Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Free radicals, natural antioxidants, and their reaction mechanisms
Satish Balasaheb Nimse*a and
Dilipkumar Palb
aInstitute for Applied Chemistry, Department of Chemistry, Hallym University, Chuncheon, 200-702, Korea. E-mail: satish_nimse@hallym.ac.kr; Fax: +82-33-256-3421; Tel: +82-33-248-2076
bInstitute of Pharmaceutical Sciences, Guru Ghasidas Vishwavidyalaya (A Central University), Koni, Bilaspur, Chhattisgarh-495009, India
First published on 12th March 2015
Abstract
The normal biochemical reactions in our body, increased exposure to the environment, and higher levels of dietary xenobiotic's result in the generation of reactive oxygen species (ROS) and reactive nitrogen species (RNS). The ROS and RNS create oxidative stress in different pathophysiological conditions. The reported chemical evidence suggests that dietary antioxidants help in disease prevention. The antioxidant compounds react in one-electron reactions with free radicals in vivo/in vitro and prevent oxidative damage. Therefore, it is very important to understand the reaction mechanism of antioxidants with the free radicals. This review elaborates the mechanism of action of the natural antioxidant compounds and assays for the evaluation of their antioxidant activities. The reaction mechanisms of the antioxidant assays are briefly discussed (165 references). Practical applications: understanding the reaction mechanisms can help in evaluating the antioxidant activity of various antioxidant compounds as well as in the development of novel antioxidants.
1. Introduction and background
Antioxidants are molecules that inhibit or quench free radical reactions and delay or inhibit cellular damage.1 Though the antioxidant defenses are different from species to species, the presence of the antioxidant defense is universal. Antioxidants exists both in enzymatic and non-enzymatic forms in the intracellular and extracellular environment.Normal biochemical reactions, increased exposure to the environment, and higher levels of dietary xenobiotics result in the generation of reactive oxygen species (ROS) and reactive nitrogen species (RNS).2 ROS and RNS are responsible for the oxidative stress in different pathophysiological conditions.3 Cellular constituents of our body are altered in oxidative stress conditions, resulting in various disease states. The oxidative stress can be effectively neutralized by enhancing cellular defenses in the form of antioxidants.4,5 Certain compounds act as in vivo antioxidants by raising the levels of endogenous antioxidant defenses. Expression of genes encoding the enzymes such as superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GSHPx) increases the level of endogenous antioxidants.6
Antioxidants can be categorized in multiple ways. Based on their activity, they can be categorized as enzymatic and non-enzymatic antioxidants. Enzymatic antioxidants work by breaking down and removing free radicals. The antioxidant enzymes convert dangerous oxidative products to hydrogen peroxide (H2O2) and then to water, in a multi-step process in presence of cofactors such as copper, zinc, manganese, and iron. Non-enzymatic antioxidants work by interrupting free radical chain reactions. Few examples of the non-enzymatic antioxidants are vitamin C, vitamin E, plant polyphenol, carotenoids, and glutathione.7
The other way of categorizing the antioxidants is based on their solubility in the water or lipids. The antioxidants can be categorized as water-soluble and lipid-soluble antioxidants. The water-soluble antioxidants (e.g. vitamin C) are present in the cellular fluids such as cytosol, or cytoplasmic matrix. The lipid-soluble antioxidants (e.g. vitamin E, carotenoids, and lipoic acid) are predominantly located in cell membranes.
The antioxidants can also be categorized according to their size, the small-molecule antioxidants and large-molecule antioxidants. The small-molecule antioxidants neutralize the ROS in a process called radical scavenging and carry them away. The main antioxidants in this category are vitamin C, vitamin E, carotenoids, and glutathione (GSH). The large-molecule antioxidants are enzymes (SOD, CAT, and GSHPx) and sacrificial proteins (albumin) that absorb ROS and prevent them from attacking other essential proteins.
To understand the mechanism of action of antioxidants, it is necessary to understand the generation of free radicals and their damaging reactions. This review elaborates the generation and damages that free radicals create, mechanism of action of the natural antioxidant compounds and assays for the evaluation of their antioxidant properties. The reaction mechanisms of the antioxidant assays are discussed. The scope of this article is limited to the natural antioxidants and the in vitro assays for evaluation of their antioxidant properties.
2. Generation of free radicals
The generation of ROS (Table 1) begins with rapid uptake of oxygen, activation of NADPH oxidase, and the production of the superoxide anion radical (O2˙−, eqn (1)),
 | (1) |
| Symbol | Name |
|---|---|
| 1O2 | Singlet oxygen |
| O2˙− | Superoxide anion radical |
| ˙OH | Hydroxyl radical |
| RO˙ | Alkoxyl radical |
| ROO˙ | Peroxyl radical |
| H2O2 | Hydrogen peroxide |
| LOOH | Lipid hydroperoxide |
The O2˙− is then rapidly converted to H2O2 (eqn (2)) by SOD
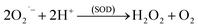 | (2) |
These ROS can act by either of the two oxygen dependent mechanisms resulting in the destruction of the microorganism or other foreign matter. The reactive species can also be generated by the myeloperoxidase–halide–H2O2 system. The enzyme myeloperoxidase (MPO) is present in the neutrophil cytoplasmic granules. In presence of the chloride ion, which is ubiquitous, H2O2 is converted to hypochlorous (HOCl, eqn (3)), a potent oxidant and antimicrobial agent.8
 | (3) |
ROS are also generated from O2˙−and H2O2 via ‘respiratory burst’ by Fenton (eqn (4)) and/or Haber–Weiss (eqn (5)) reactions.9
| H2O2 + Fe2+ → ˙OH + OH− + Fe3+ | (4) |
| O2˙− + H2O2 → ˙OH + OH− + O2 | (5) |
The enzyme nitric oxide synthase produce reactive nitrogen species (RNS), such as nitric oxide (NO˙) from arginine (eqn (6)).
| L-Arg + O2 + NADPH → NO˙ + citrulline | (6) |
An inducible nitric oxide synthase (iNOS) is capable of continuously producing large amount of NO˙, which act as a O2˙−quencher. The NO˙ and O2˙− react together to produce peroxynitrite (ONOO−, eqn (7)), a very strong oxidant, hence, each can modulate the effects of other. Although neither NO˙ nor O2˙− is a strong oxidant, peroxynitrite is a potent and versatile oxidant that can attack a wide range of biological targets.10
| NO˙ + O2˙− → ONOO− | (7) |
Peroxynitrite reacts with the aromatic amino acid residues in the enzyme resulting in the nitration of the aromatic amino acids. Such a change in the aminoacid residue can result in the enzyme inactivation. However, nitric oxide is an important cytotoxic effector molecule in the defense against tumor cells, various protozoa, fungi, helminthes, and mycobacteria.11,12 The other sources of free radical reactions are cyclooxygenation, lipooxygenation, lipid peroxidation, metabolism of xenobiotics, and ultraviolet radiations.13
3. Damaging reactions of free radicals
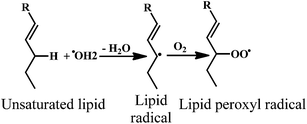
ROS (Table 1) induced oxidative stress is associated with the chronic diseases such as cancer, coronary heart disease (CHD), and osteoporosis.14 Free radicals attack all major classes of biomolecules, mainly the polyunsaturated fatty acids (PUFA) of cell membranes. The oxidative damage of PUFA, known as lipid peroxidation is particularly destructive, because it proceeds as a self-perpetuating chain reaction.15,16The general process of lipid peroxidation can be envisaged as depicted bellow (eqn (8)–(11)), where LH is the target PUFA and R˙ is the initializing, oxidizing radical. Oxidation of the PUFA generates a fatty acid radical (L˙) (eqn (8)), which rapidly adds oxygen to form a fatty acid peroxyl radical (LOO˙, eqn (9)). The peroxyl radicals are the carriers of the chain reactions. The peroxyl radicals can further oxidize PUFA molecules and initiate new chain reactions, producing lipid hydroperoxides (LOOH) (eqn (10) and (11)) that can break down to yet more radical species.17
| LH + R˙ → L˙ + RH | (8) |
| L˙ + O2 → LOO˙ | (9) |
| LOO˙ + LH → LOOH + L˙ | (10) |
| LOOH → LO˙ + LOO˙ + aldehydes | (11) |
Lipid hydroperoxides always break down to aldehydes. Many of these aldehydes are biologically active compounds, which can diffuse from the original site of attack and spread the attack to the other parts of the cell.18,19 Lipid peroxidation has been widely associated with the tissue injuries and diseases.20
Oxygen metabolism generates ˙OH, O2˙−, and the non-radical H2O2. The ˙OH is highly reactive and reacts with biological molecules such as DNAs, proteins, and lipids, which results in the chemical modifications of these molecules. There are several research reports on the oxidative damage of DNA due to the ˙OH.21–23
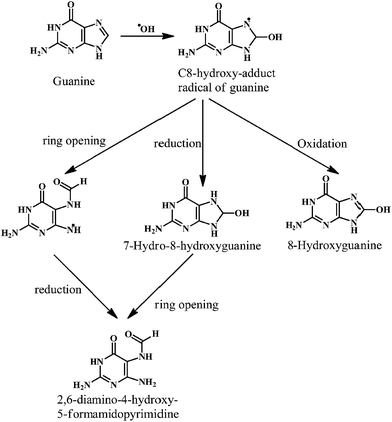
The ˙OH reacts with the basepairs of DNA, resulting in the oxidative damage of the heterocyclic moiety and the sugar moiety in the oligonucleotides by a variety of mechanisms. This type of oxidative damage to DNA is highly correlated to the physiological conditions such as mutagenesis, carcinogenesis, and aging.24,25 The addition reactions yield OH-adduct radicals of DNA bases (Scheme 1), whereas the allyl radical of thymine and carbon-centered sugar radicals (Scheme 2) are formed from the abstraction reactions.
