Hybrid cholesterol-based nanocarriers containing phosphorescent Ir complexes: in vitro imaging on glioblastoma cell line†
Maria Naddakaa,
Erica Locatellib,
David Colecchiac,
Letizia Sambrib,
Ilaria Monacob,
Andrea Baschierib,
Federica Sasdellic,
Mario Chiarielloc,
Elia Matteuccib,
Paolo Zanib and
Mauro Comes Franchini*b
aStephenson, Institute for Renewable Energy, University of Liverpool, Peach Street, L69 7ZF, Liverpool, UK
bDepartment of Industrial Chemistry “Toso Montanari”, University of Bologna, Viale Risorgimento 4, 40136, Bologna, Italy. E-mail: mauro.comesfranchini@unibo.it
cIstituto Toscano Tumori (ITT), Core Research Laboratory, Consiglio Nazionale delle Ricerche (CNR), Istituto di Fisiologia Clinica, Via Fiorentina 1, 53100, Siena, Italy
First published on 27th November 2014
Abstract
Recently the use of phosphorescent heavy-metal complexes in bioimaging techniques has been a promising research field and has been attracted increasing interest. Among these, phosphorescent iridium(III) complexes have shown many photophysical characteristics that made them promising candidates for fluorescence probes. In this study an innovative copolymer consisting of cholesterol, a natural component of biological membranes, and the well-known biocompatible Polyethylene (PEG), has been synthesized. Cholesterol–PEG amphiphilic copolymer has been used to form novel nanocarriers characterized by the incorporation and/or linkage of the phosphorescent iridium(III) derivatives through covalent or non-covalent interactions. Finally the nanocarrier's surface has been functionalized with the peptide chlorotoxin (Cltx), a targeting agent selective for glioblastoma cells (U87MG). The so obtained targeted water soluble nanocarrier has been tested for in vitro imaging on the glioblastoma cell line and has shown no toxic effect on cells.
Introduction
To date, early cancer detection remains the best possibility for increase in patients' survival rate. Indeed, the types of cancer more easily detectable with common techniques (such as ultrasound, biopsy or magnetic resonance imaging) are today the ones with best results in terms of death reduction and life prolongation. However, many forms of cancer are still presenting serious difficulties in early detection, due to their nature or location in the body, and therefore remain practically untreatable.1Commonly employed diagnostic contrast agents, present several limitations especially due to poor selectivity for the infected tissues, thus causing serious unwanted side effects and reduction in diagnosis sensitivity. By developing tumor-targeted contrast agents, based on biocompatible nanoparticle formulations, both an enhancement in sensitivity and specificity for tumor imaging may arise.2
In the past few years, there has been a rapidly growing interest in the use of phosphorescent iridium(III) complexes for cellular imaging, intracellular sensing, gene delivery, and cancer cell detection.3,4 Compared to purely organic luminophores for bioimaging, these complexes show many advantageous photophysical characteristics such as compatibility with time-gated bioimaging techniques that completely eliminate background signals due to sample-autofluorescence, and significant Stokes shifts in the visible region. In addition, the possibility to use a wide range of ligands for the fine tuning of the complexes physical and chemical properties together with intense, long-living, and environment-sensitive emission, permit their employment as reporters of their local surroundings and intracellular biological events.5,6 Unfortunately, many iridium(III) complexes suffer from high cytotoxicity, limiting their use as live-cell imaging reagents. This problem can be prevented by the incorporation and/or linkage of the luminescent derivatives into biologically compatible nanocarriers, through covalent or non-covalent interactions.7–9
Cholesterol based nanocarriers are of great potential for biomedical applications such as drug delivery and imaging.10,11 The natural origin of these nanostructures makes them superior over synthetic base biodegradable polymers in terms of availability and biocompatibility. However, fabrication of an amphiphilic structure through the attachment of a hydrophilic tail to the cholesterol hydrophobic macrostructure is necessary in order to create water dispersible nanocarriers.12–14 Polyethylene (PEG) is worldwide recognized as a biocompatible polymer and it has been frequently used as stabilizing agent for nanoparticles in aqueous media thanks to its hydrophilicity; moreover it could act as an enhancer of in vivo circulation half-life of nanoparticles thanks to its “stealth” behavior. That in mind, only limited amount of works have described the fabrication of pure PEG functionalized cholesterol particles. Unfortunately, even those were over 200 nm or needed additional surfactants for their stabilization.15–17
Glioblastoma Multiforme (GBM), a common and highly aggressive brain tumor, belongs to the so-called high-grade astrocytomas. The current standard therapy for its treatment consists of surgery followed by radiotherapy combined with chemotherapy but the median survival time is still limited to approximately 15 months.18
Following our long-standing interest in GBM, we have recently validated the use of the chlorotoxin (Cltx) peptide, as a suitable in vitro19,20 and in vivo21,22 targeting agents for glioma cell lines. Chlorotoxin, is a 36-amino acid peptide that specifically binds to Metalloproteinase 2 (MMP-2), a receptor over-expressed in brain cancer (glioma cells) but not in healthy cells,23,24 suggesting the potential use of Cltx-conjugated nanomaterials as glioma-specific markers with diagnostic and therapeutic applications.
