynamic diameters increased several to ten folds in mediums in particular the protein-containing mediums. The overall variation of sizes between the MA-AuNPs of different ligand length in the same type of mediums such as 10% FBS/DMEM was within a small range (i.e. ∼10%). The surface charge maintained negative in all types of mediums checked, but the absolute values became less negative in the sequence of PB, BSA/PB, and 10% FBS/DMEM, and became around −20 mV in 10% FBS/DMEM used for cell culture. The length of alkyl ligands on the MA-AuNPs had no significant influence on the amount of adsorbed serum proteins, but significantly affected the adsorption of adhesion mediators especially Vn which can specifically interact with cells. A549 cells took up higher amount of MA-AuNPs with longer ligands, which showed better dispersity in cytoplasm too. Therefore, the cytoplasmic distribution and cellular uptake of MA-AuNPs with different alkyl ligands are controlled by the MA-AuNPs–protein complex consisting of different amounts of adhesion mediators especially Vn, but not the bare MA-AuNPs and the total amount of adsorbed proteins. Elucidating the mechanism of cellular uptake of NPs with different length of alkyl ligands will be a valuable resource for designing alternative formulations with improved performance.4. Materials & methods
4.1 Materials
Hydrogen tetrachloroaurate hydrate (HAuCl4·4H2O), and trisodium citrate dehydrate (C6H5Na3O7·2H2O) were purchased from Sinopharm Chemical Reagent Co., Ltd. Sodium borohydride (NaBH4) was purchased from Aladdin. Bovine serum albumin (BSA), 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 3-mercaptopropionic acid (99%, 3MA), 11-mercaptoundecanoic acid (95%, 11MA), and 16-mercaptohexadecanoic acid (90%, 16MA) were obtained from Sigma-Aldrich and used without further purification. Laminin β-2 (sc-2312), fibrinogen γ (sc-18032), vitronectin 10/75 (sc-7776), fibronectin (sc-73611), and β-actin were all purchased from Santa Cruz. All other chemicals were of analytical grade and used without further treatment if not specially mentioned. The Milli-Q water was used throughout the experiments. For nanoparticles synthesis, all glasswares were cleaned by freshly prepared Aqua Regia solution (HCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HNO3, 3
HNO3, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). A549 cells were purchased from the Cell Bank of Typical Culture Collection of Chinese Academy of Sciences, and all reagents for the cell culture were used directly.
1). A549 cells were purchased from the Cell Bank of Typical Culture Collection of Chinese Academy of Sciences, and all reagents for the cell culture were used directly.
4.2 Synthesis of gold nanoparticles and surface MA-immobilization
The 5 nm gold nanoparticles (AuNPs) were synthesized by a citrate and NaBH4 reduction method.42 Briefly, 90 mL water and 1 mL HAuCl4 solution (25 mM) were added to a 250 mL Erlenmeyer flask and stirred for 1 min, and then 2 mL 38.8 mM sodium citrate was added and stirred for another 1 min, and finally 1 mL 0.075 wt% NaBH4/38.8 mM sodium citrate solution was added and stirred for 5 min.The obtained AuNPs (3MA-AuNPs, 11MA-AuNPs, and 16MA-AuNPs) were capped sufficiently with 3MA, 11MA and 16MA in the respective solutions of 3.5 mM MA/phosphate buffer (PB) at 37 °C overnight, and followed by dialysis for 2 days.
4.3 Characterization of AuNPs
The gold concentration (m, g mL−1) of the NPs solution was measured by using inductively coupled plasma mass spectrometry (ICP-MS, XSENIES, USA) and by referring to a calibration curve constructed with known concentrations of HAuCl4 at the same conditions. The size of the MA-AuNPs was determined by transmission electron microscopy (TEM). 10 μL of diluted MA-AuNPs solution was applied on a carbon-coated copper grid and dried under atmosphere overnight. The diameter of each particle (D) was measured by using Image J Software from the TEM images, with at least 500 particles counted per sample.4.4 Quantification of adsorbed proteins
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g min−1 for 120 min, the supernatant was discarded. The NPs were redispersed in PB (pH 7.4) by gentle shaking for 10 min, and centrifuged again. Totally 3 washes were performed to remove the free proteins. The proteins-adsorbed MA-AuNPs were then treated with 5% sodium dodecylsulphate (SDS) to release the protein, whose amount was quantified by using the μBCA assay regent kits.
