Design, synthesis and biological evaluation of novel 1,2,4-triazolo and 1,2,4-triazino[4,3-a]quinoxalines as potential anticancer and antimicrobial agents†
Doaa A. E. Issa *ab, Nargues S. Habib a and Abeer E. Abdel Wahab c
aDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, University of Alexandria, 21521, Alexandria, Egypt
bDepartment of Pharmaceutical Sciences, Faculty of Pharmacy, Beirut Arab University, 115020, Beirut, Lebanon. E-mail: d.issa@bau.edu.lb
cGenetic Engineering and Biotechnology Research Institute (GEBRI), City for Scientific Research and Technology Application, Borg El-Arab, 21934, Alexandria, Egypt
First published on 29th September 2014
Abstract
In an effort to find new leads as anticancer or antimicrobial agents, the present work deals with the synthesis of some novel 1-substituted 1,2,4-triazolo[4,3-a]quinoxalines 7, 9a,b, and 14–19 and 1,2,4-triazino[4,3-a]quinoxalines 10a–c as well as 2-[5-amino-3-(4-chlorophenyl)pyrazol-1-yl]-3-benzylquinoxaline 13. These were synthesized using the key intermediate 3-benzyl-2-hydrazinoquinoxaline 6 with various reagents. Ten compounds, namely 7, 9a, 10b, 11, and 13–18 were chosen by the National Cancer Institute of Bethesda (NCI) for evaluation of their anticancer activity. The results indicated that 9a was the most active and was further evaluated for in vitro five dose assay against 60 human cell lines. It was proven to possess the highest broad spectrum anticancer activity. It showed particular effectiveness towards leukemia SR, non-small cell lung cancer HOP-92, NCI-H460, colon cancer HCT-116, HCT-15, CNS cancer U251, melanoma LOX IMVI, renal cancer A498, prostate cancer PC-3, and breast cancer MDA-MB-468 cell lines (GI50 = 3.91, 3.45, 3.49, 3.21, 1.96, 5.18, 3.69, 1.80, 5.19, and 5.55 μM, respectively). All new compounds were screened for their antimicrobial activity and were very active against P. aeruginosa. Compounds 10a and 16 were twice as active as ampicillin against P. aeruginosa. Five compounds, 9a, b, 10b, 13, and 14 were equipotent to ampicillin against P. aeruginosa. In addition, compound 16 showed a broad spectrum antimicrobial activity. Furthermore, compound 9a showed dual activity as an anticancer and antimicrobial agent.
1. Introduction
The quinoxaline pharmacophore, being isosteric to purine antimetabolites, developed an appealing platform for the discovery of active chemotherapeutic agents. Several anticancer drugs containing a quinoxaline ring have been reported along with their pharmacological data, activity against solid tumours and clinical trials studies.1 The antineoplastic antibiotic quinoxaline, Triostatin A, (Fig. 1) showed considerable interest. It is characterized by cross-linked octapeptide rings bearing two quinoxalines and is stabilized at its centre by a cysteine pair (disulfhydryl covalent bonds). The two quinoxaline rings represent the planar aromatic ring structure which is a major requirement for intercalation.2 Besides the two antineoplastic quinoxalines topoisomerase II poisons, XK469 ((±)2-[4-(7-chloro-2-quinoxalinyloxy)phenoxyl]propanoic acid) and CQS (5-chloro quinoxalin-2-yl)-(4-aminobenzene sulfonamide) (Fig. 1) showed difference in DNA site specificity of topoisomerase II poisoning. This may be caused by differences in their geometry, side chains or electronic structure.3 In addition, XK469 induced apoptosis of human ovarian cancer cell line PA1.4 Several efforts have been made to search for new quinoxaline anticancer agents; thus a series of 2-alkylcarbonyl and 2-benzoyl-3-trifluromethylquinoxaline-1,4-di-N-oxide derivatives was found to inhibit the growth of Leukemia cell lines.5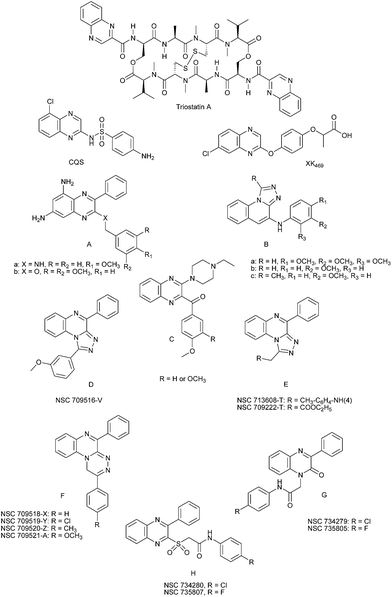 | ||
| Fig. 1 Some quinoxaline derivatives having anticancer activity. | ||
Recently, a series of 5,7-diamino-3-phenyl-2-benzylamino and 5,7-diamino-3-phenyl-2-benzyloxy, substituted quinoxalines of the general formula (A) (Fig. 1) has been synthesized. Among them two compounds showed promising anticancer activity.6 4-Substituted anilino-1,2,4-triazolo[4,3-a]quinoxalines of the general formula (B) (Fig. 1) exhibited prominent cytotoxicity against mock-infected M.T-4 cells.7 Whereas, 3-benzoyl-2-piperazinylquinoxalines of the general formula (C) (Fig. 1) showed anticancer activity against melanoma, renal and colon cancer.8 Quinoxalines are currently recognized to display good affinity to the ATP-binding site of the c-kit tyrosine protein. Their activity as antitumor agents may be due to their ability to inhibit protein tyrosine kinases.9
During our ongoing research program on quinoxaline derivatives,10–13 we were able to synthesize new lead structures. In particular: 1-(3-methoxyphenyl)-4-phenyl-1,2,4-triazolo[4,3-a]quinoxaline (D), 1-(substituted methyl)-4-phenyl-1,2,4-triazolo[4,3-a]quinoxalines (E), 2-(4-substituted phenyl)-5-phenyl-1H-1,2,4-triazino[4,3-a]quinoxalines (F),12 1-(N-arylcarbamoylmethyl)-3-phenylquinoxaline-2(1H)-ones (G),13 and 2-(N-arylcarbamoylmethylsulfonyl)-3-phenylquinoxaline (H) (Fig. 1).14 The antimicrobial activity of the triazolo and triazinoquinoxalines is well documented.10,11,13–15
Lately the incorporation of 1,2,4-triazolo fused heterocycles in anti-proliferative and anti-microbial drug design projects revealed a very interesting scaffold to find new potential agents.16,17 These findings prompted us to continue our investigations on quinoxalines having dual activity as anticancer and antimicrobial agents and to synthesize 1-substituted 4-benzyl-1,2,4-triazolo[4,3-a]quinoxalines having various pharmacophoric groups at the 1-position (7, 9a, b, and 16–19); also a series of 2-aryl-5-benzyl-1H-1,2,4-triazino[4,3-a]quinoxalines 10a–c were synthesized besides 2-[5-amino-3-(4-chlorophenyl)pyrazol-1-yl]-3-benzylquinoxaline 13 and 4-benzyl-1,2,4-triazolo[4,3-a]quinoxaline-1(2H)-one 15.