Here, we describe a simple method for the fabrication of multifunctional cholesterol–PEG based nanocarriers, possessing both carboxylic acid and amino groups on their surface, having the ability of encapsulating and protecting hydrophobic molecules as well as the possibility of further modification in the outer shell for conjugation of bioactive molecules. Thanks to this versatile novel nanocarrier, we were able to shield an iridium(III) complex, by introducing it either inside the lipophilic core or outside on the outer shell of the nanocarrier and to visualize such nanocarriers into a cancer cell line, U87MG, derived from human glioblastoma multiforme.
Experimental section
Materials and methods
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) and used as received. Polyethylene glycol with amino and carboxylic acid end groups (NH2–PEG–COOH and NH2–PEG–NH2, MW ∼ 3 kDa) were purchased from Rapp Polymere GmbH (Tübingen, Germany). All aqueous solutions were prepared with deionized water obtained using an ultrafiltration system (Milli-Q, Millipore) with a measured resistivity above 18 MΩ cm−1.UV/Vis absorption spectra were measured on a Varian Cary 4 double-beam UV-Vis spectrometer and baseline corrected. Steady-state emission spectra were recorded on an Edinburgh FLS920P spectrofluorimeter equipped with a 450 W Xenon arc lamp, double excitation and single emission monochromators and a Peltier cooled Hamamatsu R928P photomultiplier tube (185–850 nm). Emission and excitation spectra were corrected for source intensity (lamp and grating) and emission spectral response (detector and grating) by calibration curve supplied with the instrument. Emission lifetimes were determined on the same Edinburgh instrument with the Time Correlated Single Photon Counting (TCSPC) technique using pulsed picosecond LEDs (EPLED 309.6, FHWM <800 ps, repetition rates between 10 kHz and 1 MHz) as the excitation source and the above-mentioned R928P PMT as detector. The goodness of fit was assessed by minimizing the reduced χ2 function and visual inspection of the weighted residuals. The emission quantum yields were determined according to the optically dilute solutions method in aqueous solutions with reference to Ru(bpy)3Cl2 as the standard (r) according to eqn (1)25 where I refers to the area of the emission peaks of the complex and the reference, A to their absorptions and n is the refractive index of the corresponding solvents.
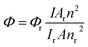 | (1) |
Iridium concentrations of the samples were measured by the inductively coupled plasma emission spectrometry (ICP-AES, Varian Liberty 51): after dissolution of the samples in concentrated nitric acid, the measurements were carried out by solution nebulization; instrumental sensitivity is 0.0013 ppm (mg L−1). DLS measurements were performed on a Malvern Zetasizernano-S working with a 532 nm laser beam. ζ potential measurements were conducted in DTS1060C-Clear disposable zeta cells at 25 °C.