As shown in the Scheme 1, the ˙OH reacts with the guanine of the DNA to produce the C-8-hydroxy-adduct radical of guanine, which is converted to the 2,6-diamino-4-hydroxy-5-formamidopyrimidine upon reduction and ring opening reactions. However, the C-8-hydroxy-adduct radical of guanine is converted to the 8-hydroxyguanine upon oxidation reaction. The ˙OH radical reacts with the heterocyclic moiety of the thymine and cytosine at C5- and C6-positions, resulting in the C5–OH and C6–OH adduct radicals, respectively. The oxidation reaction of these adduct radicals with water (followed by deprotonation) results in the formation of the cytosine glycol and thymine glycol, respectively.26 Overall, the reactions of the ˙OH with the DNA bases result in the impaired dsDNA.
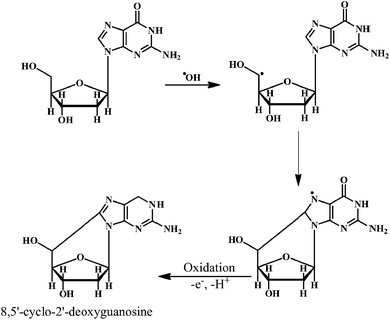
As shown in the Scheme 2, the ˙OH reacts with the sugar moiety of DNA by abstracting an H-atom from rom C5 carbon atom. One unique reaction of the C5′-centered radical of the sugar moiety in DNA is the addition to the C8-position of the purine ring in the same nucleoside (e.g. guanine). This intramolecular cyclization results in the formation of the 8,5′-cyclopurine-2′-deoxynucleosides. The reactions of carbon-centered sugar radicals result in the DNA strand breaks and base-free sites by a variety of mechanisms.
Proteins are oxidatively damaged by the combined action of activated oxygen species and the trace metal ions such as Fe2+ and Cu2+. The amino acid's lysine, proline, histidine, and arginine have been found to be the most sensitive to oxidative damage. Recent studies indicate that, a wide range of residue modifications can occur including formation of peroxides,27,28 and carbonyls.29 Generation of the carbonyl residue is a useful measure of oxidative damage to proteins. Thus, the oxidative damage to tissue results in the increased amount of oxidized protein. A detailed review by Cooke et al. provides important informations on the oxidative DNA damage, mechanisms, mutations, and related diseases.30
Low levels of antioxidants have been associated with the heart disease and cancer.31,32 Antioxidants provide protection against a number of disease processes such as aging, allergies, algesia, arthritis, asthma, atherosclerosis, autoimmune diseases, bronchopulmonary dyspepsia, cancer. The other disorders to which antioxidants provide protection are cataract, cerebral ischemia, diabetes mellitus, eczema, gastrointestinal inflammatory diseases, genetic disorders.33 Following section elaborates the mechanism of action of the radical scavenging activities of various natural antioxidant molecules.
4. Modulation of free radicals by natural antioxidants
Two types of antioxidants namely the enzymatic antioxidants and nonenzymatic antioxidants modulate the free radical reactions. Body protects itself from ROS by using enzymatic antioxidant mechanisms.34 The antioxidant enzymes reduce the levels of lipid hydroperoxide and H2O2, thus they are important in the prevention of lipid peroxidation and maintaining the structure and function of cell membranes. Examples of the enzymatic antioxidants (Fig. 1, Table 2) are CAT, GSHPx, SOD, and peroxiredoxin I–IV (I–IV).
 | (12) |
 | (13) |
| Enzymatic antioxidant | Cellular location | Substrate | Reaction |
|---|---|---|---|
| Mn/Cu/Zn SOD | Mitochondrial matrix (Mn SOD) cytosol (Cu/Zn SOD) | O2˙− | O2˙− → H2O2 |
| CAT | Peroxisomes cytosol | H2O2 | 2H2O2 → O2 + H2O |
| GSHPx | Cytosol | H2O2 | H2O2 + GSH → GSSG + H2O |
| Prx–I | Cytosol | H2O2 | H2O2 + TrxS2 → Trx(SH)2 + H2O |
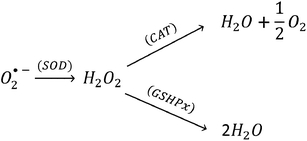
SOD's located in the cytosol and mitochondria, catalytically convert the O2˙− into oxygen and H2O2 in presence of the metal ion cofactors such as copper (Cu), zinc (Zn), or manganese (Mn).35 The enzyme CAT present in the peroxisome, converts H2O2 to water and oxygen (eqn (12)).36,37 GSHPx are found both in the cytoplasm and extracellularly in almost every human tissue. GSHPx convert the H2O2 into the water (Table 2). The enzyme GSHPx has strong activity towards both H2O2 and fatty acid hydroperoxides (eqn (13)).38,39 The enzyme peroxyredoxin catalyze the reduction of H2O2, organic hydroperoxides and the peroxynitrite (ONOO−). The different expression profiles, subcellular locations, and substrates of the antioxidant enzymes reveal the complex nature of the ROS biology. Clearly, the antioxidant enzymes play a major role in the prevention of oxidative damage. As demonstrated in the Scheme 3, CAT, GSHPx, and SOD show synergistic effect in the scavenging of O2˙−.
The enzymatic antioxidants and their mechanism of antioxidant activity has been explained in details in several review articles.40–42 Therefore, this article focuses mainly on the nonenzymatic antioxidants of natural origin.
The nonenzymatic antioxidants are of two types, the natural antioxidants and the synthetic antioxidants. However, the scope of this article is limited to the natural antioxidants; hence the synthetic antioxidants will not be considered for the discussion.
4.1. Vitamins
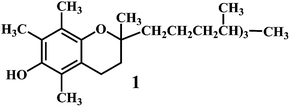
Vitamin E 1,43 vitamin C 2,44 vitamin A 3.Vitamin E (α-tocopherol) 1, is an efficient lipid soluble antioxidant that functions as a ‘chain breaker’ during lipid peroxidation in cell membranes and various lipid particles including low-density lipoprotein (LDL). It functions to intercept lipid peroxyl radicals (LOO˙) and to terminate the lipid peroxidation chain reactions (eqn (14)).
| LOO˙ + α-tocopherol–OH → LOOH + α-tocopherol–O˙ | (14) |
The resultant tocopheroxyl radical is relatively stable and in normal circumstances, insufficiently reactive to initiate lipid peroxidation itself, which is an essential criterion of a good antioxidant.45–47 It should be noted that, vitamin E exerts antioxidant effects by scavenging lipid peroxyl radicals in vivo as well as in vitro systems. However, vitamin E is not an efficient scavenger of ˙OH and alkoxyl radicals (˙OR) in vivo.48
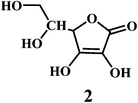
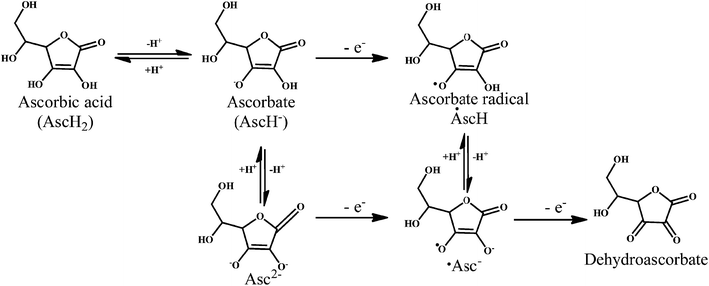
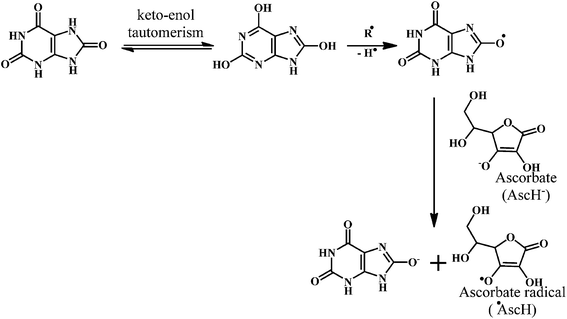
Vitamin C or ascorbic acid 2, is a water-soluble free radical scavenger. Moreover, it regenerates vitamin E in cell membranes in combination with GSH or compounds capable of donating reducing equivalents.49–51 Vitamin C, changes to the ascorbate radical (Scheme 4) by donating an electron to the lipid radical in order to terminate the lipid peroxidation chain reaction. The pairs of ascorbate radicals react rapidly to produce one molecule of ascorbate and one molecule of dehydroascorbate. The dehydroascorbate does not have any antioxidant capacity. Hence, dehydroascorbate is converted back into the ascorbate by the addition of two electrons. The last stage of the addition of two electrons to the dehydroascorbate has been proposed to be carried out by oxidoreductase.