2. Results and discussion
2.1. Chemistry
Target 1,2,4-triazolo[4,3-a]quinoxalines 7, 9a, b, 14–19 and 1,2,4-triazino[4,3-a]quinoxalines 10a–c as well as 2-[5-amino-3-(4-chlorophenyl)pyrazol-1-yl]-3-benzylquinoxaline 13 were synthesized by the reactions depicted in Schemes 1–3.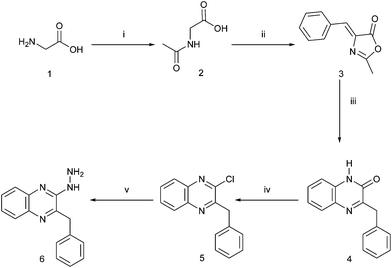 | ||
| Scheme 1 Synthesis of key intermediate 6. Reagents and conditions: (i) (CH3CO)2O·H2O, rt, 2 h, 91%; (ii) C6H5CHO, (CH3CO)2O, CH3CO2Na, 100 °C, 2 h, 86%; (iii) o-C6H4(NH2)2, CH3CH2OH, reflux, 5 h, 79%; (iv) POCl3, 100 °C, 2 h, 75%; (v) NH2NH2·H2O, CH3CH2OH, reflux, 2 h, 92%. | ||
 | ||
| Scheme 2 Synthesis of the target compounds 7–13. Reagents and conditions: (i) (CH3CO)2O or 2-acetylbuterolactone, dry xylene, reflux, 2–3 h, 55–77%; (ii) 2-Cl or 4-NO2C6H4COOH, POCl3, 100 °C, 2 h, 92–95%; (iii) 4-RC6H4COCH2Br, dry dioxane, reflux, 1 h, 64–73%; (iv) CH3COCH2COOC2H5, 160–170 °C, 1 h, 69%; (v) 4-ClC6H4COCH2CN, C2H5OH, CH3COOH, reflux, 2 h, 97%. | ||
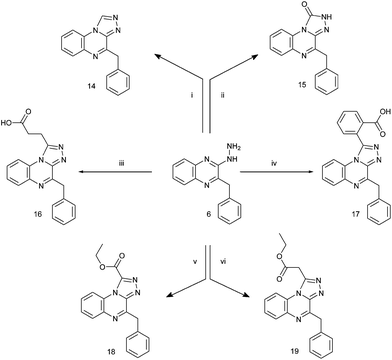 | ||
| Scheme 3 Synthesis of the target compounds 14–19. Reagents and conditions: (i) HCOOH, reflux, 3 h, 57%; (ii) ClCOOC2H5, C2H5OH, reflux, 3 h, 69%; (iii) succinic anhydride, gl AcOH, reflux, 5 h, 57%; (iv) phthalic anhydride, gl AcOH, reflux, 5 h, 59%; (v) (COOC2H5)2, dry xylene, reflux, 3 h, 68%; (vi) CH2(COOC2H5)2, 160–170 °C, 1 h, 87%. | ||
Preparation of the hydrazino key intermediate 6 was accomplished according to the sequence of reactions of Scheme 1. The azlactone of α-acetamidocinnamic acid 3 has been prepared according to the literature.18,19 Its condensation with o-phenylenediamine achieved ring closure to quinoxalinone 4 in good yield, the product was identical to that previously described by Moffitt and Schultz obtained from the hydrolysis and decarboxylation of 3-(α-carboxamido)benzyl-2(1H)-quinoxalinone.20 Earlier, 3-benzyl-1H-quinoxalin-2-one 4 has been differently prepared from o-phenylenediamine and phenylpyruvic acid.8 Treatment of 4 with phosphorus oxychloride gave the chloroquinoxaline derivative 5. Replacement of the chlorine atom of 5 with the hydrazine moiety resulted in the key intermediate 6.
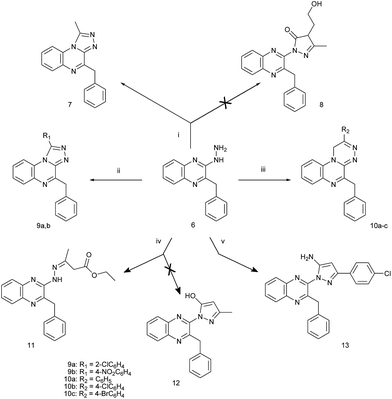
In the present work we studied the reaction of 6 with a variety of carbonyl compounds. The synthetic routes adopted to obtain the newly synthesized compounds are depicted in Schemes 2 and 3. Treatment of 6 with 2-acetylbutyrolactone failed to afford the corresponding 2-(pyrazol-1-yl)-3-benzylquinoxaline derivative 8 as reported for the preparation of related compounds.21 Instead, 4-benzyl-1-methyl-1,2,4-triazolo[4,3-a]quinoxaline 7 was obtained. The structure of 7 was confirmed by IR and 1H-NMR spectra and chemically through its parallel synthesis from 6 and acetic anhydride.15 Cyclization of 6 using aromatic acids and phosphorus oxychloride yielded 1-aryl-4-benzyl-1,2,4-triazolo[4,3-a]quinoxaline 9a,b where 9a is reported to be differently prepared.22 The 1,2,4-triazino[4,3-a]quinoxalines 10a–c were obtained through the reaction of 6 with phenacyl bromides. Fusion of a mixture of 6 and ethyl acetoacetate failed to afford 2-[5-hydroxy-3-methylpyrazol-1-yl]-3-benzylquinoxaline 12 as previously reported for analogous compounds.11 It yielded ethyl 3-[(3-benzylquinoxalin-2-yl)hydrazono]butyrate 11. Reacting 6 with 4-chlorophenyl-ω-cyanoacetophenone yielded 2-[5-amino-3-(4-chlorophenyl)pyrazol-1-yl]-3-benzylquinoxaline 13.
Compounds 14 and 15 were obtained by the reaction of 6 with formic acid and ethyl chloroformate, respectively. Compound 14 was previously reported to be synthesized from 2-benzyl-3-(2-methylenehydrazinyl) quinoxaline by its pyrolysis in dimethylformamide or its acylation with acetic anhydride in pyridine.22 Compound 15 was reported to be synthesized through the reaction of 2-chIoro-3-benzylquinoxaline with semicarbazide hydrochloride.15 Compounds 16 and 17 were obtained by cyclization of 6 using succinic or phthalic anhydride, respectively, in glacial acetic acid. Furthermore reacting 6 with diethyl oxalate or diethyl malonate in boiling dry xylene gave compounds 18 and 19 respectively as described for the synthesis of analogous compounds.13
2.2. Biological evaluation
| Cpd | NSC no. | Panel | Subpanel tumor cell lines (% growth inhibitory activity) |
|---|---|---|---|
| a The data obtained from NCI in vitro disease-oriented human tumor cell screen at 10 μM concentration. | |||
| 7 | 761 669 | Non-small cell lung cancer | HOP-92 (17.65); NCI-H226 (15.11) |
| CNS cancer | SNB-75 (21.94) | ||
| Ovarian cancer | NCI/ADR-RES (12.55) | ||
| 9a | 761 676 | Leukemia | HL-60(TB) (46.36); K-562 (78.62); MOLT-4 (60.41); RPMI-8226 (63.81); SR (84.89) |
| Non-small cell lung cancer | EKVX (48.06); HOP-92 (50.11); NCI-H460 (84.44) | ||
| Colon cancer | HCT-116 (65.87); HCT-15 (55.37); KM12 (51.07) | ||
| CNS cancer | SF-268 (47.55); U251 (57.23) | ||
| Melanoma | LOX IMVI (60.62) | ||
| Renal cancer | A498 (61.07) | ||
| Prostate cancer | PC-3 (60.87) | ||
| 10b | 761 674 | Leukemia | HL-60(TB) (74.81); K-562 (69.87); RPMI-8226 (50.59); SR (74.57) |
| Non-small cell lung cancer | NCI-H522 (61.57) | ||
| Colon cancer | HCT-116 (46.86) | ||
| Renal cancer | CAKI-1 (44.63) | ||
| 11 | 761 673 | Renal cancer | A498 (17.75); UO-31 (15.85) |
| 13 | 761 675 | Non-small cell lung cancer | A549/ATCC (21.96); EKVX (21.36); NCI-H226 (15.73); NCI-H23 (18.49) |
| Colon cancer | HCT-116 (27.18); HCT-15 (25.56); HT29 (30.32) | ||
| CNS cancer | SNB-75 (15.58) | ||
| Renal cancer | CAKI-1 (19.3); RXF 393 (21.29) | ||
| Breast cancer | MDA-MB-231/ATCC (24.88) | ||
| 14 | 761 678 | Non-small cell lung cancer | NCI-H226 (15.47) |
| CNS cancer | SNB-75 (23.49) | ||
| Renal cancer | A498 (38.71); UO-31 (22.68) | ||
| Prostate cancer | PC-3 (14.24) | ||
| 15 | 761 677 | Renal cancer | A498 (28.58); UO-31 (35.5) |