Nanotechnology
Cell culture
U87MG cells were purchased by ATCC (HTB14) and maintained in Eagle's minimal essential medium (EMEM; PAA, E15-024) supplemented with 10% fetal bovine serum (PAA, A15-151), 2 mM L-glutamine and 100 units per mL penicillin–streptomycin at 37 °C in an atmosphere of 5% CO2/air. For cell viability experiment 105 cells were seeded in 6-well plates 24 hours before treatments. For internalization experiments, 2.5 × 104 cells were seeded on coverslips placed in 12-well plates.Cell viability assays
Cells were seeded in 6-well plates at 105 cells per well in triplicate. After 24 hours cells were incubated with Chol-NPs at the indicated concentration for 24 hours, then, cell were washed in PBS and harvested. Cell number of each sample was determined by counting the viable cells in a hemocytometer by trypan blue dye exclusion assay in triplicate. Data about cell viability were plotted in Prism 6 software (GraphPad) to draw dose-response curve and to calculate IC50.Internalization experiments
After incubation with Chol-NPs, cells were washed three times with PBS, then fixed with 4% paraformaldehyde in PBS for 20 min. Cells were incubated with Wheat germ agglutinin Alexa Fluor 647-conjugated (WGA-647 Life Technologies, W32466) for 20 minutes and then washed three times with PBS. Nuclei were stained with a solution of 1.5 μM of 4′,6-diamidino-2-phenylindole (DAPI; Sigma Aldrich, D9542) in PBS for 5 minutes. Coverslips were mounted in Fluorescence Mounting Medium (Dako, S3023). Samples were visualized on a TSC SP5 confocal microscope (Leica) installed on an inverted LEICA DMI 6000CS microscope and equipped with an oil immersion PlanApo 63X 1.4 NA objective. Images were acquired using the LAS AF acquisition software (Leica).Results and discussion
The synthesis of complex 4 is reported in Scheme 1 and described in ESI.† The commercial bromide 1 was transformed into the corresponding azide that was further reacted with 2-ethynyl pyridine through a Huisgen cycloaddition26,27 to give the bidentate compound 2. Pyridyl-triazole 2 was then employed as ancillary ligand to give the Ir(III) complex 3 by reaction with the dimer [(ppy)2Ir(μ-Cl)]2 (with ppy = 2-phenylpyridine). A further ester saponification gave the desired Ir(III) complex 4 able to be linked to the nanoparticles through an amide bond.Two covalently-linked amphiphilic macrostructures, based on a cholesterol unit and a PEG portion, were prepared; they differentiate only in terminal residual groups of the PEG portion, presenting either a carboxylic acid or an amino group. In both cases cholesteryl chloroformate was reacted with the ending amino group of the PEG polymer: differently from the synthesis of Chol–PEG–COOH copolymer, where the two reagents are used in stoichiometric ratio, for the preparation of Chol–PEG–NH2 a large excess of NH2–PEG–NH2 is necessary in order to avoid the coupling of both of the two amino groups of PEG. 1H-NMR confirmed the success of the synthesis (see ESI†).
The nature of the cholesterol–PEG amphiphilic macrostructure allowed their self-assembly into the corresponding nanocarriers using the nanoprecipitation technique, creating in this way nanocarriers that can host hydrophobic molecules in their core.28 For the synthesis of both the empty nanocarrier (Chol-NPs) and the iridium(III) complex-containing nanocarrier (Ircomp@Chol-NPs) we used a mixture of carboxylic acid and amino terminated cholesterol–PEG copolymers in order to have both of the orthogonal functionalities on the surface of the nanocarriers for further modifications; a 20% (w/w) amount of free cholesteryl hemisuccinate was found to be added to all the preparations in order to increase the lipophilic core portion otherwise too small in comparison to the PEG block, thus allowing formation of nanoparticles (Scheme 2).
Characterization data for all four types of nanocarriers is summarized in Table 1. The obtained empty nanocarrier (Chol-NPs) showed a small hydrodynamic diameter of around 50 nm with relatively narrow size distribution and ζ potential of around 20 mV (Table 1, entry 1). The negative, not too high, values of the ζ potential could be ascribed to the presence of free carboxylic acid groups, which are usually deprotonated at physiological pH, and of amino groups, which are generally not protonated at physiological pH, present on the nanocarriers' outer shell. Similar ζ potential was measured also for the iridium(III) complex-containing nanocarrier (Ircomp@Chol-NPs), however as expected the incorporation of the iridium(III) complex in the cholesterol core brings to an increase in the mean diameter by more than 10 nm (Table 1, entry 2), though remaining suitable for biological and in vivo applications. Functionalization of Chol-NPs on their outer shell with iridium(III) complex and/or targeting peptide chlorotoxin (Cltx) was achieved by using EDC-based chemistry. Firstly, a blank nanocarrier functionalized only with Ircomp were prepared (Chol-NPs@Ircomp) which showed a hydrodynamic diameter around 50 nm with a narrow size distribution and a negative ζ potential value (Table 1, entry 3); the conjugation reaction did not affected the hydrodynamic diameter as it is also possible to see from the resulting Chol-NPs@Cltx/Ircomp nanocarriers that remained to be around 50 nm (Table 1, entry 5). The success of the conjugation reactions is supported by the increase in the ζ potential to above −30 mV due to change in nanocarrier's surface functionality. Ircomp was connected through the reaction of its carboxylic acids with the amino groups on the surface of the nanocarriers, while Cltx could be connected to both functionalities as it possesses both amino and carboxylic acid terminus.