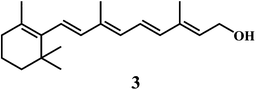
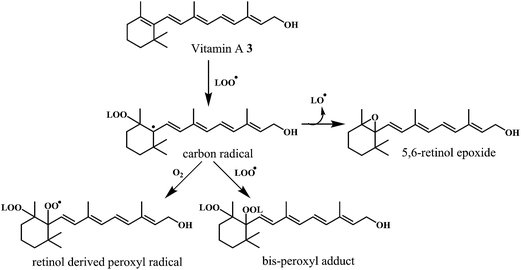
Antioxidant potential of vitamin A 3 was first described by Monaghan and Schmitt,52 who reported that vitamin A can protect lipids against rancidity. Several reviews have appeared to outline the basic structural and metabolic characteristics of vitamin A and information about its potential as antioxidants in relation to the heart diseases.53,54 Vitamin A has a vital antioxidant contribution in protecting human LDL against copper-stimulated oxidation (Scheme 5).55,56
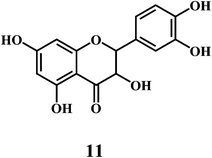
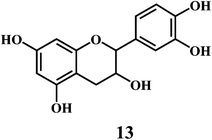
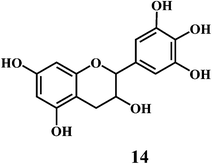
4.2. Bioflavonoids
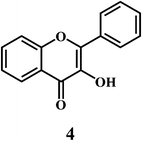
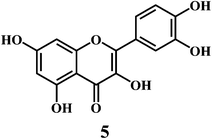
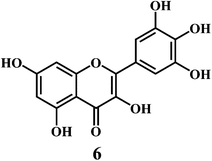
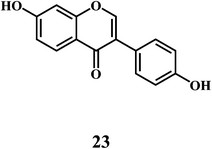
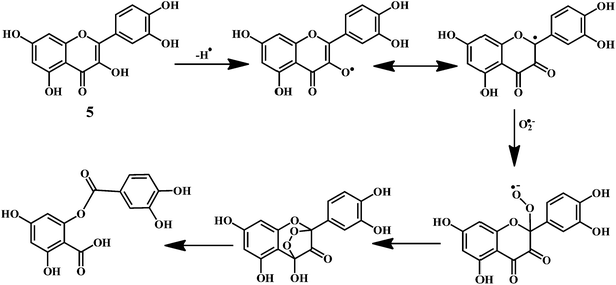
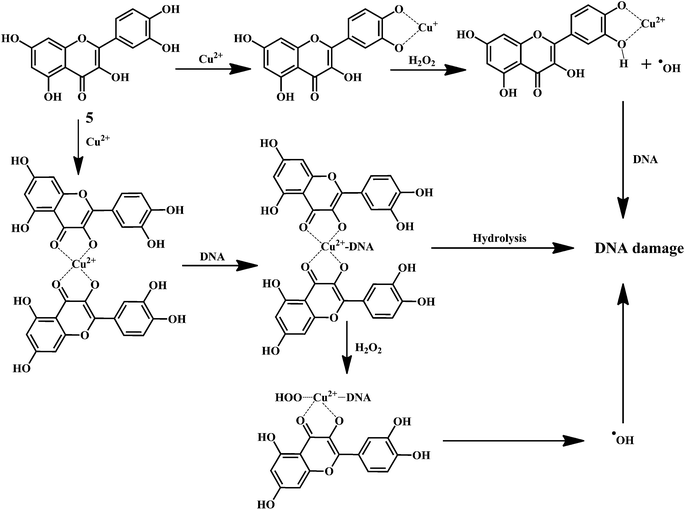
Flavonol 4 (e.g. quercetin 5, myricetin 6), flavone 7 (e.g. apigenin 8, luteoline 9), flavonolols 10 (e.g. taxifolin 11), flavan-3-ols 12 (e.g. catechin 13, epigallocatechin 14), flavonone 15 (e.g. hesperetin 16, naringenin 17), anthocyanidin 18 (e.g. cynidin 19, delphidin 20), isoflavone 21 (e.g. genistein 22, daidzein 23).57,58Bioflavonoids are a group of natural benzo-γ-pyran derivatives (4–23) and are found to possess strong antioxidant activities.59,60 Bioflavonoids widely distributed in fruits and vegetables, are reported to exert multiple biological effects including free radical-scavenging activity. It has been reported that the bioflavonoids have a protective effect on the DNA damage induced by the hydroxyl radicals.61 One of the mechanisms that explains the protective effect of the flavonoids on the DNA is the involvement of the chelating metal ions, such as copper or iron. The flavonoids complexed with the copper or iron prevent the generation of the ROS.62–64
Quercetin 5 is a flavonol, known to protect DNA from oxidative damage resulting from the attack of ˙OH, H2O2, and O2˙− on the DNA oligonucleotides (Scheme 6).65 On the contrary, quercetin is also reported to be carcinogenic agent.66,67 According to the reports, quercetin has opposite effects on DNA damage induced by cupric ion depending on the concentration of cupric ion (Scheme 7).68,69 At the low concentration of cupric ions (≤25 μM), quercetin exhibit a protective role. While, at higher concentration of cupric ion (≥25 μM), quercetin enhances the damage to DNA by ROS. Therefore, it is very important to consider the concentration of the chelating metal ions, such as copper or iron while evaluating the protective or degenerative effects of quercetin and other bioflavonoids.
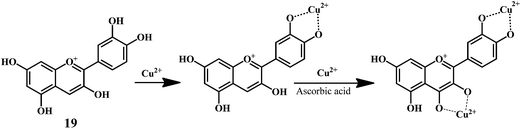
Anthocynidine, a class of flavonoids are potential antioxidants and their effectiveness in the inhibition of the lipid oxidation is related to their metal ion-chelating activity (Scheme 8) and free-radical scavenging activity (Scheme 9). Three structural groups are important determinants of the radical-scavenging activity of anthocynidines 18–20.70 First, the ortho-dihydroxy structure in the B-ring. Second, the 2,3 double bond in conjugation. Third, the 4-oxofunction in the C-ring. Flavonoids form complexes with the metal ions by using the 3- or 5-hydroxyl and 4-ketosubstituents or hydroxyl groups in ortho position in the B-ring.71
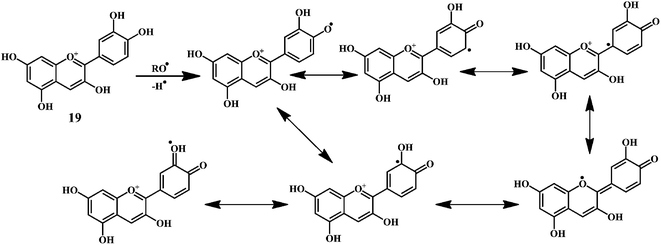
As shown in the Scheme 9, the anthocynidins (cynidin 19) can donate an electron (accompanied by a hydrogen nucleus) to a free radical from –OH groups attached to the phenolic rings.72–74 This electron stabilizes and inactivates the free radical. In this process, the polyphenolic reducing agent changes to an aroxyl radical, which is comparatively more stable due to resonance than the free radical that it has reduced. The overall result is the termination of damaging oxidative chain reactions.
4.3. Carotenoids75,76
(i) Carotines: lycopene 26, β-carotene 27,(ii) Xanthophyll: zeaxanthine 28, lutein 29.
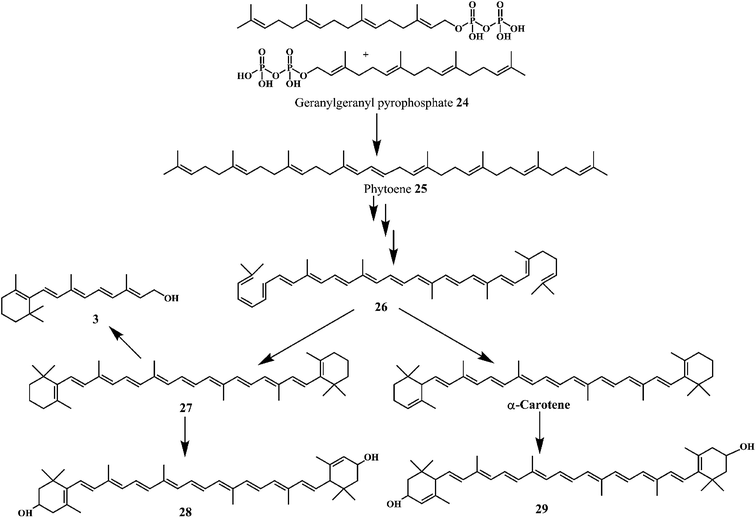
Carotenoids are among the most common lipid soluble phytonutrients. Lycopene 24 and β-carotene 25 are the prominent carotenoids among other 600 different compounds.77 The biosynthetic pathway as shown in Scheme 10 demonstrates the synthesis of carotenoids 26–29 from phytoene 25, which is synthesized from two molecules of geranylgeranyl pyrophosphate 24. Carotenoids are well known to scavenge the peroxyl radicals more efficiently as compared to any other ROS. The peroxyl radicals generated in the process of lipid peroxidation can damage the lipids in the cell wall. Scavenging of peroxyl radicals can disrupt the reaction sequence and prevent the damage to cellular lipids. The long unsaturated alkyl chains in carotenoids make them highly lipophilic. Carotenoids are known to play an important role in the protection of cellular membranes and lipoproteins against the ROS due to their peroxyl radical scavenging activity.78,79 Carotenoids deactivate the peroxyl radicals by reacting with them to form resonance stabilized carbon-centered radical adducts.
Lycopene 24, is the most potent antioxidant naturally present in many fruits and vegetables. The high number of conjugated double bonds in lycopen endows it the singlet oxygen quenching ability. Lycopene demonstrate the strong singlet oxygen quenching ability as compared to the α-tocopherol 1 or β-carotene 25.80 β-Carotene 12 is a naturally occurring orange-colored carotenoid, abundantly found in the yellow-orange fruits and in dark-green leafy vegetables.81,82 β-Carotene demonstrates potential antioxidant property due to its chemical structure and the interaction with biological membranes.83 It is well-known that, the β-carotene quenches singlet oxygen with higher efficiency as compared to the α-tocopherol.84 In addition, it is also known that the (Z)-isomers of the β-carotene possess antioxidant activity in vitro.85,86 Furthermore, the β-carotene can be converted into the two molecules of vitamin A by the β-carotene-15,15′-dioxygenase catalyzed cleavage.
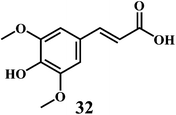
4.4. Hydroxycinnamates
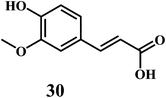
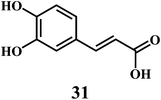
The examples are ferulic acid 30, caffeic acid 31, sinapic acid 32, p-coumaric acid 33.It is widely accepted that, the dietary antioxidants that protect LDL from oxidation can prevent the atherosclerosis and coronary heart disease. Hydroxycinnamic acids 30–33 and their conjugates prevent oxidative damage to the LDL.87 The in vitro studies involving human LDL as the oxidizing substrate showed that the hydroxycinnamic acids have higher antioxidant activity as compared to the corresponding hydroxybenzoic acids.88 The antioxidant activity of the derivatives of the hydroxycinnamates is clearly correlated with the hydroxylation and methylation patterns of the aromatic ring. The antioxidant efficiency of the free hydroxycinnamates on the human LDL oxidation in vitro, decreases in the order of caffeic acid 31 > sinapic acid 32 > ferulic acid 30 > p-coumaric acid 33.
The presence of the o-dihydroxy group in the phenolic ring (as in caffeic acid) enhances the antioxidant activity of hydroxycinnamic acids toward human LDL oxidation in vitro.89 The radical scavenging antioxidant mechanism of the hydroxycinnamic acids are similar to that of the flavanoids because of their ability to donate an hydroxyl hydrogen and resonance stabilization of the resulting antioxidant radicals. The o-dihydroxy substituents also allow the metal ion chelation similar to that of flavanoids.