000g min−1 for 120 min, the supernatant was discarded. The NPs were redispersed in PB (pH 7.4) by gentle shaking for 10 min, and centrifuged again. Totally 3 washes were performed to remove the free proteins. The proteins-adsorbed MA-AuNPs were then treated with 5% sodium dodecylsulphate (SDS) to release the protein, whose amount was quantified by using the μBCA assay regent kits.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g min−1 at 4 °C for 120 min, the supernatant was discarded. The NPs were redispersed in PB by gentle shaking, and centrifuged again. Totally 3 washes were performed to remove the free proteins. The proteins-conjugated MA-AuNPs were completely homogenized in radio immunoprecipitation assay buffer (RIPA) with protease inhibitors. The lysates were centrifuged at 20
000g min−1 at 4 °C for 120 min, the supernatant was discarded. The NPs were redispersed in PB by gentle shaking, and centrifuged again. Totally 3 washes were performed to remove the free proteins. The proteins-conjugated MA-AuNPs were completely homogenized in radio immunoprecipitation assay buffer (RIPA) with protease inhibitors. The lysates were centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g min−1 at 4 °C for 120 min, and the supernatants were analyzed on a SDS-PAGE. After being transferred to a poly(vinylidene fluoride) (PVDF) membrane (Millipore, MA, USA), the transferred membranes containing lysates were incubated overnight with antibodies (laminin β-2 (sc-2312), fibrinogen γ (sc-18032), vitronectin 10/75 (sc-7776), and fibronectin (sc-73611)), and detected using an enhanced chemiluminescence (ECL Western Blotting Substrate, Pierce, USA) system. The integral optical density (IOD) was determined using the software Bandscan 5.0.
000g min−1 at 4 °C for 120 min, and the supernatants were analyzed on a SDS-PAGE. After being transferred to a poly(vinylidene fluoride) (PVDF) membrane (Millipore, MA, USA), the transferred membranes containing lysates were incubated overnight with antibodies (laminin β-2 (sc-2312), fibrinogen γ (sc-18032), vitronectin 10/75 (sc-7776), and fibronectin (sc-73611)), and detected using an enhanced chemiluminescence (ECL Western Blotting Substrate, Pierce, USA) system. The integral optical density (IOD) was determined using the software Bandscan 5.0.4.5 Dynamic light scattering (DLS) and zeta potential measurement
The Z-average hydrodynamic diameter of the MA-AuNPs was determined at 37 °C in different concentrations of NaCl solutions (10, 30 and 50 mM), PB (10 mM, pH 7.4), 4 mg mL−1 BSA/PB solution, and 10% FBS/DMEM (pH 5 and 7) solution by using DLS with a high performance particle analyzer (Zetasizer Nano, Malvern) equipped with a 633 nm wavelength laser. The scattering intensity was recorded at a 173° angle in kilo counts per second.The zeta potential of the MA-AuNPs was measured at pH 7.4 and 37 °C by using the same machine in PB, 4 mg mL−1 BSA/PB, and 10% FBS/DMEM solutions.