| Entry | Name | Diam. [nm] | PDI | ζ pot [mV] | Total conc. [mg mL−1] | Ir conc. [mg L−1] |
|---|---|---|---|---|---|---|
| 1 | Chol-NPs | 52.4 ± 2.0 | 0.281 ± 0.018 | −23.3 | 10.4 | — |
| 2 | Ircomp@Chol-NPs | 63.9 ± 1.3 | 0.257 ± 0.015 | −20.4 | 9.4 | 28.3 |
| 3 | Chol-NPs@Ircomp | 54.9 ± 0.85 | 0.261 ± 0.014 | −33.3 | 6.8 | 9.4 |
| 4 | Ircomp@Chol-NPs@Cltx | 75.9 ± 0.5 | 0.207 ± 0.004 | −33.2 | 9.9 | 20.4 |
| 5 | Chol-NPs@Cltx/Ircomp | 53.8 ± 2.1 | 0.274 ± 0.014 | −32.3 | 7.2 | 19.3 |
Similar ζ potential was also measured upon the conjugation of Cltx to Ircomp@Chol-NPs to yield Ircomp@Chol-NPs@Cltx however after this conjugation reaction also the hydrodynamic diameter have increased to above 75 nm, this can be an indication for a slight destabilization of the nanocarrier system (Table 1, entry 4). The concentration of all five nanocarriers in the final solution was estimated by weighting the residual organic matter after solvent evaporation (gravimetric analysis) and was found to be in the range 7–10 mg mL−1. Ir concentration was measured using ICP and was found to be in the range 10–30 mg L−1 for all types of Ir containing samples. All the obtained particles showed stability upon two months if store in darkness and at +4 °C, as assessed by DLS analysis which revealed no aggregation and no significant ζ potential variations, and because no iridium leaching was observed.
The photophysical properties of Chol-NPs@Cltx/Ircomp and Ircomp@Chol-NPs@Cltx in air equilibrated water solution were investigated. The absorption spectra (Fig. 1) resemble those of the free Ir(III) complex 4 and those previously reported in the literature for analogous complexes.29 The bands in the 250–300 nm region are assigned to spin allowed ligand-centered transitions 1LC (π–π*) localizing on the triazole–pyridine and phenyl–pyridine ligands. The weaker absorption bands in the 350–420 nm region belong to spin allowed metal-to-ligand charge-transfer 1MLCT and spin forbidden 3MLCT transitions. As observed for free 4, the Chol-NPs@Cltx/Ircomp and Ircomp@Chol-NPs@Cltx exhibited intense phosphorescence with maxima at 484 and 512 nm and with quantum yields (Φ) of 23% (Ircomp@Chol-NPs@Cltx) and 31% (Chol-NPs@Cltx/Ircomp) and lifetimes of 1.41 ns (Ircomp@Chol-NPs@Cltx) and 1.46 ns (Chol-NPs@Cltx/Ircomp). These results indicate that the Ir(III) complex exhibit its phosphorescent properties even when embedded in Chol-NPs, therefore the obtained Ir-functionalized nano-carriers can be suitable candidates for bio-imaging studies.