4.5. Other natural antioxidants
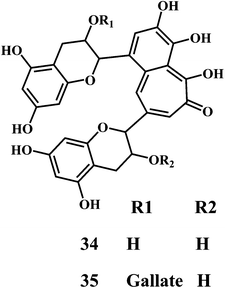
Theaflavin 34, theaflavin-3-gallate 35, allicin 36, piperine 37, curcumin 38.90,91Theaflavin 34 and theaflavin-3-gallate 35 possesses in vitro antioxidative properties against lipid peroxidation in the erythrocyte membranes and microsomes. They also suppress the mutagenic effects induced by H2O2.92 Theaflavins inhibit the H2O2 induced cleavage and mutagenicity of the DNA single-strand.93,94 In general, theaflavins scavenge the free radicals to produce antioxidative and antimutagenic effects. Apart from the aromatic hydroxyl groups of theaflavins, the gallic acid moiety is essential for their antioxidant activity. The theaflavin-3-gallate 35 is a stronger antioxidant than that of theaflavin 34. Moreover, the digallate derivatives of theaflavin demonstrate the increased antioxidant activity.
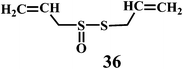
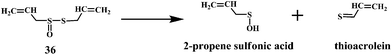
Allicin (diallyl thiosulfinate) 36 is the biologically active compound mainly found in the garlic extracts. Allicin is known to possess various biological activities including the antibacterial, antifungal, and inhibition of cancer promotion.95 Moreover, allicin is known to reduce serum cholesterol and triglyceride levels as well as atherosclerotic plaque formation and platelet aggregation.96 Until now, a variety of biological effects of allicin were attributed to antioxidant activity.97 However, recently it has been found that the active ingredients responsible for the antioxidant property of garlic is 2-propenesulfenic acid and not the allicin.98 Thiosulfinates undergoes Cope elimination to form sulfenic acids, thioaldehydes or thioketones. The S–S bond in the thiosulfinate is much weaker than the S–C bond in a sulfoxide. Hence, this process can occur at room temperature. Cope elimination is even more susceptible for the allyl (and benzyl) thiosulfinates, such as allicin 36, because of the weak β C–H bond of the allyl moiety. Allicin is known to undergo Cope elimination at room temperature to give 2-propenesulfenic acid and thioacrolein as shown in the Scheme 11.99,100
The Scheme 12a demonstrate the mechanism of the radical-scavenging activity of the allicin. The radical-scavenging activity of allicin involves H-atom transfer to a peroxyl radical from the methylene of the allyl group on the divalent sulfur. Scheme 12b demonstrate an alternative mechanism, where the radical-scavenging activity of allicin can be accounted for 2-propenesulfenic acid, which is produced from allicin by Cope elimination.101 2-Propenesulfenic acid is reported to be over 1000 times more reactive toward ˙OOH radicals than allicin (2.60 × 107 vs. 7.38 × 103 L mol−1 s−1, at 298 K).102
 | ||
| Scheme 12 Mechanism for the radical-trapping activity of (a) allicin and (b) 2-propenesulfenic acid. | ||
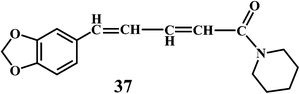
Piperine (1-piperoylpiperidine) 37, is an alkaloid present in fruits of black pepper (Piper nigrum), long pepper (Piper longum), and other piper species (family: Piperaceae). Piperine possesses many pharmacological activities, including anti-inflammatory and analgesic effect,103 anti-ulcer activities,104 antidepressant effect,105 cognitive enhancing effect,106 cytoprotective effect, and antioxidant activity.107 It is interesting to notice that, the higher concentration of piperine results in the increased production of the ˙OH. Whereas, in low concentrations piperine acts as an antioxidant.108 Piperine demonstrates synergistic antioxidant activity by doubling the absorption of dietary curcumin 38.109
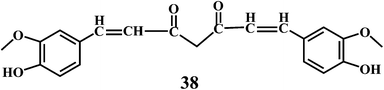
Curcumin 38, a lipid soluble active principle of turmeric is a bis-α, β-unsaturated β-diketone that exhibit's keto–enol tautomerism.110 Curcumin 38, shows remarkable antioxidant activity, and it has been found to be an excellent free radical scavenger.111 Curcumin has a chain breaking antioxidant ability comparable to that of the vitamin E.
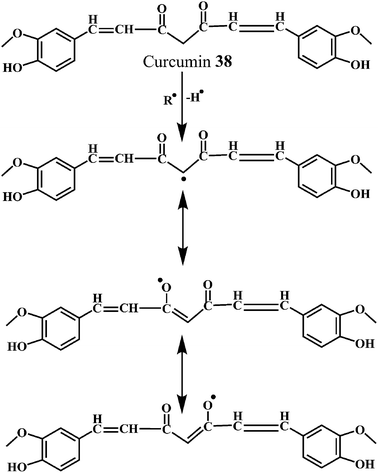
As shown in the Schemes 13 and 14, the free radical scavenging activity of curcumin is correlated to the phenolic OH group and the CH2 group of the β-diketone moiety. The free radical can undergo electron transfer or abstract H-atom from either of these two sites. However, pulse radiolysis and other biochemical methods credited the antioxidant activity of curcumin to its phenolic OH group.112
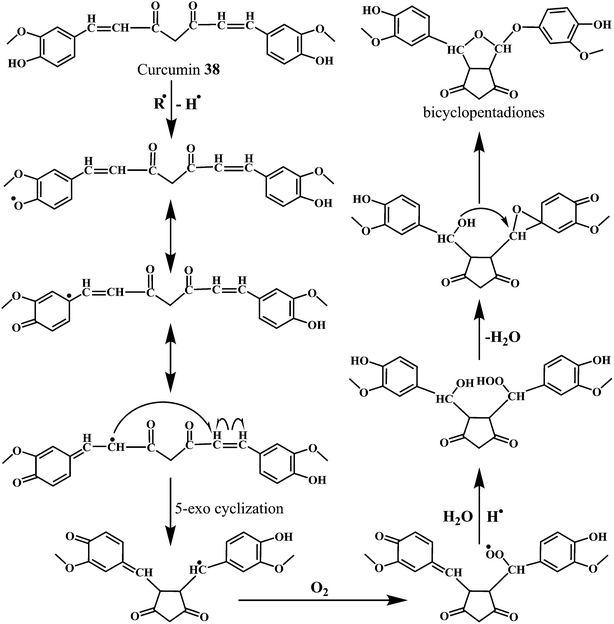
The Scheme 14 depicts the mechanism for the autoxidation of curcumin initiated by hydrogen abstraction from one of the phenolic hydroxyl groups.113 The phenoxyl radical moves into the carbon chain leaving a quinone methide that is eventually quenched by the water molecules. The methide radical performs a 5-exo-cyclization with the double bond to give the cyclopentadione ring and generating the carbon-centered radical.
The reaction of curcumin with the molecular oxygen (O2) results in the peroxyl radical. The peroxyl radical is then reduced to the hydroperoxide by abstracting a hydrogen atom from another curcumin molecule, propagating the autoxidation chain reaction. Subsequently, the hydroperoxide loses water and rearranges into the spiro-epoxide. The hydrolysis of the epoxide by the (water-derived) hydroxyl group results in the formation of the final bicyclopentadione product.114 It has been found that the copper complex of curcumin (curcumin–Cu(II)) show promising SOD activity, with improved antioxidant efficacy.115,116
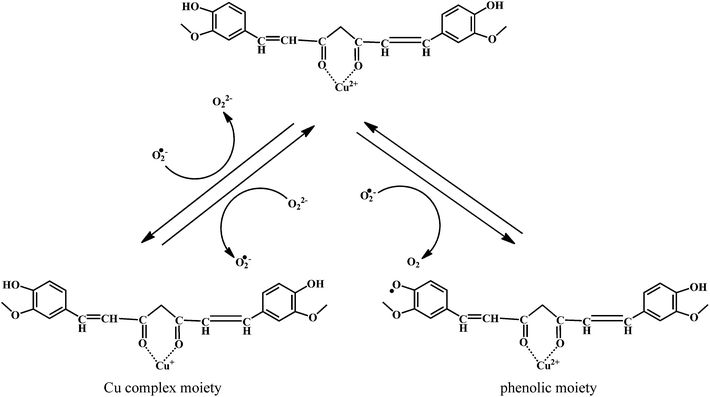
The mechanism of the O2˙− scavenging activity of the curcumin–Cu(II) complex is depicted in the Scheme 15. When O2˙− are allowed react with the curcumin–Cu(II) complex, a major fraction of O2˙− reacts with Cu2+ moiety, while only a small fraction reacts with curcumin. The reaction causes reduction of Cu2+ to Cu+. The Cu+ undergoes subsequent oxidation by another molecule of O2˙−, thereby regenerating the parent complex.
Therefore, the catalytic activity comes mainly from the reversible redox reactions within the Cu2+/Cu+ couple in the complex. However, in presence of the excess O2˙−, the phenolic moiety undergoes oxidation resulting in the production of the phenoxyl radicals. Then these phenoxyl radicals can generate new products or react with reduced copper ions of the complex resulting in the regeneration of the complex.
4.6. Physisological antioxidants
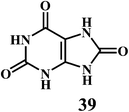
Uric acid 39 in plasma, and GSH 40.Uric acid 39 in plasma possesses strong radical scavenging activity.117,118 Uric acid is the most abundant aqueous antioxidant found in humans. It contributes for as much as two-thirds of all free radical scavenging activities in the plasma.119 Uric acid is a powerful scavenger of carbon-centered radicals and peroxyl radicals in the hydrophilic environment. However, it loses it's radical scavenging activity within lipid membranes.120
Uric acid is an exceptional scavenger of peroxynitrite (ONOO−) in the extracellular fluid.121 However, it is important to note that the uric acid cannot scavenge the O2˙−. Moreover, uric acid requires the presence of ascorbic acid (Scheme 16) and thiols for the complete scavenging of peroxynitrites. Neither of these antioxidants (ascorbic acid, thiols) alone can prevent reaction of peroxynitrite with tetrahydrobiopterin, which leads to uncoupling of nitric oxide (NO˙) synthase.122 This indicates that the uric acid plays a crucial role in the scavenging of the peroxynitrite.