4.6 UV-vis absorbance measurements
The effect of proteins on the plasmon excitation wavelength of MA-AuNPs was determined by recording the absorbance of gold NPs (400–700 nm) in the absence and presence of proteins (4 mg mL−1 BSA/PB or 10% FBS/DMEM) on a UV-vis spectrophotometer (Shimadzu UV2550). Each spectrum was an average of 3 individual samples recorded twice. When the spectra of MA-AuNPs were recorded in cell culture medium, DMEM supplemented with 10% FBS without phenol red was used to exclude the strong absorbance of phenol red.4.7 Cell viability and proliferation
To determine the A549 viability, the cells were plated at a density of 5 × 104 cells per cm2 in a 24-well plate and cultured for 24 h. The medium was replaced with fresh one containing the MA-AuNPs with an Au atomic concentration of 50 μg mL−1. After treatment for a given time, 100 μL 5 mg mL−1 MTT solution was added to each well, and the cells were further cultured at 37 °C for 3 h. The dark blue formazan crystals generated by the mitochondria dehydrogenase in live cells were dissolved with dimethyl sulfoxide to measure the absorbance at 570 nm by a microplate reader (MODEL 680, Bio Rad).4.8 Cellular uptake of AuNPs
The amount of MA-AuNPs internalized by A549 cells was determined by ICP-MS. Briefly, the cells were seeded on a 12-well plate at a density of 5 × 104 cells per cm2 and allowed to attach for 24 h. The medium was then replaced with 10% FBS containing 3MA-AuNPs, 11MA-AuNPs, and 16MA-AuNPs with an Au concentration of 50 μg mL−1, respectively. After 24 h, the plates were washed 5 times with phosphate buffered saline (10 mM, PBS) to remove the free MA-AuNPs. The cells were harvested by trypsinization, and the cellular uptake amount of MA-AuNPs was quantitatively determined by ICP-MS.For TEM cell section analysis, the cells were seeded on a 6-well plate at a density of 5 × 104 cells per cm2. After cultured for 24 h, the cells were further incubated with the MA-AuNPs (Au concentration 50 μg mL−1) for 24 h. At a determined time interval, the cells were washed 5 times with PBS, trypsinized, centrifuged, and then fixed with 2.5% glutaraldehyde. After 2 h of fixation at 4 °C, the samples were washed with PBS (0.1 M, pH 7.0) 3 times. Then the samples were fixed with 1% perosmic oxide for 2 h at 4 °C. After being washed in water, the samples were dehydrated in a series of alcohol solutions with increased concentrations, embedded, and sliced with a thickness between 50 and 70 nm. The morphology and structure of the obtained samples were examined by a transmission electron microscope (TEM, H-7650, Hitachi) equipped with X-ray energy dispersion spectrum (EDS, GENESIS XM2).
Acknowledgements
This study is financially supported by the Natural Science Foundation of China (51120135001), the National Basic Research Program of China (2011CB606203), the Ph.D. Programs Foundation of Ministry of Education of China (20110101130005), and Open Project of State Key Laboratory of Supramolecular Structure and Materials (sklssm201412).References
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nat. Nanotechnol., 2007, 2, 751–760 CrossRef CAS PubMed.
- M. De, P. S. Ghosh and V. M. Rotello, Adv. Mater., 2008, 20, 4225–4241 CrossRef CAS.
- D. A. Giljohann, D. S. Seferos, W. L. Daniel, M. D. Massich, P. C. Patel and C. A. Mirkin, Angew. Chem., Int. Ed., 2010, 49, 3280–3294 CrossRef CAS PubMed.
- E. Boisselier and D. Astruc, Chem. Soc. Rev., 2009, 38, 1759–1782 RSC.
- P. Ghosh, G. Han, M. De, C. K. Kim and V. M. Rotello, Adv. Drug Delivery Rev., 2008, 60, 1307–1315 CrossRef CAS PubMed.
- A. Verma, J. M. Simard, J. W. E. Worrall and V. M. Rotello, J. Am. Chem. Soc., 2004, 126, 13987–13991 CrossRef CAS PubMed.
- B. D. Chithrani, A. A. Ghazani and W. C. Chan, Nano Lett., 2006, 6, 662–668 CrossRef CAS PubMed.
- W. Jiang, B. Y. Kim, J. T. Rutka and W. C. Chan, Nat. Nanotechnol., 2008, 3, 145–150 CrossRef CAS PubMed.
- T. S. Hauck, A. A. Ghazani and W. C. Chan, Small, 2008, 1, 153–159 CrossRef PubMed.
- M. Bartneck, H. A. Keul, S. Singh, K. Czaja, J. Bornemann, M. Bockstaller, M. Moeller, G. Zwadlo-Klarwasser and J. Groll, ACS Nano, 2010, 4, 3073–3086 CrossRef CAS PubMed.
- E. A. Sykes, J. Chen, G. Zheng and W. C. W. Chan, ACS Nano, 2014, 8, 5696–5706 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed.
- C. D. Walkey and W. C. Chan, Chem. Soc. Rev., 2012, 41, 2780–2799 RSC.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- J. Deng, M. Sun, J. Zhu and C. Gao, Nanoscale, 2013, 5, 8130–8137 RSC.
- J. E. Gagner, M. D. Lopez, J. S. Dordick and R. W. Siegel, Biomaterials, 2011, 32, 7241–7252 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS PubMed.