 | ||
| Fig. 1 Absorption and normalized emission spectra (λexc = 386 nm) of Ircomp@Chol-NPs@Cltx and Chol-NPs@Cltx/Ircomp in air equilibrated water solution at room temperature. | ||
Matrix metalloproteinase 2 (MMP-2) is a protein involved in the breakdown of extracellular matrix during physiological as well as disease processes.24 It is over-expressed in glioma cells as compared to healthy cells and can be recognized and bound with elevated specificity by the Cltx peptide. In particular, we have recently demonstrated cell-specific recognition, by Cltx-bound PNPs, of U87MG cell lines, in comparison to Balb/3T3 (mouse embryonic fibroblast cell line).19 We have, therefore, investigated the ability of our nanocarriers, containing the iridium(III) complex either inside the lipophilic core or outside, on the outer shell, to visualize U87MG human cells from Glioblastoma Multiforme. Indeed, by taking advantage of iridium(III) phosphorescent emission spectra, we demonstrated that both Chol-NPs@Cltx/Ircomp and Ircomp@Chol-NPs@Cltx were actively internalized into these cells, upon 24 h treatment (Fig. 2A), at concentrations well below their measured IC50 (Fig. 2B). By analyzing the images, it could not be said without doubt which of the two nanosystems is more effective or selective toward U87MG. However it appeared that the Ircomp@Chol-NPs@Cltx nanosystem showed, together with signals corresponding to internalization, also some green dots detected outside the cells (Fig. 2, right bottom, Marge image). This can be explained by lower stability of this nanocarriers due to physical entrapment of the Ircomp in comparison to its covalent linkage in the case of Chol-NPs@Cltx/Ircomp. Thus leading to diffusion of the Ircomp from the nanocarrier as was already suspected due to the increase in the hydrodynamic diameter of these nanocarriers upon functionalization with Cltx.
Importantly, upon substitution of the Cltx moiety with alternative targeting agents (e.g. antibodies and peptides), this system will allow high versatility to be successfully applied for diagnostic purposes to a large range of human tumors. Based on the low toxicity presented by nanocarriers with iridium outside the superficial shell as well as entrapped, we will be also able to choose the best and more appropriate molecular “skeleton” to build multifunctional structures able to contemporary diagnose and treat both primary and metastatic cancer.
Conclusions
In conclusion we have reported the synthesis and characterization of a novel nanosystem made of cholesterol–PEG copolymer with a phosphorescent iridium(III) complex inside. Due to the continuous search for newly and highly biocompatible nanosystems, we have shown toxicity studies on a GBM cell line and these results were satisfactory. At the same time, we have verified the importance of linking and/or entrapping the phosphorescent iridium complexes to the nanomicelle, Ir(III) complexes are indeed very powerful to make imaging itself but they would be hardly targeted to specific sites without the concept of drug delivery.The application of this innovative nanosystem in nanomedicine, and in particular in drug delivery, is currently studied in our laboratories.
Acknowledgements
University of Bologna is gratefully acknowledged.Notes and references
- D. M. Parkin, F. Bray, J. Ferlay and P. Pisani, Ca-Cancer J. Clin., 2005, 55, 74–108 CrossRef.
- X. Wang, L. Yang, Z. Chen and D. M. Shin, Ca-Cancer J. Clin., 2008, 58, 97–110 CrossRef PubMed.
- Y. You, Curr. Opin. Chem. Biol., 2013, 17, 699–707 CrossRef CAS PubMed.
- K. K. Lo and K. Y. Zhang, RSC Adv., 2012, 2, 12069–12083 RSC.
- I. M. Dixon, J. P. Collin, J. P. Sauvage, L. Flamigni, S. Encinas and F. Barigelletti, Chem. Soc. Rev., 2000, 29, 385–391 RSC.
- Y. You and P. S. Young, Dalton Trans., 2009, 1267–1282 RSC.
- H. Wu, T. Yang, Q. Zhao, J. Zhou, C. Li and F. Li, Dalton Trans., 2011, 1969–1976 RSC.
- C. W. Lai, Y.-H. Wang, C.-H. Lai, M.-J. Yang, C.-Y. Chen, P.-T. Chou, C.-S. Chan, Y. Chi, Y.-C. Chen and J.-K. Hsiao, Small, 2008, 4, 218–224 CrossRef CAS PubMed.
- Z. Zhou, D. Li, H. Yang, Y. Zhub and S. Yang, Dalton Trans., 2011, 11941–11944 RSC.
- R. Cavalli, A. Bisazza, R. Bussano, M. Trotta, A. Civra, D. Lembo, E. Ranucci and P. Ferruti, J. Drug Delivery, 2011, 587604–587610 CAS.