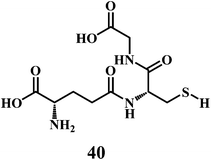
GSH 40 in cell cytosol, together with its related enzymes, comprises a system that maintains the intracellular reducing environment, which acts as primary defense against excessive generation of harmful ROS.123,124 The oxygen radical scavenging activity of GSH directly expedites the ROS neutralization and the repair of ROS-induced damage.125
As shown in the Scheme 17, three groups of enzymes can be identified in the GSH catalytic cycle: glutathione oxidase, glutathione reductase, and GSHPx. Glutathione oxidase and GSHPx catalyze the oxidation of GSH to GSH disulfide (GSSG). Whereas, glutathione reductase is responsible for the regeneration of GSH from GSSG in an NADPH-dependent process.126 Cells can produce GSSG or convert it to GSH by using NADPH in the presence of the glutathione reductase. However, the de nova synthesis of glutathione from its amino acid constituents is required for the elevation of glutathione as an adaptive response to oxidative stress. The presence of the sulfhydryl group in glutathione allows it to serve as an antioxidant.
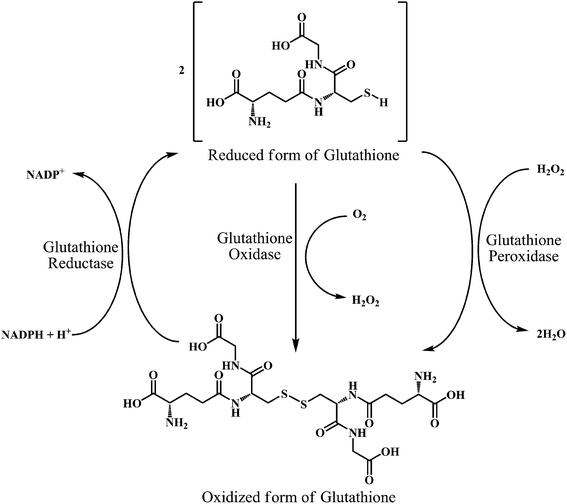 | ||
| Scheme 17 Interconversion of glutathione in its reduced form (GSH) and oxidized form (GSSG) by the action of glutathione oxidase, glutathione reductase, and glutathione peroxidase enzymes. | ||
4.7. Fungal antioxidants
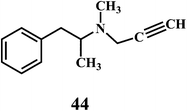
The microorganisms such as Ganoderma lucidum,127 Ganoderma applanatum, Meripilus giganteus, Flammulina velutipes, and Endophytic Fungi128,129 possess a very efficient antioxidative system consisting of enzymatic (peroxidases, laccase, catalase, and superoxide dismutase) and nonenzymatic elements (phenolic derivatives or polysaccharides).The synthetic antioxidants are the second type of nonenzymatic antioxidants. Cinnamic acid derivatives130,131 41, 42, melatonin 43, selegiline 44, are the few examples of the synthetic antioxidants.132,133
5. In vitro methods for evaluation of antioxidant activity
Various in vitro methods are available for the evaluation of antioxidant activity of different compounds.134–1375.1. Assay of superoxide anion radical scavenging activity
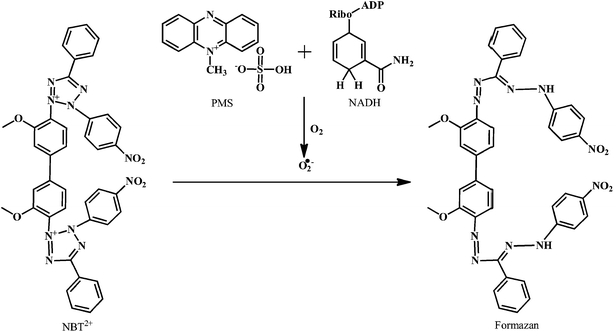
SOD is an antioxidant enzyme involved in scavenging the ROS.138 SOD converts the O2˙− to H2O2. The H2O2 is then converted to the O2 and H2O in the reaction catalyzed by GSHPx and CAT.139 There are several classes of SOD, which include intracellular copper, zinc SOD (Cu, Zn SOD/SOD1), mitochondrial manganese SOD (Mn SOD/SOD2), and extracellular Cu, Zn SOD (EC SOD/SOD3).The method for the evaluation of the O2˙− scavenging activity of antioxidants is explained here by using PMS–NADH–NBT system, which is composed of N-methylphenazine methosulphate (PMS), nitroblue tetrazolium chloride (NBT), and NADH (a reduced form of nicotineamide-adenine-dinucleotide).
As shown in the Scheme 18, the O2˙− produced in the coupling reaction of PMS–NADH in presence of dissolved oxygen reduces NBT. The decrease of absorbance at 560 nm with antioxidant indicates the consumption of O2˙− in the reaction mixture. The O2˙− scavenging activity can be measured as described by Robak and Gryglewski.140 Gallic acid, BHA, ascorbic acid, α-tocopherol, and curcumin can be used as positive controls in this assay.
5.2. Assay of 2,2-diphenyl-1-picrylhydrazyl (DPPH) free radical scavenging activity
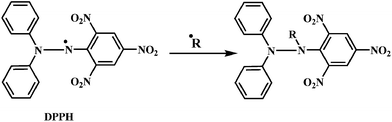
Evaluation of the antioxidant activity of any compound can be carried out either by in vitro or in vivo models.141,142 DPPH is a stable free radical that can accept an electron or hydrogen radical to become a stable diamagnetic molecule.Due to its odd electron, the methanolic solution of DPPH shows a strong absorption band at 517 nm. As shown in the Scheme 19, the DPPH radical reacts with suitable reducing agent producing new bond, thus changing the color of solution. The solution loses color with the increase in the concentration of antioxidant as the electrons taken up by DPPH radical from the antioxidant.143,144 Such reactivity has been used to test the ability of compounds/plant extracts to act as free radical scavengers.145 Reduction of the DPPH radicals can be monitored spectrophotometrically by the decrease in absorbance at 517 nm.
5.3. Assay for total reactive oxygen potential (TRAP) and total antioxidant reactivity (TAR)
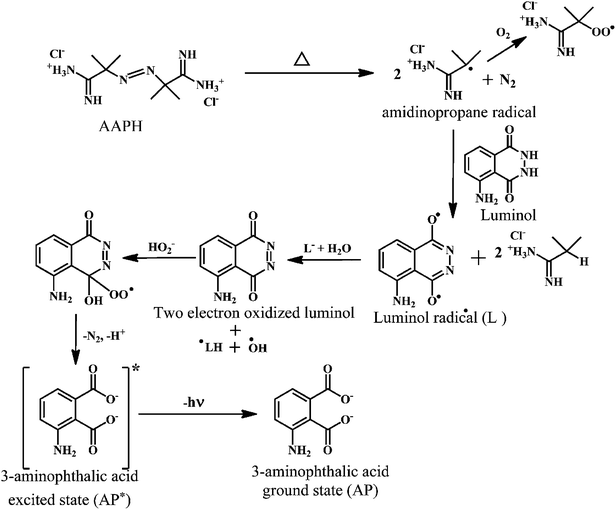
Luminol enhanced chemiluminescence is used to measure TRAP and TAR.146,147 When the luminol is allowed to react with the free radical source, a steady chemiluminescence is observed that can be directly correlated to the rate of luminol oxidation.148 The addition of free radical scavengers reduces the chemiluminescence intensity.149 The effect of antioxidants on the induced chemiluminescence intensity of luminol by radicals derived from the thermolysis of 2,2′-azobis-2-amidinopropanedihydrochloride (AAPH) can be employed to monitor the TRAP and TAR levels.150As shown in the Scheme 20, the AAPH undergoes thermal decomposition in solution to produce two carbon-centered amidino propane (AP) radicals, which can add O2 to form peroxyl radicals. However, the carbon-centered radicals usually predominate.151 The amidino propane (AP) radical takes up a proton from luminol to produce a luminol radical. The luminol radical reacts with de-protonated H2O2 to yield a short-lived hydroperoxide intermediate (LO2H−), which rapidly decomposes into the excited state 3-aminophthalic acid (AP*). The AP* loses energy in the form of chemiluminescence to give ground state 3-aminophthalic acid.152
5.4. In vitro antioxidant evaluation by phospholipids peroxidation
Lipid peroxidation is an oxidative degradation of lipids.153 In this process, the free radical takes up the electrons from the lipids in cell membranes, which results in the cell damage.154,155 The tentative mechanism for this free radical chain reaction involved in the phospholipid peroxidation is depicted in the Scheme 21.The activity of test compound to inhibit peroxidation of membrane lipids at pH 7.4 is tested using phospholipids. The interference of the test drug with color development is determined by adding a previously determined concentration of the test compound to the TBA reagents and used to determine the extent of peroxidation of animal phospholipids.156,157 In this assay, the antioxidant activity is a measure of concentration-dependent inhibition of a phospholipid peroxidation.
5.5. In vitro antioxidant evaluation by deoxyribose assay
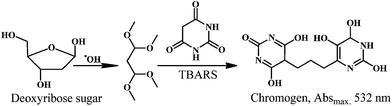
The ˙OH in presence of ascorbic acid attack the sugar deoxyribose to generate the product that on heating with thiobarbituric acid (TBA) or thiobarbiturate reactive substances (TBARS), at low pH, yield a chromogen. Therefore, the deoxyribose assay can be used to detect ˙OH scavenging activity of test compounds.The reaction of deoxyribose and ˙OH has been discussed extensively in the literature.158,159 The ˙OH attack deoxyribose to form products that react with TBA upon heating at low pH and yield a pink chromogen. Scheme 22 depicts the proposed mechanism of chromogen formation from reaction of deoxyribose and ˙OH followed by reaction with TBARS.