| 16 | 761 671 | Non-small cell lung cancer | HOP-92 (18.87) |
| CNS cancer | SNB-75 (17.71) | ||
| Renal cancer | UO-31 (12.22) | ||
| 17 | 761 670 | Non-small cell lung cancer | NCI-H522 (14.55) |
| CNS cancer | SF-268 (14.1); SNB-75 (24.41) | ||
| Ovarian cancer | OVCAR-3 (20.22) | ||
| Renal cancer | UO-31 (18.01) | ||
| Prostate cancer | PC-3 (43.92) | ||
| 18 | 761 672 | CNS cancer | SNB-75 (10.34) |
| Renal cancer | UO-31 (15.06) | ||
| Panel | Subpanel cell lines (cytotoxicity GI50 μM) |
|---|---|
| a Data obtained from NCI in vitro disease-oriented human cell screen. | |
| Leukemia | CCRF-CEM (7.11); HL-60(TB) (32.5); K-562 (5.55); MOLT-4 (9.04); RPMI-8226 (5.64); SR (3.91) |
| Non-small cell lung cancer | A549/ATCC (8.77); EKVX (5.96); HOP-62 (12.4); HOP-92 (3.45); NCI-H226 (9.96); NCI-H322M (53.9); NCI-H460 (3.49); NCI-H522 (53.9) |
| Colon cancer | COLO 205 (8.77); HCC-2998 (42.8); HCT-116 (3.21); HCT-15 (1.96); HT29 (9.15); KM12 (9.48); SW-620 (43.4) |
| CNS cancer | SF-295 (12.3); SF-539 (24.2); SNB-19 (27.8); SNB-75 (6.12); U251 (5.18) |
| Melanoma | LOX IMVI (3.69); MALME-3M (23.8); M14 (36.7); SK-MEL-2 (55.8); SK-MEL-5 (5.90); UACC-257 (34.3); UACC-62 (7.96) |
| Ovarian cancer | IGROV1 (27.7); OVCAR-3 (21.7); OVCAR-4 (8.01); OVCAR-8 (7.21); NCI/ADR-RES (7.04); SK-OV-3 (23.9) |
| Renal cancer | 786-0 (10.4); A498 (1.80); ACHN (34.1); CAKI-1 (9.71); RXF 393 (8.45); TK-10 (37.4); UO-31 (25.0) |
| Prostate cancer | PC-3 (5.19); DU-145 (44.2) |
| Breast cancer | MCF7 (22.2); MDA-MB-231/ATCC (6.24); HS 578T (9.66); BT-549 (6.72); T-47D (6.82); MDA-MB-468 (5.55) |
As revealed from Table 1 showing the percentage growth inhibition (GI%) caused by the test compounds, compound 9a, having the 2-chlorophenyl moiety at the 1-position of the triazoloquinoxaline, was the most active compound. It showed a broad spectrum anticancer activity against most cell lines, namely Leukemia HL-60(TB), K-562, MOLT-4, RPMI-8226 and SR cell lines with a growth inhibition of 46.36%, 78.62%, 60.41%, 63.81% and 84.89%, respectively. Compound 9a was remarkably active against non-small cell lung cancer EKVX, HOP-92 and NCI-H460 cell lines with a growth inhibition of 48.06%, 50.11% and 84.44%, respectively. The colon cancer HCT-116, HCT-15 and KM12 cell lines showed a growth inhibition of 65.87%, 55.37%, and 51.07%, respectively. In addition CNS cancer SF-268 and U251 cell lines were reasonably inhibited by compound 9a with a growth inhibition of 47.55% and 57.23%, respectively. Also compound 9a manifested appreciable activity against melanoma LOX IMVI, renal cancer A498 and prostate cancer PC-3 cell lines with a growth inhibition of 60.62%, 61.07% and 60.87%, respectively.
Replacement of 2-chlorophenyl with 2-carboxyphenyl at the 1-position of the triazoloquinoxaline decreased the activity where compound 17 showed only activity against CNS cancer SNB-75, ovarian cancer OVCAR-3 and prostate cancer PC-3 cell lines with a growth inhibition of 24.41%, 20.22% and 43.92%, respectively. The unsubstituted triazoloquinoxaline 14 displayed moderate activity against CNS cancer SNB-75, renal cancer A498 and UO-31 cell lines with a growth inhibition of 23.49%, 38.71% and 22.68% respectively. Compound 15 having a carbonyl group at the 1-position of the triazoloquinoxaline was fairly active against renal cancer A498 and UO-31 cell lines with 28.58% and 35.5% growth inhibition, respectively. The 1-methyltriazoloquinoxaline 7 showed weak activity against the CNS cancer SNB-75 cell line with a growth inhibition of 21.94%. Conversely the triazinoquinoxaline 10b having 4-chloro substitution demonstrated a significant growth inhibition of 74.81%, 69.87%, 50.59%, 74.57%, 61.57%, 46.86% and 44.63% against leukemia HL-60(TB), K-562, RPMI-8226, SR, non-small cell lung cancer NCI-H522, colon cancer HCT-116 and renal cancer CAKI-1 cell lines, respectively.
Furthermore the pyrazolylquinoxaline 13 showed some activity against non-small cell lung cancer, colon cancer, renal cancer and breast cancer specifically non-small cell lung cancer A549/ATCC, EKVX, colon cancer HCT-116, HCT-15, HT29, renal cancer RXF 393 and breast cancer MDA-MB-231/ATCC cell lines with a growth inhibition of 21.96%, 21.36%, 27.18%, 25.56%, 30.32%, 21.29% and 24.88%, respectively.
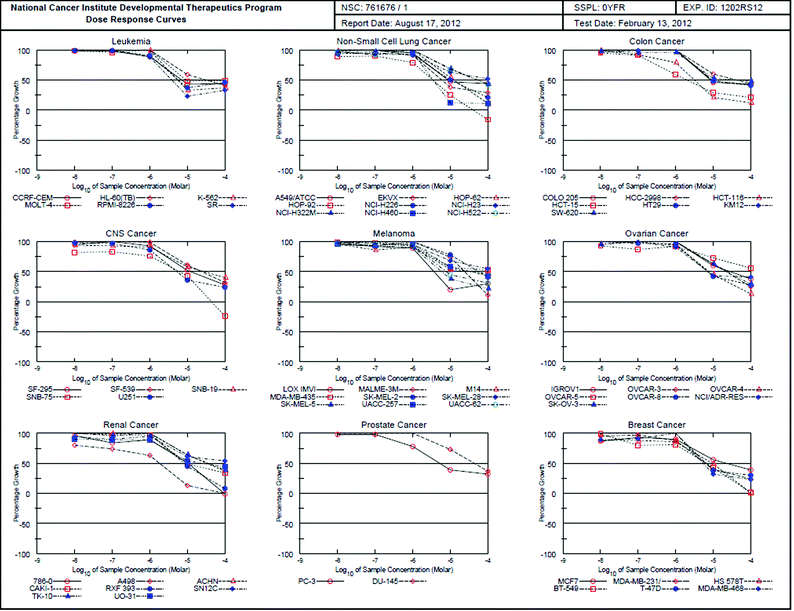
Only compound 9a fulfilled the requirements of selection for five-dose assay. Further interpretation of the five-dose screening data for compound 9a (Table 2) revealed that it was the most active lead in this study with a broad spectrum anticancer activity against most of the tested subpanel tumor cell lines with particular effectiveness against leukemia CCRF-CEM K-562, MOLT-4, RPMI-8226 and SR (GI50 = 7.11, 5.55, 9.04, 5.64 and 3.91 μM respectively). The compound also showed activity against non-small cell lung cancer A549/ATCC, EKVX, HOP-92, NCI-H226 and NCI-H460 (GI50 = 8.77, 5.96, 3.45, 9.96 and 3.49 μM, respectively), while TGI against HOP-92 was 41.8 μM. It also illustrated significant activity against colon cancer COLO 205, HCT-116, HCT-15, HT29 and KM12 (GI50 = 8.77, 3.21, 1.96, 9.15 and 9.48 μM, respectively). The compound displayed moderate activity against CNS Cancer SNB-75 and U251 (GI50 = 6.12 and 5.18 μM respectively), while TGI against SNB-75 was 43.6 μM. Its GI50 against Melanoma LOX IMVI, SK-MEL-5 and UACC-62 was equal to 3.69, 5.90 and 7.96 μM, respectively. Compound 9a demonstrated moderate activity against ovarian cancer OVCAR-4, OVCAR-8 and NCI/ADR-RES (GI50 = 8.01, 7.21 and 7.04 μM, respectively). The compound revealed promising activity against renal cancer A498, CAKI-1, RXF 393, prostate cancer PC-3, breast cancer MDA-MB-231/ATCC, HS 578T, BT-549, T-47D and MDA-MB-468 (GI50 = 1.80, 9.71, 8.45, 5.19, 6.24, 9.66, 6.72, 6.82 and 5.55 μM, respectively). The dose response curve of compound 9a is illustrated in Fig. 2.