- Y. Kuang, J. Liu, Z. Liu and R. Zhuo, Biomaterials, 2012, 33, 1596–1606 CrossRef CAS PubMed.
- Y. Wang, H. Wang, G. Liu, X. Liu, Q. Jin and J. Ji, Macromol. Biosci., 2013, 13, 1084–1091 CrossRef CAS PubMed.
- J.-H. Lee, S.-W. Jung, I.-S. Kim, Y.-I. Jeong, Y.-H. Kim and S.-H. Kim, Int. J. Pharm., 2003, 251, 23–32 CrossRef CAS.
- B. Luppi, T. Cerchiara, F. Bigucci, R. Basile and V. Zecchi, J. Pharm. Pharmacol., 2004, 56, 407–411 CrossRef CAS PubMed.
- Y. Maitani, A. Nakamura, T. Tanaka and Y. Aso, Int. J. Pharm., 2012, 427, 372–378 CrossRef CAS PubMed.
- M. A. Maslov, T. O. Kabilova, I. A. Petukhov, N. G. Morozova, G. A. Serebrennikova, V. V. Vlassov and M. A. Zenkova, J. Controlled Release, 2012, 160, 182–193 CrossRef CAS PubMed.
- S. Winzen, M. Bernhardt, D. Schaeffel, A. Koch, M. Kappl, K. Koynov, K. Landfester and A. Kroeger, Soft Matter, 2013, 9, 5883–5890 RSC.
- M. L. Bondy, M. E. Scheurer, B. Malmer, J. S. Barnholtz-Sloan, F. G. Davis, D. Il'yasova, C. Kruchko, B. J. McCarthy, P. Rajaraman, J. A. Schwartzbaum, S. Sadetzki, B. Schlehofer, T. Tihan, J. L. Wiemels, M. Wrensch and P. A. Buffler, Cancer, 2008, 1(113, 7 suppl), 1953–1968 CrossRef PubMed.
- E. Locatelli, F. Broggi, J. Ponti, P. Marmorato, F. Franchini, S. Lena and M. Comes Franchini, Adv. Healthcare Mater., 2012, 1, 342–347 CrossRef CAS PubMed.
- A. Pucci, E. Locatelli, J. Ponti, C. Uboldi, V. Molinari and M. Comes Franchini, J. Nanopart. Res., 2013, 15, 1818–1824 CrossRef.
- E. Locatelli, W. Bost, M. Fournelle, J. Llop, L. Gil, F. Arena, V. Lorusso and M. Comes Franchini, J. Nanopart. Res., 2014, 16, 2304–2312 CrossRef.
- E. Locatelli, M. Naddaka, C. Uboldi, G. Loudos, E. Fragogeorgi, V. Molinari, A. Pucci, T. Tsotakos, D. Psimadas, J. Ponti and M. Comes Franchini, Nanomedicine, 2014, 839–849 CrossRef CAS PubMed.
- O. Veiseh, C. Sun, J. Gunn, N. Kohler, P. Gabikian, D. Lee, N. Bhattarai, R. Ellenbogen, R. Sze, A. Hallahan, J. Olson and M. Zhang, Nano Lett., 2005, 5, 1003–1008 CrossRef CAS PubMed.
- J. Deshane, C. C. Garner and H. Sontheimer, J. Biol. Chem., 2003, 278, 4135–4144 CrossRef CAS PubMed.
- K. Binnemans, Chem. Rev., 2009, 109, 4283–4374 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., 2002, 114, 2708–2711 CrossRef.
- R. Huisgen, in 1,3-Dipolar cycloaddition chemistry, ed. A. Padwa, Wiley, New York, 1984, vol. 1, p. 1 Search PubMed.
- E. Locatelli and M. Comes Franchini, J. Nanopart. Res., 2012, 14, 1316–1333 CrossRef.
- E. Orselli, R. Q. Albuquerque, P. M. Fransen, R. Fröhlich, H. M. Janssenc and L. De Cola, J. Mater. Chem., 2008, 18, 4579–4590 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis of Ir complex. See DOI: 10.1039/c4ra12936a |
| This journal is © The Royal Society of Chemistry 2015 |