In general, the in vivo assays for testing potential antioxidants are more expensive because they require complex cellular testing systems or full clinical trials. However, it is very important to proceed to cellular assays after screening antioxidant activity with an in vitro method in order to obtain information on some aspects like uptake, bioavailability, and metabolism.160 The new definition of an antioxidant, a redox-active compound or mixture able to modulate the redox status of the cell, makes it critical to use in vivo assays in order to evaluate the antioxidant activity of a compound.161 There are several reports on the in vivo assays for the evaluation of the antioxidant activity.162,163 However, we have limited the scope of this review to the in vitro assays for the evaluation of the antioxidant activities of natural antioxidants. There are several other reports, which elaborate the advantages and disadvantages of various methods for the evaluation of antioxidant activity.164,165
6. Current trends and future directions
In recent years, there has been upsurge in the novel approaches for the study of free radicals and antioxidants in relation with the improvement of human health. Multiple studies have showed that the neuronal and behavioral changes occur with ageing, even in the absence of degenerative disease. Recent studies have found the association between the lower status of dietary antioxidants and decline in the cognitive function. The evidences from the experimental, clinical, and epidemiological studies indicate that the consumption of foods containing high levels of dietary antioxidants may prevent or reduce the risk of cognitive deterioration. Tempol, an example of a new class of SOD mimetic drugs, alleviates acute and chronic pain. These drugs substantially reduce the tissue damage incurred by inflammation. The speculations of the relations between radical damage and disease conditions need to the support of by more secure data. The knowledge on the mechanisms of various physiological radical reactions and the mechanisms of the antioxidants in scavenging those free radicals will open up the path for more potent drug molecules.Many investigators found that, increasing the level of defense mechanisms against oxidative stress could extend an organism's health span. Therefore, few setbacks in the antioxidant research with the molecules showing strong antioxidant activity in vitro and non-antioxidant effects in cells and tissues should not discourage the important research in this field. Finally, the collective effort is must be undertaken for the understanding of the mechanisms in the free radical scavenging activities of known antioxidants to derive the potent antioxidants.
7. Conclusion
ROS, the radical derivatives of oxygen are the most important free radical in biological systems. The ROS are the harmful byproducts generated during the normal cellular functions. Increasing intake of natural antioxidants may help to maintain a tolerable antioxidant status, perhaps the normal physiological functioning. The reported chemical evidence suggests that the dietary antioxidants help in the disease prevention. The antioxidant compounds react in one-electron reactions with free radicals in vitro and prevent the oxidative damage. Therefore, it is very important to understand the reaction mechanism of antioxidant with the free radicals. The reaction mechanisms can be used to evaluate the antioxidant activity of various naturally occurring antioxidant compounds. This review elaborates the mechanism of action of the natural antioxidant compounds and assays for the evaluation of their antioxidant activities. The reaction mechanisms of the antioxidant assays are briefly discussed (165 references). The scope of this article is limited to the natural antioxidants and the in vitro assays for evaluation of their antioxidant properties.Conflicts of interest
The author declares no conflict of interest.Abbreviations
| ROS | Reactive oxygen species |
| RNS | Reactive nitrogen species |
| SOD | Superoxide dismutase |
| CAT | Catalase |
| GSHPx | Glutathione peroxidase |
| MPO | Myeloperoxidase |
| O2˙− | Superoxide anion radical |
| H2O2 | Hydrogen peroxide |
| ˙OH | Hydroxyl radical |
| EDRF | Endothelium derived relaxation factor |
| CHD | Coronary heart disease |
| PUFA | Polyunsaturated fatty acids |
| DNA | Deoxyribonucleic acid |
| Prx | Peroxiredoxin |
| LDL | Low density lipoprotein |
| GSH | Glutathione |
| GSSG | Glutathione disulfide |
| PMS | N-Methylphenazine methosulphate |
| NBT | Nitroblue tetrazolium chloride |
| DPPH | 2,2-Diphenyl-1-picrylhydrazyl |
| TRAP | Total reactive oxygen potential |
| TAR | Total antioxidant reactivity |
| AAPH | 2,2′-Azobis-2-amidinopropanedihydrochloride |
| TBA | Thiobarbituric acid |
| TBARS | Thiobarbiturate reactive substances |
Acknowledgements
This research was supported by the Hallym University Research Fund (HRF-201411-014).Notes and references
- I. S. Young and J. V. Woodside, J. Clin. Pathol., 2001, 54, 176–186 CrossRef CAS.
- K. Bagchi and S. Puri, East. Meditrr. Health. J., 1998, 4, 350–360 Search PubMed.
- Y. W. Kim and T. V. Byzova, Blood, 2014, 123, 625–631 CrossRef CAS PubMed.
- H. Sies, Exp. Physiol., 1997, 82, 291–295 CrossRef CAS.
- D. K. Pal and S. B. Nimse, Asian J. Chem., 2006, 13, 3004–3008 Search PubMed.
- C. E. Thomas and B. Kalyanaraman, in Oxygen Radical and the Disease Process, Harvard Academic Publishers, Netherlands, 1997 Search PubMed.
- F. Shahidi and Y. Zhong, Eur. J. Lipid Sci. Technol., 2010, 112, 930–940 CrossRef CAS.
- B. M. Babior, Blood, 1999, 93, 1464–1476 CAS.
- J. A. Knight, in Free radicals, antioxidants, aging and disease, AACC Press, Washington, 1999 Search PubMed.
- L. Zhu, C. Gunn and J. S. Beckman, Arch. Biochem. Biophys., 1992, 298, 452–457 CrossRef CAS.
- S. Moncanda, R. M. J. Palmer and E. A. Higgs, Pharmacol. Rev., 1991, 43, 109–142 Search PubMed.
- C. F. Nathan and J. B. Hibbs Jr, Curr. Opin. Immunol., 1991, 3, 65–70 CrossRef CAS.
- F. Shahidi and Y. Zhong, Chem. Soc. Rev., 2010, 39, 4067–4079 RSC.
- A. V. Rao and S. Agarwal, Nutr. Res., 1999, 19, 305–326 CrossRef CAS.
- A. M. Jenkinson, A. R. Collins, S. J. Duthie, K. W. J. Wahle and G. G. Duthie, FASEB J., 1999, 13, 2138–2142 CAS.
- Y. Park, S. Nam, H. J. Yi, H. J. Hong and M. Lee, Nutr. Res., 2009, 29, 812–818 CrossRef CAS PubMed.
- H. Esterbauer, M. Dieber-Rotheneder, G. Waeg, G. Striegl and G. Juergens, Chem. Res. Toxicol., 1990, 3, 77–92 CrossRef CAS.
- W. A. Pryor and N. A. Porter, Free Radical Biol. Med., 1990, 8, 541–543 CrossRef CAS.
- T. P. A. Devasgayam, K. K. Boloor and T. Ramasarma, Indian J Biochem. Biophys., 2003, 40, 300–308 Search PubMed.
- H. Esterbauer, R. J. Schanr and H. Zollen, Free Radical Biol. Med., 1991, 11, 81–121 CrossRef CAS.
- M. Dizdaroglu, P. Jaruga, M. Birincioglu and H. Rodriguez, Free Radical Biol. Med., 2002, 32, 1102–1115 CrossRef CAS.
- B. Halliwell and J. M. C. Gutteridge, in Free radicals in biology and medicine, Oxford University Press, Oxford, 4th edn, 2007 Search PubMed.
- C. von Sonntag, in Free-radical-induced DNA damage and its repair, Springer, Hiedelberg, 2006 Search PubMed.
- M. Dizdaroglu, Mutat. Res., 1992, 275, 331–342 CAS.
- A. P. Breen and J. A. Murphy, Free Radical Biol. Med., 1995, 18, 1033–1077 CrossRef CAS.
- M. Dizdaroglu and P. Jaruga, Free Radical Res., 2012, 46, 382–419 CrossRef CAS PubMed.
- J. A. Simpson, S. Narita, S. Gieseg, S. Gebicki, J. M. Gebicki and R. T. Dean, Biochem. J., 1992, 282, 621–624 CAS.
- S. Gieseg, S. Duggan and J. M. Gebicki, Biochem. J., 2000, 350, 215–218 CrossRef CAS.
- E. R. Stadtman, Free Radical Biol. Med., 1990, 9, 315–325 CrossRef CAS.
- M. S. Cooke, M. D. Evans, M. Dizdaroglu and J. Lunec, FASEB J., 2003, 17, 1195–1214 CrossRef CAS PubMed.
- P. M. Kris-Etherton, K. D. Hecker, A. Bonanome, S. M. Coval, A. E. Binkoski, K. F. Hilpert, A. E. Griel and T. D. Etherton, Am. J. Med., 2002, 113, 71–88 CrossRef.
- D. Pal, S. Banerjee and A. Ghosh, J. Adv. Pharm. Technol. Res., 2012, 3, 16–24 CAS.
- E. A. Lissi, M. Salim-Hanna, C. Pascual and M. Del-Castillo, Free Radical Biol. Med., 1995, 18, 153–158 CrossRef CAS.
- M. Koruk, S. Taysi, M. C. Savas, O. Yilmaz, F. Akcay and M. Karakok, Ann. Clin. Lab. Sci., 2004, 34, 57–62 CAS.
- D. R. Gough and T. G. Cotter, Cell Death Dis., 2011, 2, e213 CrossRef CAS PubMed.
- C. D. Zhan, R. K. Sindhu, J. Pang, A. Ehdaie and N. D. Vaziri, J. Hypertens., 2004, 22, 2025–2033 CrossRef CAS PubMed.
- J. R. Stone and S. Yang, Antioxid. Redox Signaling, 2006, 8, 243–270 CrossRef CAS PubMed.
- E. Cabiscol, J. Tamarit and J. Ros, Int. Microbiol., 2000, 3, 3–8 CAS.
- J. R. Arthur, Cell Mol. Life Sci., 2000, 57, 1825–1835 CrossRef CAS.
- J. M. Matés, Toxicology, 2000, 153, 83–104 CrossRef.
- M. Valko, C. J. Rhodes, J. Moncol, M. Izakovic and M. Mazur, Chem.–Biol. Interact., 2006, 160, 1–40 CrossRef CAS PubMed.
- J. M. Matés, C. Perez-Gomez and I. N. De Castro, Clin. Biochem., 1999, 32, 595–603 CrossRef.
- S. Tafazoli, J. S. Wright and P. J. O'Brien, Chem. Res. Toxicol., 2005, 18, 1567–1574 CrossRef CAS PubMed.
