Curcumin associated poly(allylamine hydrochloride)-phosphate self-assembled hierarchically ordered nanocapsules: size dependent investigation on release and DPPH scavenging activity of curcumin†
Abstract
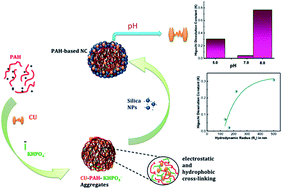
Combination of a cationic polyamine with a multivalent anionic salt results in the spontaneous generation of ionically crosslinked capsules. Here we report curcumin associated poly(allylamine hydrochloride) crosslinks with dipotassium phosphate and subsequently congregates with silica nanoparticles to form hierarchically ordered nanocapsule structures. The capsule sizes vary depending on the concentration of dipotassium phosphate. SEM data ascertain the spherical shape of the nanocapsules and TEM analysis demonstrates that the outer layer made up of silica has a thickness of ∼50–250 nm. The fluorescence images confirm that curcumin are present all over the capsules. Strong interaction between nanocapsules and curcumin is evident from spectroscopic analysis and TGA data. Release of curcumin from the nanocapsules is found to be triggered by pH where basic environment trigger the maximum release compared to acidic and neutral conditions. The drug release profile of curcumin from the nanocapsules follows the Higuchi model and depends on the size of the nanocapsules. 2,2-Diphenyl-1-picrylhydrazyl (DPPH) scavenging activity of the encapsulated curcumin decreases exponentially with a decrease in size of the nanocapsules, suggesting not only weight percentage of curcumin in the nanocapsules plays a role but also availability of β-diketone group of curcumin for H-donation is vital. Thus, in larger nanocapsules more curcumin per surface area of the nanocapsules are exposed on the outside for scavenging activity, compared to smaller sized nanocapsules where a substantial percentage of curcumin are buried/encapsulated inside the capsules without exposing themselves for scavenging activity.

 Please wait while we load your content...
Please wait while we load your content...