The ratio obtained by dividing the compound fullpanel MG-MID (μM) by its individual subpanel MG-MID (μM) is considered as a measure of compound selectivity (Table 3). Ratios between 3 and 6 refer to moderate selectivity, ratios >6 indicate high selectivity toward the corresponding cell line, while compounds meeting neither of these criteria are rated non-selective.23 Accordingly, compound 9a was proven to be non-selective.
| MG-MIDa | Subpanel tumor cell linesb GI50 MG-MID (μM) (SI)c | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | IX | |
| a GI50: full panel mean-graph midpoint (μM). b I: leukemia; II: non-small cell lung cancer; III: colon cancer; IV: CNS cancer; V: melanoma; VI: ovarian cancer; VII: renal cancer; VIII: prostate cancer; IX: breast cancer. c SI: selectivity index. | |||||||||
| 16.80 | 10.62 (1.58) | 18.98 (0.89) | 16.97 (0.99) | 15.12 (1.11) | 24.02 (0.70) | 15.93 (1.05) | 18.12 (0.93) | 24.69 (0.68) | 9.53 (1.76) |
As revealed from Tables 4 and 5, regarding the antimicrobial activity against S. aureus, the tested compounds showed weak activity IZ (12–17 mm). Only compound 16 showed one fifth the activity of ampicillin. However, the tested compound demonstrated better activity against B. subtilis IZ (12–16 mm). Compound 16 was the most active compound in this respect. It was equipotent to ampicillin (MIC = 12.5 μg mL−1). Compounds 9a and 10a exhibited half the potency of ampicillin (MIC = 25 μg mL−1). The inhibition zone of the tested compounds against P. aeruginosa was 11–14 mm, two compounds 10a and 16 were double as active as ampicillin (MIC = 25 μg mL−1), and five compounds 9a, b, 10b, 13, and 14 were equipotent to ampicillin (MIC = 50 μg mL−1). The tested compounds displayed activity against E. coli; the inhibition zones ranging 12–16 mm, only compound 16 was nearly equipotent to ampicillin (MIC = 12.5 μg mL−1). On the other hand, the inhibition zone against C. albicans was 12–17 mm. Compound 10a showed nearly half the potency of clotrimazole and compounds 9b, 10b, 17, 18, and 19 showed one fifth the activity of clotrimazole.
| Cpd | S. aureus | B. subtilis | P. aeruginosa | E. coli | C. albicans |
|---|---|---|---|---|---|
| 7 | 12 | 13 | 11 | 12 | 14 |
| 9a | 12 | 12 | 14 | 10 | 16 |
| 9b | 14 | 12 | 12 | 14 | 15 |
| 10a | 13 | 14 | 12 | 16 | 14 |
| 10b | 15 | 16 | 14 | 15 | 15 |
| 10c | 12 | 12 | 12 | 14 | 14 |
| 11 | 15 | 12 | 12 | 14 | 14 |
| 13 | 14 | 15 | 12 | 16 | 16 |
| 14 | 17 | 12 | 14 | 12 | 17 |
| 15 | 16 | 16 | 12 | 10 | 14 |
| 16 | 13 | 12 | 12 | 15 | 16 |
| 17 | 14 | 16 | 12 | 15 | 14 |
| 18 | 14 | 13 | 10 | 12 | 15 |
| 19 | 14 | 14 | 14 | 15 | 12 |
| Cpd | S. aureus | B. subtilis | P. aeruginosa | E. coli | C. albicans | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| 7 | 50 | 100 | 50 | 100 | 100 | 100 | 50 | 50 | 100 | 200 |
| 9a | 100 | 200 | 25 | 50 | 50 | 100 | 25 | 50 | 100 | 200 |
| 9b | 50 | 100 | 100 | 100 | 50 | 50 | 100 | 100 | 25 | 50 |
| 10a | 50 | 50 | 25 | 50 | 25 | 50 | 50 | 50 | 12.5 | 25 |
| 10b | 50 | 100 | 100 | 200 | 50 | 100 | 50 | 50 | 25 | 25 |
| 10c | 50 | 50 | 50 | 50 | 100 | 100 | 50 | 50 | 50 | 50 |
| 11 | 100 | 100 | 50 | 50 | 100 | 100 | 50 | 100 | 50 | 100 |
| 13 | 100 | 100 | 100 | 100 | 50 | 50 | 100 | 100 | 50 | 50 |
| 14 | 50 | 100 | 50 | 50 | 50 | 100 | 25 | 25 | 100 | 200 |
| 15 | 100 | 200 | 100 | 200 | 100 | 100 | 100 | 100 | 50 | 100 |
| 16 | 25 | 50 | 12.5 | 25 | 25 | 25 | 12.5 | 25 | 50 | 100 |
| 17 | 100 | 200 | 100 | 100 | 100 | 100 | 25 | 50 | 25 | 50 |
| 18 | 50 | 50 | 50 | 100 | 100 | 100 | 50 | 50 | 25 | 50 |
| 19 | 50 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 25 | 25 |
| Ampicillin | 5 | 12.5 | 50 | 10 | ||||||
| Clotrimazole | 5 | |||||||||
It can be concluded that the tested compounds were weakly active against S. aureus and C. albicans. Compounds 10a and 16 were double as active as ampicillin against P. aeruginosa and five compounds 9a, b, 10b, 13, and 14 had similar activity to ampicillin, i.e. the tested compounds were very active against P. aeruginosa. The tested compounds showed medium activity against B. subtilis and E. coli. It is worth mentioning that compound 16 possesses a broad spectrum antimicrobial activity.
3. Conclusions
Preliminary in vitro anticancer screening revealed that compound 9a was the most active. It was proven to possess the highest broad spectrum anticancer activity after its further evaluation for in vitro five dose assay against 60 human cell lines. It showed particular effectiveness towards leukemia SR, non-small cell lung cancer HOP-92, NCI-H460, colon cancer HCT-116, HCT-15, CNS cancer U251, melanoma LOX IMVI, renal cancer A498, prostate cancer PC-3, and breast cancer MDA-MB-468 cell lines (GI50 = 3.91, 3.45, 3.49, 3.21, 1.96, 5.18, 3.69, 1.80, 5.19, and 5.55 μM, respectively). From the antimicrobial screening it was found that the most active compounds were 10a and 16. They showed twice the activity of ampicillin against P. aeruginosa. Moreover five compounds, namely 9a, b, 10b, 13 and 14 were equipotent to ampicillin against P. aeruginosa.In conclusion, the compound 4-benzyl-1-(2-chlorophenyl)-1,2,4-triazolo[4,3-a]quinoxaline 9a proved to possess dual effects as a broad spectrum anticancer and antimicrobial agent against P. aeruginosa.