- A. Vojdani, M. Bazargan, E. Vojdani and J. Wright, Cancer. Detect. Prev., 2000, 24, 508–523 CAS.
- P. K. Witting, J. M. Upston and R. Stocker, Biochemistry, 1997, 36, 1251–1258 CrossRef CAS PubMed.
- P. Morlière, L. K. Patterson, C. M. M. Santos, A. M. S. Silva, J. Mazière, P. Filipe, A. Gomes, E. Fernandes, M. B. Q. Garciah and R. Santusc, Org. Biomol. Chem., 2012, 10, 2068–2076 Search PubMed.
- R. Stocker, V. W. Boury and B. Frei, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 1646–1650 CrossRef CAS.
- E. Niki, Free Radical Biol. Med., 2014, 66, 3–12 CrossRef CAS PubMed.
- E. Niki, Am. J. Clin. Nutr., 1991, 54, 1119S–1124S CAS.
- K. L. Retsky, M. W. Freeman and B. Frei, J. Biol. Chem., 1993, 268, 1304–1309 CAS.
- C. Oh, M. Li, E. Kim, J. S. Park, J. Lee and S. W. Ham, Bull. Korean Chem. Soc., 2010, 31, 3513–3514 CrossRef CAS.
- B. R. Monaghan and F. O. Schmitt, J. Biol. Chem., 1932, 96, 387–395 CAS.
- R. S. Parker, FASEB J., 1996, 10, 542–551 CAS.
- A. V. Vieira, W. J. Schneider and P. M. Vieira, J. Endocrinol., 1995, 146, 201–207 CrossRef CAS.
- M. A. Livrea, L. Tesoriere, A. Bongiorno, A. M. Pintaudi, M. Ciaccio and A. Riccio, Free Radical Biol. Med., 1995, 18, 401–409 CrossRef CAS.
- L. Tesoriere, D. D'arpa, R. Re and M. A. Livrea, Arch. Biochem. Biophys., 1997, 343, 13–18 CrossRef CAS PubMed.
- P. G. Pietta, J. Nat. Prod., 2000, 63, 1035–1042 CrossRef CAS PubMed.
- C. A. Rice-Evans, N. J. Miller and G. Paganga, Free Radical Biol. Med., 1996, 20, 933–956 CrossRef CAS.
- P. Pietta, J. Nat. Prod., 2000, 63, 1035–1042 CrossRef CAS PubMed.
- R. J. Nijveldt, E. van Nood, D. E. C. van Hoorn, P. G. Boelens, K. van Norren and P. A. van Leeuwen, Am. J. Clin. Nutr., 2001, 74, 418–425 CAS.
- A. Russo, R. Acquaviva, A. Campisi, V. Sorrenti, C. D. Giacomo, G. Virgata, M. L. Barcellona and A. Vanella, Cell Biol. Toxicol., 2000, 16, 91–98 CrossRef CAS.
- F. V. Rubens de Souza and F. W. De Giovani, Redox Rep., 2004, 9, 97–104 CrossRef PubMed.
- T. Armida, T. Maurizio, T. Andrea and B. Sergio, J. Mol. Struct., 2005, 44, 759–766 Search PubMed.
- J. Zhou, L. F. Wang, J. Y. Wang and N. Tang, Transition Met. Chem., 2001, 26, 57–93 CrossRef CAS.
- V. Krishnamachari, L. H. Levine and P. W. Pareá, J. Agric. Food Chem., 2002, 50, 4357–4363 CrossRef CAS PubMed.
- A. Das, J. H. Wang and E. J. Lien, Prog. Drug Res., 1994, 42, 133–166 CrossRef CAS.
- S. J. Duthie, J. W. ohnson and V. L. Dobson, Mutat. Res., 1997, 390, 141–151 CAS.
- T. Jun, Z. Liancai and W. Bochu, Int. J. Pharmacol., 2007, 3, 19–26 Search PubMed.
- D. Galaris and A. Evangelou, Crit. Rev. Oncol. Hematol., 2002, 42, 93–103 CrossRef.
- S. S. Pekkarinen, I. M. Heinonen and A. I. Hopia, J. Sci. Food Agric., 1999, 79, 499–506 CrossRef CAS.
- M. G. Miguel, J. Appl. Pharm. Sci., 2011, 01, 07–15 Search PubMed.
- A. Castañeda-Ovando, M. L. Pacheco-Hernández, M. E. Páez-Hernández, J. A. Rodríguez and G.-C. Vidal, Food Chem., 2009, 113, 859–871 CrossRef PubMed.
- R. J. Nijveldt, E. van Nood, D. E. C. van Hoorn, P. G. Boelens, K. van Norren and P. A. M. van Leeuwen, Am. J. Clin. Nutr., 2001, 74, 418–425 CAS.
- S. A. van Acker, D. J. van den Berg, M. N. Tromp, D. H. Griffioen, W. P. van Bennekom, W. J. van der Vijgh and A. Bast, Free Radical Biol. Med., 1996, 20, 331–342 CrossRef CAS.
- W. Stahl and H. Sies, Mol. Aspects Med., 2003, 24, 345–351 CrossRef CAS.
- L. Mueller and V. Boehm, Molecules, 2011, 16, 1055–1069 CrossRef PubMed.
- J. A. Olson and N. I. Krinsky, FASEB J., 1995, 9, 1547–1550 CAS.
- H. Sies and W. Stahl, Am. J. Clin. Nutr., 1995, 62, 1315S–1321S CAS.
- W. Stahl and H. Sies, Mol. Aspects Med., 2003, 24, 345–351 CrossRef CAS.
- J. G. Erhardt, C. Meisner, J. C. Bode and C. Bode, Am. J. Clin. Nutr., 2003, 78, 1219–1224 CAS.
- A. V. Rao and L. G. Rao, Pharmacol. Res., 2007, 55, 207–216 CrossRef CAS PubMed.
- N. I. Krinsky and E. J. Johnson, Mol. Aspects Med., 2005, 26, 459–516 CrossRef CAS PubMed.
- G. Riccioni, Curr. Atheroscler. Rep., 2009, 11, 434–439 CrossRef CAS.
- P. Di Mascio, S. Kaiser and H. Sies, Arch. Biochem. Biophys., 1989, 274, 532–538 CrossRef CAS.
- G. Levin, M. Yeshurun and S. Mokady, Nutr. Cancer, 1997, 27, 293–297 CrossRef CAS PubMed.
- V. Böhm, N. L. Puspitasari-Nienaber, M. G. Ferruzzi and S. J. Schwartz, J. Agric. Food Chem., 2002, 50, 221–226 CrossRef PubMed.
- M. F. Andreasen, A. Landbo, L. P. Christensen, Å. Hansen and A. S. Meyer, J. Agric. Food Chem., 2001, 49, 4090–4096 CrossRef CAS PubMed.
- F. Natella, M. Nardini, M. Di Filici and C. Scaccini, J. Agric. Food Chem., 1999, 47, 1453–1459 CrossRef CAS PubMed.
- A. S. Meyer, J. L. Donovan, D. A. Pearson, A. L. Waterhouse and E. N. Frankel, J. Agric. Food Chem., 1998, 46, 1783–1787 CrossRef CAS.
- V. P. Menon and A. R. Sudheer, Adv. Exp. Med. Biol., 2007, 595, 105–125 CrossRef.
- J. S. Wright, J. Mol. Struct.: THEOCHEM, 2002, 59, 207–217 CrossRef.
- N. J. Miller, C. Castelluccio, L. Tijburg and C. Rice-Evans, FEBS Lett., 1996, 392, 40–44 CrossRef CAS.
- M. Shiraki, Y. Hara, T. Osawa, H. Kumon, T. Nakayama and S. Kawakishi, Mutat. Res., 1994, 323, 29–34 CAS.
- L. K. Leung, Y. Su, R. Chen, Z. Zhang, Y. Huang and Z. Y. Chen, J. Nutr., 2001, 131, 2248–2251 CAS.
- S. Ankri, T. Miron, A. Rabinkov, M. Wilchek and D. Mirelman, Antimicrob. Agents Chemother., 1997, 41, 2286–2288 CAS.
- K. C. Agarwal, Med. Res. Rev., 1996, 16, 111–124 CrossRef CAS.
- K. Prasad, V. A. Laxdal, M. Yu and B. L. Raney, Mol. Cell. Biochem., 1995, 148, 183–189 CrossRef CAS.
- V. Vaidya, K. U. Ingold and D. A. Pratt, Angew. Chem., Int. Ed., 2009, 48, 157–159 CrossRef CAS PubMed.
- E. Block, Angew. Chem., Int. Ed., 1992, 31, 1135–1178 CrossRef.
- P. T. Lynett, K. Butts, V. Vaidya, G. E. Garretta and D. A. Pratt, Org. Biomol. Chem., 2011, 9, 3320–3330 CAS.
- R. Amorati, P. T. Lynett, L. Valgimigli and D. A. Pratt, Chem.–Eur. J., 2012, 18, 6370–6379 CrossRef CAS PubMed.
- A. Galano and M. Francisco-Marquez, J. Phys. Chem. B, 2009, 113, 16077–16081 CrossRef CAS PubMed.
- S. K. Gupta, P. Bansal, R. K. Bhardwaj and T. Velpandian, Pharmacol. Res., 2000, 41, 657–662 CrossRef CAS PubMed.
- Y. F. Bai and H. Xu, Acta Pharmacol. Sin., 2000, 21, 357–359 CAS.
- S. A. Lee, S. S. Hong, X. H. Han, J. S. Hwang, G. J. Oh, K. S. Lee, M. K. Lee, B. Y. Hwang and J. S. Ro, Chem. Pharm. Bull., 2005, 53, 832–835 CrossRef CAS.
- J. Wattanathorn, P. Chonpathompikunlert, S. Muchimapura, A. Priprem and O. Tankamnerdthai, Food Chem. Toxicol., 2008, 46, 3106–3110 CrossRef CAS PubMed.
- K. Selvendiran, J. P. Singh, K. B. Krishnan and D. Sakthisekaran, Fitoterapia, 2003, 74, 109–115 CrossRef CAS.
- G. Shoba, D. Joy, T. Joseph, M. Majeed, R. Rajendran and P. S. Srinivas, Planta Med., 1998, 64, 353–356 CrossRef CAS PubMed.