- M. P. Calatayud, B. Sanz, V. Raffa, C. Riggio, M. R. Ibarra and G. F. Goya, Biomaterials, 2014, 35, 6389–6399 CrossRef CAS PubMed.
- J. C. Y. Kah, C. Grabinski, E. Untener, C. Garrett, J. Chen, D. Zhu, S. M. Hussain and K. Hamad-Schifferli, ACS Nano, 2014, 8, 4608–4620 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. Chan, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef CAS PubMed.
- R. Ayala, C. Zhang, D. Yang, Y. Hwang, A. Aung, S. S. Shroff, F. T. Arce, R. Lal, G. Arya and S. Varghese, Biomaterials, 2011, 32, 3700–3711 CrossRef CAS PubMed.
- M. Kruger, M. Coetzee, M. Haag and H. Weiler, Prog. Lipid Res., 2010, 49, 438–449 CrossRef CAS PubMed.
- R. Rai, T. Keshavarz, J. Roether, A. R. Boccaccini and I. Roy, Mater. Sci. Eng., R, 2011, 72, 29–47 CrossRef PubMed.
- T. J. Daou, L. Li, P. Reiss, V. Josserand and I. Texier, Langmuir, 2009, 25, 3040–3044 CrossRef CAS PubMed.
- L. Treuel, X. Jiang and G. U. Nienhaus, J. R. Soc., Interface, 2013, 10, 0939–0943 CrossRef PubMed.
- O. Lunov, T. Syrovets, C. Röcker, K. Tron, G. Ulrich Nienhaus, V. Rasche, V. Mailänder, K. Landfester and T. Simmet, Biomaterials, 2010, 31, 9015–9022 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi and C. Reinhardt, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS PubMed.
- D. Yang, X. Lü, Y. Hong, T. Xi and D. Zhang, Biomaterials, 2013, 34, 5747–5758 CrossRef CAS PubMed.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, J. Am. Chem. Soc., 2010, 132, 5761–5768 CrossRef CAS PubMed.
- A. Lesniak, A. Salvati, M. J. Santos-Martinez, M. W. Radomski, K. A. Dawson and C. Åberg, J. Am. Chem. Soc., 2013, 135, 1438–1444 CrossRef CAS PubMed.
- R. Tedja, M. Lim, R. Amal and C. Marquis, ACS Nano, 2012, 6, 4083–4093 CrossRef CAS PubMed.
- D. E. Owens III and N. A. Peppas, Int. J. Pharm., 2006, 307, 93–102 CrossRef PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39–44 CrossRef CAS PubMed.
- F. Rosso, G. Marino, A. Grimaldi, G. Cafiero, E. Chiellini, F. Chiellini, M. Barbarisi and A. Barbarisi, BioMed. Res. Int., 2013, 2013, 509348–509447 Search PubMed.
- T. Dos Santos, J. Varela, I. Lynch, A. Salvati and K. A. Dawson, Small, 2011, 7, 3341–3349 CrossRef CAS PubMed.
- D. Yu, Y. Zhang, X. Zhou, Z. Mao and C. Gao, Biomacromolecules, 2012, 13, 3272–3282 CrossRef CAS PubMed.
- Z. J. Zhu, T. Posati, D. F. Moyano, R. Tang, B. Yan, R. W. Vachet and V. M. Rotello, Small, 2012, 8, 2659–2663 CrossRef CAS PubMed.
- W. Pitt, T. Grasel and S. Cooper, Biomaterials, 1988, 9, 36–46 CrossRef CAS.
- C. A. Scotchford, C. P. Gilmore, E. Cooper, G. J. Leggett and S. Downes, J. Biomed. Mater. Res., 2002, 59, 84–99 CrossRef CAS PubMed.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Åberg, K. A. Dawson and A. Salvati, ACS Nano, 2012, 6, 5845–5857 CrossRef CAS PubMed.
- R. Tedja, M. Lim, R. Amal and C. Marquis, ACS Nano, 2012, 6, 4083–4093 CrossRef CAS PubMed.
- A. Albanese and W. C. Chan, ACS Nano, 2011, 5, 5478–5489 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra15960h |
| This journal is © The Royal Society of Chemistry 2015 |