4. Experimental section
4.1. Chemistry
All reagents and solvents were purchased from commercial suppliers and were dried and purified when necessary by standard techniques. All melting points were determined in open glass capillaries on a Gallenkamp melting point apparatus and are uncorrected. The IR spectra were recorded using KBr discs on a Perkin-Elmer 1430 spectrophotometer. 1H-NMR (δ ppm) spectra were recorded on a JNM-LA 400 FT NMR system (400 MHz) and on a Jeol (500 MHz) spectrometer (both JEOL, Tokyo, Japan). 13C-NMR spectra were run on Jeol spectrometer using TMS as the internal standard and DMSO-d6 as a solvent. Mass spectra were run on a Finnigan mass spectrometer model SSQ/7000 (70 eV). The microanalyses were performed at the Microanalytical Laboratory, National Research Center, Cairo, Egypt and the data were within ±0.4% of the theoretical values. Following up of the reactions and checking the homogeneity of the compounds were made by TLC on silica gel aluminum sheets (Type 60 GF254, Merck, Darmstadt, Germany) and the spots were detected by exposure to a UV-lamp at λ 254 nm for few seconds.Method A. The title compound was prepared by refluxing a solution of 3-benzyl-2-hydrazinoquinoxaline 6 (0.5 g, 2 mmol) in acetic anhydride (2.5 mL) for 2 h. The reaction mixture was poured onto ice water under stirring and the solid obtained was collected by filtration, washed with water and recrystallized from ethanol as yellowish needles (0.42 g, 76.6%), m.p. 182–183 °C; reported m.p.176–178 °C.15
Method B. A mixture of 6 (0.5 g, 2 mmol) and 2-acetylbutyrolactone (0.28 g, 2.2 mmol) in dry xylene (5 mL) was refluxed for 3 h. The reaction mixture was cooled; the obtained crystalline product was filtered, dried, and recrystallized from ethanol to yield the desired compound (0.3 g, 54.8%). The products obtained from method A and B were identical in IR (KBr, cm−1): 1677 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1605, 1495 (C
N); 1605, 1495 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 3.08 (s, 3H, CH3), 4.54 (s, 2H, CH2–C6H5), 7.20 (t, 1H, J = 7.4 Hz, C6H5–C4–H), 7.31 (t, 2H, J = 7.4 Hz, C6H5–C3,5–H), 7.46 (d, 2H, J = 7.4 Hz, C6H5–C2,6–H), 7.59–7.78 (m, 2H, triazoloquinox. C7,8–H), 8.04 (ddd, 1H, J = 7.2, 4.65, 2.1 Hz, triazoloquinox. C6–H), 8.30 (dd, 1H, J = 9, 4.5 Hz, triazoloquinox. C9–H). Anal. calcd for C17H14N4 (274.32): C, 74.43; H, 5.14; N, 20.42. Found: C, 74.29; H, 5.27; N, 20.74.
C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 3.08 (s, 3H, CH3), 4.54 (s, 2H, CH2–C6H5), 7.20 (t, 1H, J = 7.4 Hz, C6H5–C4–H), 7.31 (t, 2H, J = 7.4 Hz, C6H5–C3,5–H), 7.46 (d, 2H, J = 7.4 Hz, C6H5–C2,6–H), 7.59–7.78 (m, 2H, triazoloquinox. C7,8–H), 8.04 (ddd, 1H, J = 7.2, 4.65, 2.1 Hz, triazoloquinox. C6–H), 8.30 (dd, 1H, J = 9, 4.5 Hz, triazoloquinox. C9–H). Anal. calcd for C17H14N4 (274.32): C, 74.43; H, 5.14; N, 20.42. Found: C, 74.29; H, 5.27; N, 20.74. 4.1.2.1. 4-Benzyl-1-(2-chlorophenyl)-1,2,4-triazolo[4,3-a]quinoxaline (9a). Reddish-orange crystals (0.7 g, 94.5%), m.p. 200–201 °C; reported m.p. 194–195 °C.22 IR (KBr, cm−1): 1643 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1597, 1489 (C
N); 1597, 1489 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 767 (C–Cl). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.61, 4.70 (2d, each 1H, J = 14.1 Hz, CH2–C6H5), 7.10 (d, 1H, J = 7.5 Hz, C6H5–C2–H), 7.26 (d, 1H, J = 7.5 Hz, C6H5–C6–H), 7.34 (t, 2H, J = 7.5 Hz, C6H5–C3,5–H), 7.49–7.55 (m, 3H, C6H5–C4–H and chlorophenyl C4,5–H), 7.62–7.71 (m, 2H, triazoloquinox. C7,8–H), 7.78–7.86 (m, 3H, chlorophenyl C3,6–H and triazoloquinox. C6–H), 8.09 (dd, 1H, J = 8.1, 1.5 Hz triazoloquinox. C9–H). 13C-NMR (500 MHz, DMSO-d6, δ ppm): 42.3 (CH2–C6H5), 73.2 (triazoloquinox. C1), 114.6 (triazoloquinox. C9), 115 (triazoloquinox. C7), 125 (triazoloquinox. C6), 126.7 (benz. C1), 128.0, 128.1, 129.0, 129.1, 129.7, 129.9, 130.1, 132.4, 133.2, 133.8, 135.7, 136.5, 153.6, 209.6. Anal. calcd for C22H15ClN4 (370.84): C, 71.25; H, 4.08; N, 15.11. Found: C, 71.42; H, 4.04; N, 14.86.
C); 767 (C–Cl). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.61, 4.70 (2d, each 1H, J = 14.1 Hz, CH2–C6H5), 7.10 (d, 1H, J = 7.5 Hz, C6H5–C2–H), 7.26 (d, 1H, J = 7.5 Hz, C6H5–C6–H), 7.34 (t, 2H, J = 7.5 Hz, C6H5–C3,5–H), 7.49–7.55 (m, 3H, C6H5–C4–H and chlorophenyl C4,5–H), 7.62–7.71 (m, 2H, triazoloquinox. C7,8–H), 7.78–7.86 (m, 3H, chlorophenyl C3,6–H and triazoloquinox. C6–H), 8.09 (dd, 1H, J = 8.1, 1.5 Hz triazoloquinox. C9–H). 13C-NMR (500 MHz, DMSO-d6, δ ppm): 42.3 (CH2–C6H5), 73.2 (triazoloquinox. C1), 114.6 (triazoloquinox. C9), 115 (triazoloquinox. C7), 125 (triazoloquinox. C6), 126.7 (benz. C1), 128.0, 128.1, 129.0, 129.1, 129.7, 129.9, 130.1, 132.4, 133.2, 133.8, 135.7, 136.5, 153.6, 209.6. Anal. calcd for C22H15ClN4 (370.84): C, 71.25; H, 4.08; N, 15.11. Found: C, 71.42; H, 4.04; N, 14.86. 4.1.2.2. 4-Benzyl-1-(4-nitrophenyl)-1,2,4-triazolo[4,3-a]quinoxaline (9b). Brownish clusters of needles (0.7 g, 91.9%), m.p. 212–213 °C. IR (KBr, cm−1): 1630 (sh C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1601 (C
N); 1601 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1527, 1348 (NO2). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.64 (s, 2H, CH2–C6H5), 7.23 (t, 1H, J = 7.65 Hz, C6H5–C4–H), 7.33 (t, 2H, J = 7.65 Hz, C6H5–C3,5–H), 7.36 (d, 1H, J = 7.65 Hz, C6H5–C2–H), 7.47 (d, 1H, J = 7.65 Hz, C6H5–C6–H), 7.51–7.60 (m, 2H, triazoloquinox. C7,8–H), 7.64 (dd, 1H, J = 7.8, 1.2 Hz, triazoloquinox. C6–H), 8.1 (dd, 1H, J = 8.6, 1.5 Hz triazoloquinox. C9–H), 8.12 (d, 2H, J = 8.7 Hz, nitrophenyl C2,6–H), 8.51 (d, 2H, J = 8.7 Hz, nitrophenyl C3,5–H). Mass spectrum m/z (%): 381 (M+˙) (100), 380 (98), 334 (25), 233 (61), 232 (95), 205 (25), 102 (35), 91(87), 77 (25), 65 (45). Anal. calcd for C22H15N5O2 (381.39): C, 69.28; H, 3.96; N, 8.39. Found: C, 68.99; H, 4.09; N, 8.21.