- P. Anand, A. B. Kunnumakkara, R. A. Newman and B. B. Aggarwal, Mol. Pharm., 2007, 4, 807–818 CrossRef CAS PubMed.
- P. Ananda, S. G. Thomas, A. B. Kunnumakkara, C. Sundaram, K. B. Harikumar, B. Sung, S. T. Tharakan, K. Misra, I. K. Priyadarsini, K. N. Rajasekharan and B. B. Aggarwal, Biochem. Pharmacol., 2008, 76, 1590–1611 CrossRef PubMed.
- K. I. Priyadarsini, D. K. Maity, G. H. Naik, M. Sudheer, M. K. Unnikrishnan, J. G. Satav and H. Mohan, Free Radical Biol. Med., 2003, 35, 475–484 CrossRef CAS.
- M. Salem, S. Rohanib and E. R. Gillies, RSC Adv., 2014, 4, 10815–10829 RSC.
- O. N. Gordon and C. Schneider, Trends Mol. Med., 2012, 18, 361–363 CrossRef CAS PubMed.
- M. Griesser, V. Pistis, T. Suzuki, N. Tejera, D. A. Pratt and C. Schneider, J. Biol. Chem., 2011, 286, 1114–1124 CrossRef CAS PubMed.
- N. Sreejayan, M. N. A. Rao, K. I. Priyadarsini and T. P. A. Devasagayam, Int. J. Pharmacol., 1997, 151, 127–130 CrossRef CAS.
- A. Barik, B. Mishra, L. Shen, H. Mohan, R. M. Kadam, S. Dutta, H. Zhang and K. I. Priyadarsini, Free Radical Biol. Med., 2005, 39, 811–822 CrossRef CAS PubMed.
- B. Stinefelt, S. S. Leonard, K. P. Blemings, X. Shi and H. Klandorf, Ann. Clin. Lab. Sci., 2005, 35, 37–45 CAS.
- A. Galano and J. R. Alvarez-Idaboy, RSC Adv., 2011, 1, 1763–1771 RSC.
- W. S. Warning, QJM: An International Journal of Medicine, 2002, 95, 691–693 CrossRef.
- S. Muraoka and T. Miura, Pharmacol. Toxicol., 2003, 93, 284–289 CAS.
- G. L. Squadrito, R. Cueto, A. E. Splenser, A. Valavanidis, H. Zhang, R. M. Uppu and W. A. Pryor, Arch. Biochem. Biophys., 2000, 376, 333–337 CrossRef CAS PubMed.
- N. Kuzkaya, N. Weissmann, D. G. Harrison and S. Dikalov, Biochem. Pharmacol., 2005, 70, 343–354 CrossRef CAS PubMed.
- P. Ahmadpoor, E. Eftekhar, J. Nourooz-Zadeh, H. Servat, K. Makhdoomi and A. Ghafari, Iran J. Kidney Dis., 2009, 3, 22–27 Search PubMed.
- R. B. Pereira, C. Sousa, A. Costa, P. B. Andrade and P. Valentão, Molecules, 2013, 18, 8858–8872 CrossRef CAS PubMed.
- A. Pastore, G. Federici, E. Bertini and F. Piemonte, Clin. Chim. Acta, 2003, 333, 19–39 CrossRef CAS.
- P. C. Huber and W. P. Almeida, Quim. Nova, 2008, 31, 1170–1179 CrossRef CAS PubMed.
- M. A. Karaman, N. M. Mimica-Dukić and M. N. Matavulj, Arch. Biol. Sci., 2005, 57, 93–100 CrossRef.
- R. B. Jalgaonwala, B. V. Mohite and R. T. Mahajan, J. Microbiol. Biotechnol. Res., 2011, 1, 21–32 Search PubMed.
- N. Sadrati, H. Daoud, A. Zerroug, S. Dahamna and S. Bouharati, J. Plant Prot. Res., 2013, 53, 128–136 Search PubMed.
- F. Vélez-González, D. Ortegón-Reyna, A. A. Ramos-Organillo, A. L. Peraza-Campos, M. T. Sumaya-Martínez, R. Tapia-Benavides and F. J. Martínez-Martíneza, ARKIVOC, 2008, V, 55–64 Search PubMed.
- S. Lee, J. Han, H. Kim, E. Kim, T. Jeong, W. S. Lee and K. Cho, Bioorg. Med. Chem. Lett., 2004, 14, 4677–4681 CrossRef CAS PubMed.
- F. Natella, M. Nardini, M. D. Felice and C. Scaccini, J. Agric. Food Chem., 1999, 47, 1453–1459 CrossRef CAS PubMed.
- D. Point, P. Coudert, F. Leal, C. Rubat, V. Sautou-Miranda, J. Chopineau and J. Couquelet, Il Farmaco, 1998, 53, 85–88 CrossRef CAS.
- D. Krishnaiah, R. Sarbatly and R. Nithyanandam, Food Bioprod. Process., 2011, 89, 217–233 CrossRef CAS PubMed.
- D. Pal and S. Mitra, J. Adv. Pharm. Technol. Res., 2010, 1, 268–272 Search PubMed.
- M. N. Alam, N. J. Bristi and M. Rafiquzzaman, Saudi Pharm. J., 2013, 21, 143–152 CrossRef PubMed.
- M. Carocho and I. C. F. R. Ferreira, Food Chem. Toxicol., 2013, 51, 15–25 CrossRef CAS PubMed.
- J. A. Imlay, Nat. Rev. Microbiol., 2013, 11, 443–454 CrossRef CAS PubMed.
- A. Meyer-Isaksen, Trends Food Sci. Technol., 1995, 6, 300–304 CrossRef.
- J. Robak and R. J. Gryglewski, Flavonoids are scavengers of superoxide anions, Biochem. Pharmacol., 1988, 37, 837–841 CrossRef CAS.
- C. W. Choi, S. C. Kim, S. S. Hwang, B. K. Choi, H. J. Ahn, M. Y. Lee, S. H. Park and S. K. Kim, Plant Sci., 2002, 163, 1161–1168 CrossRef CAS.
- C. Sánchez-Moreno, Food Sci. Technol. Int., 2002, 8, 121–137 CrossRef PubMed.
- C. A. Calliste, P. Trouillas, D. P. Allais, A. Simon and J. L. Duroux, J. Agric. Food Chem., 2001, 49, 3321–3327 CrossRef CAS PubMed.
- C. A. Calliste, D. Kozlowski, J. L. Duroux, Y. Champavier, A. J. Chulia and P. Trouillas, Food Chem., 2010, 118, 489–496 CrossRef CAS PubMed.
- H. Chang, Y. Ho, M. Sheu, Y. Lin, M. Tseng, S. Wu, G. Huang and Y. Chang, Bot. Stud., 2007, 48, 407–417 CAS.
- P. Evelson, M. Travacio, M. Repetto, J. Escobar, S. Llesuy and E. A. Lissi, Arch. Biochem. Biophys., 2001, 388, 261–266 CrossRef CAS PubMed.
- H. Leontowicz, S. Gorinstein, A. Lojek, M. Leontowicz, M. Ciz, R. Soliva-Fortuny, Y. Park and S. Jung, J. Nutr. Biochem., 2002, 13, 603–610 CrossRef CAS.
- M. G. Repetto and S. F. Llesuy, Braz. J. Med. Biol. Res., 2002, 35, 523–534 CAS.
- P. Miciński, K. Pawlicki, E. Wielgus, M. Bochenek, P. Gogol and B. Ficek, Reprod. Biol., 2011, 11, 135–144 CrossRef.
- R. He, Y. Li, X. Li, R. Yi, X. Wang, B. Tsoi, K. K. H. Lee, K. Abe, X. Yang and H. Kurihara, PLoS One, 2013, 8, e57732 CAS.
- F. S. Hosseinian, A. D. Muir, N. D. Westcott and E. S. Krol, Org. Biomol. Chem., 2007, 5, 644–654 CAS.
- S. Kohtani, K. Yoshida, T. Maekawa, A. Iwase, A. Kudo, H. Miyabe and R. Nakagaki, Phys. Chem. Chem. Phys., 2008, 10, 2986–2992 RSC.
- H. Esterbauer, J. H. Gebicki and G. Jürgens, Free Radical Biol. Med., 1992, 13, 341–390 CrossRef CAS.
- E. Niki, Y. Yoshida, Y. Saito and N. Noguchi, Biochem. Biophys. Res. Commun., 2005, 338, 668–676 CrossRef CAS PubMed.
- T. A. Dix and J. Aikens, Chem. Res. Toxicol., 1993, 6, 2–18 CrossRef CAS.
- V. Mišík, K. Ondriaš and P. Balgavý, Gen. Physiol. Biophys., 1992, 11, 317–325 Search PubMed.
- K. Marshall, R. J. Reiter, B. Poeggeler, O. I. Aruoma and B. Halliwell, Free Radical Biol. Med., 1996, 21, 307–315 CrossRef CAS.
- O. I. Aruoma, in Free radicals in tropical diseases, Harwood Academic Publishers, 1993 Search PubMed.
- O. I. Aruoma, Free Radical Biol. Med., 1996, 20, 675–705 CrossRef CAS.
- C. L. Alarcón and A. Denicola, Anal. Chim. Acta, 2013, 763, 1–10 CrossRef PubMed.
- K. L. Wolfe and R. H. Liu, J. Agric. Food Chem., 2007, 55, 8896–8907 CrossRef CAS PubMed.
- D. Honzel, S. G. Carter, K. A. Redman, A. G. Schauss, J. R. Endres and G. S. Jensen, J. Agric. Food Chem., 2008, 56, 8319–8325 CrossRef CAS PubMed.
- J. A. Royall and H. Ischiropoulos, Arch. Biochem. Biophys., 1993, 302, 348–355 CrossRef CAS PubMed.
- M. Antolovich, P. D. Prenzler, E. Patsalides, S. McDonald and K. Robards, Analyst, 2002, 127, 183–198 RSC.
- R. Apak, S. Gorinstein, V. Böhm, K. M. Schaich, M. Özyürek and K. Güçlü, Pure Appl. Chem., 2013, 85, 957–998 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2015 |