C); 1527, 1348 (NO2). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.64 (s, 2H, CH2–C6H5), 7.23 (t, 1H, J = 7.65 Hz, C6H5–C4–H), 7.33 (t, 2H, J = 7.65 Hz, C6H5–C3,5–H), 7.36 (d, 1H, J = 7.65 Hz, C6H5–C2–H), 7.47 (d, 1H, J = 7.65 Hz, C6H5–C6–H), 7.51–7.60 (m, 2H, triazoloquinox. C7,8–H), 7.64 (dd, 1H, J = 7.8, 1.2 Hz, triazoloquinox. C6–H), 8.1 (dd, 1H, J = 8.6, 1.5 Hz triazoloquinox. C9–H), 8.12 (d, 2H, J = 8.7 Hz, nitrophenyl C2,6–H), 8.51 (d, 2H, J = 8.7 Hz, nitrophenyl C3,5–H). Mass spectrum m/z (%): 381 (M+˙) (100), 380 (98), 334 (25), 233 (61), 232 (95), 205 (25), 102 (35), 91(87), 77 (25), 65 (45). Anal. calcd for C22H15N5O2 (381.39): C, 69.28; H, 3.96; N, 8.39. Found: C, 68.99; H, 4.09; N, 8.21. 4.1.3.1. 5-Benzyl-2-phenyl-1H-[1,2,4]triazino[4,3-a]quinoxaline (10a). Yellow clusters of needles (0.51 g, 72.9%), m.p. 239–241 °C (ethanol/water). IR (KBr, cm−1): 3430 (NH); 1635 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1592 (C
N); 1592 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1546 (δNH). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.49 (s, 2H, CH2–C6H5), 5.60 (s, 2H, triazinoquinox. C1–H2), 7.27–7.08 (m, 10H, Ar–H), 8.02–8.08 (m, 2H, triazinoquinox. C8,9–H), 8.17 (dd, 1H, J = 7.8, 2 Hz, triazinoquinox. C7–H), 8.35 (d, 1H, J = 6.3 Hz, triazinoquinox. C10–H). Mass spectrum m/z (%): 351 (12), 350 (M+˙) (50), 349 (24), 247 (86), 246 (72), 219 (79), 218 (32), 116 (18), 103 (67), 102 (28), 91 (68), 77 (100), 65 (34), 51 (43). Anal. calcd for C23H18N4 (350.42): C, 78.83; H, 5.18; N, 15.99. Found: C, 78.67; H, 4.96; N, 16.15.
C); 1546 (δNH). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.49 (s, 2H, CH2–C6H5), 5.60 (s, 2H, triazinoquinox. C1–H2), 7.27–7.08 (m, 10H, Ar–H), 8.02–8.08 (m, 2H, triazinoquinox. C8,9–H), 8.17 (dd, 1H, J = 7.8, 2 Hz, triazinoquinox. C7–H), 8.35 (d, 1H, J = 6.3 Hz, triazinoquinox. C10–H). Mass spectrum m/z (%): 351 (12), 350 (M+˙) (50), 349 (24), 247 (86), 246 (72), 219 (79), 218 (32), 116 (18), 103 (67), 102 (28), 91 (68), 77 (100), 65 (34), 51 (43). Anal. calcd for C23H18N4 (350.42): C, 78.83; H, 5.18; N, 15.99. Found: C, 78.67; H, 4.96; N, 16.15. 4.1.3.2. 5-Benzyl-2-(4-chlorophenyl)-1H-[1,2,4]triazino[4,3-a]quinoxaline (10b). Orange-yellow fine needles (0.49 g, 63.7%), m.p. 244–245 °C (Ethanol). IR (KBr, cm−1): 3431 (NH); 1632 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1592 (C
N); 1592 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1541 (δNH); 830 (C–Cl). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.49 (s, 2H, CH2–C6H5), 5.58 (s, 2H, triazinoquinox. C1–H2), 7.29–7.40 (m, 5H, CH2–C6H5), 7.67–7.86 (m, 2H, triazinoquinox. C8,9–H), 7.72 (d, 2H, J = 8.9 Hz, chlorophenyl C2,6–H), 8.05 (dd, 1H, J = 8.25, 7.8 Hz triazinoquinox. C7–H), 8.19 (d, 2H, J = 8.9 Hz, chlorophenyl C3,5–H), 8.34 (d, 1H, J = 8.4 Hz, triazinoquinox. C10–H). 13C-NMR (500 MHz, DMSO-d6, δ ppm): 38.9 (CH2–C6H5), 62.3 (triazinoquinox. C1), 114.2 (triazinoquinox. C10), 117.8 (triazinoquinox. C8), 122.9 (triazinoquinox. C7), 125.2 (phenyl C4), 127.1 (chlorophenyl C4), 127.8 (phenyl C3,5), 128.5 (chlorophenyl C3,5), 129.0 (phenyl C2,6), 129.3 (chlorophenyl C2,6), 130.4, 132.5, 134.8, 136.2, 137.1, 138.2, 138.9, 147.0, 164.5, 172.5, 177.5. Anal. calcd for C23H17ClN4 (384.87): C, 71.78; H, 4.45; N, 14.56. Found: C, 72.06; H, 4.32; N, 14.68.
C); 1541 (δNH); 830 (C–Cl). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.49 (s, 2H, CH2–C6H5), 5.58 (s, 2H, triazinoquinox. C1–H2), 7.29–7.40 (m, 5H, CH2–C6H5), 7.67–7.86 (m, 2H, triazinoquinox. C8,9–H), 7.72 (d, 2H, J = 8.9 Hz, chlorophenyl C2,6–H), 8.05 (dd, 1H, J = 8.25, 7.8 Hz triazinoquinox. C7–H), 8.19 (d, 2H, J = 8.9 Hz, chlorophenyl C3,5–H), 8.34 (d, 1H, J = 8.4 Hz, triazinoquinox. C10–H). 13C-NMR (500 MHz, DMSO-d6, δ ppm): 38.9 (CH2–C6H5), 62.3 (triazinoquinox. C1), 114.2 (triazinoquinox. C10), 117.8 (triazinoquinox. C8), 122.9 (triazinoquinox. C7), 125.2 (phenyl C4), 127.1 (chlorophenyl C4), 127.8 (phenyl C3,5), 128.5 (chlorophenyl C3,5), 129.0 (phenyl C2,6), 129.3 (chlorophenyl C2,6), 130.4, 132.5, 134.8, 136.2, 137.1, 138.2, 138.9, 147.0, 164.5, 172.5, 177.5. Anal. calcd for C23H17ClN4 (384.87): C, 71.78; H, 4.45; N, 14.56. Found: C, 72.06; H, 4.32; N, 14.68. 4.1.3.3. 5-Benzyl-2-(4-bromophenyl)-1H-[1,2,4]triazino[4,3-a]quinoxaline (10c). Yellowish crystals (0.58 g, 67.6%), m.p. 248–249 °C (Ethanol). IR (KBr, cm−1): 3430 (NH); 1631 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1587 (C
N); 1587 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1539 (δNH); 762 (C–Br). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.49 (s, 2H, CH2–C6H5), 5.57 (s, 2H, triazinoquinox. C1–H2), 7.31–7.42 (m, 5H, CH2–C6H5), 7.77–8.05 (m, 2H, triazinoquinox. C8,9–H), 7.86 (d, 2H, J = 8.3 Hz, bromophenyl C2,6–H), 8.08–8.14 (m, 1H, triazinoquinox. C7–H), 8.10 (d, 2H, J = 8.3 Hz, bromophenyl C3,5–H), 8.33 (d, 1H, J = 8.4 Hz, triazinoquinox. C10–H). Anal. calcd for C23H17BrN4 (429.32): C, 64.35; H, 3.99; N, 13.05. Found: C, 64.17; H, 3.72; N, 13.26.
C); 1539 (δNH); 762 (C–Br). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.49 (s, 2H, CH2–C6H5), 5.57 (s, 2H, triazinoquinox. C1–H2), 7.31–7.42 (m, 5H, CH2–C6H5), 7.77–8.05 (m, 2H, triazinoquinox. C8,9–H), 7.86 (d, 2H, J = 8.3 Hz, bromophenyl C2,6–H), 8.08–8.14 (m, 1H, triazinoquinox. C7–H), 8.10 (d, 2H, J = 8.3 Hz, bromophenyl C3,5–H), 8.33 (d, 1H, J = 8.4 Hz, triazinoquinox. C10–H). Anal. calcd for C23H17BrN4 (429.32): C, 64.35; H, 3.99; N, 13.05. Found: C, 64.17; H, 3.72; N, 13.26. ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O ester); 1630 (C
O ester); 1630 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1599, 1510 (C
N); 1599, 1510 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1287, 1193, 1090 (C–O–C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 1.43 (t, 3H, J = 6.6 Hz, CH2CH3), 2.50 (s, 3H, N
C); 1287, 1193, 1090 (C–O–C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 1.43 (t, 3H, J = 6.6 Hz, CH2CH3), 2.50 (s, 3H, N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH3), 3.34 (s, 2H, CH2), 4.57 (q, 2H, J = 6.6 Hz, CH2CH3), 4.65 (s, 2H, CH2–C6H5), 7.22 (t, 1H, J = 7.4 Hz, C6H5–C4–H), 7.30 (t, 2H, J = 7.4 Hz, C6H5–C3,5–H), 7.47 (d, 2H, J = 7.4 Hz, C6H5–C2,6–H), 7.74–7.78 (m, 2H, quinox. C6,7–H), 8.09 (ddd, 1H, J = 7.4, 3.8, 2.1 Hz, quinox. C8–H), 8.75 (ddd, 1H, J = 7.5, 3.8, 2.1 Hz quinox. C5–H). 13C-NMR (500 MHz, δ ppm): 13.80 (2 × CH3), 40 (CH2–C6H5), 63.11(COOCH2CH3), 118.9 (quinox-C6), 124.85 (quinox-C8), 126.69 (phenyl-C4), 128.43 (phenyl-C3,5), 128.59 (quinox-C5), 129.03 (quinox-C7), 129.21 (phenyl-C2,6), 129.61 (phenyl-C1), 136.3 (quinox-C8a), 136.59 (quinox-C4a), 142.52 (quinox-C3), 144.53 (quinox-C2), 153.23 (N
C–CH3), 3.34 (s, 2H, CH2), 4.57 (q, 2H, J = 6.6 Hz, CH2CH3), 4.65 (s, 2H, CH2–C6H5), 7.22 (t, 1H, J = 7.4 Hz, C6H5–C4–H), 7.30 (t, 2H, J = 7.4 Hz, C6H5–C3,5–H), 7.47 (d, 2H, J = 7.4 Hz, C6H5–C2,6–H), 7.74–7.78 (m, 2H, quinox. C6,7–H), 8.09 (ddd, 1H, J = 7.4, 3.8, 2.1 Hz, quinox. C8–H), 8.75 (ddd, 1H, J = 7.5, 3.8, 2.1 Hz quinox. C5–H). 13C-NMR (500 MHz, δ ppm): 13.80 (2 × CH3), 40 (CH2–C6H5), 63.11(COOCH2CH3), 118.9 (quinox-C6), 124.85 (quinox-C8), 126.69 (phenyl-C4), 128.43 (phenyl-C3,5), 128.59 (quinox-C5), 129.03 (quinox-C7), 129.21 (phenyl-C2,6), 129.61 (phenyl-C1), 136.3 (quinox-C8a), 136.59 (quinox-C4a), 142.52 (quinox-C3), 144.53 (quinox-C2), 153.23 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 158.61(COOCH2CH3). Anal. calcd for C21H22N4O2 (362.17): C, 69.59; H, 6.12; N, 15.46. Found: C, 69.28; H, 6.20; N, 15.29.
C), 158.61(COOCH2CH3). Anal. calcd for C21H22N4O2 (362.17): C, 69.59; H, 6.12; N, 15.46. Found: C, 69.28; H, 6.20; N, 15.29. ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C
N, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 761 (C–Cl). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.75 (s, 2H, CH2–C6H5), 5.94 (s, 1H, pyrazole C4–H), 6.13 (s, 2H, NH2, D2O exchangeable), 7.06–7.17 (m, 5H, C6H5), 7.48 (d, 2H, J = 8.1 Hz, chlorophenyl C2,6–H), 7.80–7.88 (m, 3H, quinox. C5,6,7–H), 7.81 (d, 2H, J = 8.1 Hz, chlorophenyl C3,5–H), 8.11 (dd, 1H, J = 8.4, 3.6 Hz, quinox. C8–H). Mass spectrum m/z (%): 412 (36), 411 (M+˙) (100), 334 (10), 320 (15), 273 (50), 227 (19), 218 (21), 197 (15), 138 (15), 116 (25), 102 (16), 91(44). Anal. calcd for C24H18ClN5 (411.89): C, 69.99; H, 4.40; N, 17.00. Found: C, 70.24; H, 4.38; N, 17.16.
C); 761 (C–Cl). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.75 (s, 2H, CH2–C6H5), 5.94 (s, 1H, pyrazole C4–H), 6.13 (s, 2H, NH2, D2O exchangeable), 7.06–7.17 (m, 5H, C6H5), 7.48 (d, 2H, J = 8.1 Hz, chlorophenyl C2,6–H), 7.80–7.88 (m, 3H, quinox. C5,6,7–H), 7.81 (d, 2H, J = 8.1 Hz, chlorophenyl C3,5–H), 8.11 (dd, 1H, J = 8.4, 3.6 Hz, quinox. C8–H). Mass spectrum m/z (%): 412 (36), 411 (M+˙) (100), 334 (10), 320 (15), 273 (50), 227 (19), 218 (21), 197 (15), 138 (15), 116 (25), 102 (16), 91(44). Anal. calcd for C24H18ClN5 (411.89): C, 69.99; H, 4.40; N, 17.00. Found: C, 70.24; H, 4.38; N, 17.16. ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1539 (C
N); 1539 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.60 (s, 2H, CH2–C6H5), 7.60 (t, 2H, J = 7.35 Hz, C6H5–C3,5–H), 7.78 (ddd, 2H, J = 7.35, 7.3, 1.5 Hz, C6H5–C2,6–H), 7.95 (t, 1H, J = 7.35 Hz, C6H5–C4–H), 8.07–8.10 (m, 2H, triazoloquinox. C7,8–H), 8.17 (dd, 1H, J = 8.4, 1.5 Hz, triazoloquinox. C6–H), 8.54 (dd, 1H, J = 8.4, 1.5 Hz triazoloquinox. C9–H), 10.27 (s, 1H, triazoloquinox. C1–H). Anal. calcd for C16H12N4 (260.30): C, 73.83; H, 4.65; N, 21.52. Found: C, 73.80; H, 4.59; N, 21.67.
C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 4.60 (s, 2H, CH2–C6H5), 7.60 (t, 2H, J = 7.35 Hz, C6H5–C3,5–H), 7.78 (ddd, 2H, J = 7.35, 7.3, 1.5 Hz, C6H5–C2,6–H), 7.95 (t, 1H, J = 7.35 Hz, C6H5–C4–H), 8.07–8.10 (m, 2H, triazoloquinox. C7,8–H), 8.17 (dd, 1H, J = 8.4, 1.5 Hz, triazoloquinox. C6–H), 8.54 (dd, 1H, J = 8.4, 1.5 Hz triazoloquinox. C9–H), 10.27 (s, 1H, triazoloquinox. C1–H). Anal. calcd for C16H12N4 (260.30): C, 73.83; H, 4.65; N, 21.52. Found: C, 73.80; H, 4.59; N, 21.67. ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1658 (C
O); 1658 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1629 (C
N); 1629 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1450, 1372 (C–N lactam). Anal. calcd for C16H11N4O, (275.29): C, 69.81; H, 4.03; N, 20.35. Found: C, 69.63; H, 4.16; N, 20.09.
C); 1450, 1372 (C–N lactam). Anal. calcd for C16H11N4O, (275.29): C, 69.81; H, 4.03; N, 20.35. Found: C, 69.63; H, 4.16; N, 20.09. ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1665 (C
O); 1665 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 1.05 (t, 2H, J = 6.6 Hz, CH2, 3-propanoic), 3.38 (t, 2H, J = 6.6 Hz, CH2, 2-propanoic), 4.41, 4.51 (2d, 2H, J = 14 Hz, CH2–C6H5), 7.35–7.42 (m, 3H, C6H5–C2,4,6–H), 7.44 (t, 1H, J = 7.2 Hz, triazoloquinox. C8–H), 7.57 (t, 2H, J = 7.8 Hz, C6H5–C3,5–H), 7.66 (ddd, 1H, J = 10, 7.4, 1.5 Hz triazoloquinox. C7–H), 7.83 (dd, 1H, J = 8.1, 1.2 Hz, triazoloquinox. C6–H), 7.97 (d, 1H, J = 7.2 Hz, triazoloquinox. C9–H), 12.87 (s, 1H, OH, D2O exchangeable). Anal. calcd for C19H16N4O2 (332.36): C, 68.66; H, 4.85; N, 16.86. Found: C, 68.50; H, 4.93; N, 17.04.
N). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 1.05 (t, 2H, J = 6.6 Hz, CH2, 3-propanoic), 3.38 (t, 2H, J = 6.6 Hz, CH2, 2-propanoic), 4.41, 4.51 (2d, 2H, J = 14 Hz, CH2–C6H5), 7.35–7.42 (m, 3H, C6H5–C2,4,6–H), 7.44 (t, 1H, J = 7.2 Hz, triazoloquinox. C8–H), 7.57 (t, 2H, J = 7.8 Hz, C6H5–C3,5–H), 7.66 (ddd, 1H, J = 10, 7.4, 1.5 Hz triazoloquinox. C7–H), 7.83 (dd, 1H, J = 8.1, 1.2 Hz, triazoloquinox. C6–H), 7.97 (d, 1H, J = 7.2 Hz, triazoloquinox. C9–H), 12.87 (s, 1H, OH, D2O exchangeable). Anal. calcd for C19H16N4O2 (332.36): C, 68.66; H, 4.85; N, 16.86. Found: C, 68.50; H, 4.93; N, 17.04. ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1689 (C
O); 1689 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1521 (C
N); 1521 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 5.60 (s, 2H, CH2–C6H5), 7.37–8.10 (m, 11H, Ar–H, triazoloquinox. C7,8–H), 8.08 (dd, 1H, J = 6.6, 5.1 Hz, triazoloquinox. C6–H), 8.14 (ddd, 1H, J = 9.6, 6.9, 1.8 Hz, triazoloquinox. C9–H), 10.74 (s, 1H, OH, D2O exchangeable). Mass spectrum m/z (%): 382 (M+˙ + 2), 380 (M+˙) (5), 247 (20), 235 (10), 219 (13), 206 (31), 205 (100), 102 (35), 90 (10), 77 (47). Anal. calcd for C23H16N4O2 (380.41): C, 72.62; H, 4.24; N, 14.73. Found: C, 72.47; H, 4.09; N, 14.61.
C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 5.60 (s, 2H, CH2–C6H5), 7.37–8.10 (m, 11H, Ar–H, triazoloquinox. C7,8–H), 8.08 (dd, 1H, J = 6.6, 5.1 Hz, triazoloquinox. C6–H), 8.14 (ddd, 1H, J = 9.6, 6.9, 1.8 Hz, triazoloquinox. C9–H), 10.74 (s, 1H, OH, D2O exchangeable). Mass spectrum m/z (%): 382 (M+˙ + 2), 380 (M+˙) (5), 247 (20), 235 (10), 219 (13), 206 (31), 205 (100), 102 (35), 90 (10), 77 (47). Anal. calcd for C23H16N4O2 (380.41): C, 72.62; H, 4.24; N, 14.73. Found: C, 72.47; H, 4.09; N, 14.61. ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O ester); 1599, 1510 (C
O ester); 1599, 1510 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N,C
N,C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1278, 1193, 1091 (C–O–C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 1.42 (t, 3H, J = 7.4 Hz, CH2CH3), 4.58 (q, 2H, J = 7.4 Hz, CH2CH3), 4.65 (s, 2H, CH2–C6H5), 7.22 (t, 1H, J = 7.33 Hz, C6H5–C4–H), 7.30 (t, 2H, J = 7.33 Hz, C6H5–C3,5–H), 7.46 (d, 2H, J = 7.33 Hz, C6H5–C2,6–H), 7.74–7.79 (m, 2H, triazoloquinox. C7,8–H), 8.10 (ddd, 1H, J = 7.5, 3.8, 2.4 Hz, triazoloquinox. C6–H), 8.76 (ddd, 1H, J = 7.5, 3.8, 2.4 Hz triazoloquinox. C9–H). Anal. calcd for C19H16N4O2 (332.36): C, 68.66; H, 4.85; N, 16.86. Found: C, 68.82; H, 4.68; N, 16.63.
C); 1278, 1193, 1091 (C–O–C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 1.42 (t, 3H, J = 7.4 Hz, CH2CH3), 4.58 (q, 2H, J = 7.4 Hz, CH2CH3), 4.65 (s, 2H, CH2–C6H5), 7.22 (t, 1H, J = 7.33 Hz, C6H5–C4–H), 7.30 (t, 2H, J = 7.33 Hz, C6H5–C3,5–H), 7.46 (d, 2H, J = 7.33 Hz, C6H5–C2,6–H), 7.74–7.79 (m, 2H, triazoloquinox. C7,8–H), 8.10 (ddd, 1H, J = 7.5, 3.8, 2.4 Hz, triazoloquinox. C6–H), 8.76 (ddd, 1H, J = 7.5, 3.8, 2.4 Hz triazoloquinox. C9–H). Anal. calcd for C19H16N4O2 (332.36): C, 68.66; H, 4.85; N, 16.86. Found: C, 68.82; H, 4.68; N, 16.63. ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O ester); 1630 (C
O ester); 1630 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1606, 1516, 1489 (C
N); 1606, 1516, 1489 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1241, 1071 (C–O–C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 1.44 (t, 3H, J = 7.4 Hz, CH2CH3), 4.53 (s, 2H, CH2–C6H5), 5.12 (q, 2H, J = 7.4 Hz, CH2CH3), 5.83 (s, 2H, CH2–CO), 7.11 (t, 1H, J = 7.33 Hz, C6H5–C4–H), 7.17 (t, 2H, J = 7.33 Hz, C6H5–C3,5–H), 7.42 (d, 2H, J = 7.33 Hz, C6H5–C2,6–H), 7.45–7.63 (m, 2H, triazoloquinox. C7,8–H), 8.04 (d, 1H, J = 7.5 Hz, triazoloquinox. C6–H), 8.18 (d, 1H, J = 7.5 Hz triazoloquinox. C9–H). Anal. calcd for C20H18N4O2 (346.39): C, 69.35; H, 5.24; N, 16.17. Found: C, 69.42; H, 5.28; N, 16.02.
C); 1241, 1071 (C–O–C). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 1.44 (t, 3H, J = 7.4 Hz, CH2CH3), 4.53 (s, 2H, CH2–C6H5), 5.12 (q, 2H, J = 7.4 Hz, CH2CH3), 5.83 (s, 2H, CH2–CO), 7.11 (t, 1H, J = 7.33 Hz, C6H5–C4–H), 7.17 (t, 2H, J = 7.33 Hz, C6H5–C3,5–H), 7.42 (d, 2H, J = 7.33 Hz, C6H5–C2,6–H), 7.45–7.63 (m, 2H, triazoloquinox. C7,8–H), 8.04 (d, 1H, J = 7.5 Hz, triazoloquinox. C6–H), 8.18 (d, 1H, J = 7.5 Hz triazoloquinox. C9–H). Anal. calcd for C20H18N4O2 (346.39): C, 69.35; H, 5.24; N, 16.17. Found: C, 69.42; H, 5.28; N, 16.02. 4.2. Biological evaluation methodology
4.2.2.1. Inhibition-zone measurements. All synthesized compounds were evaluated for their antimicrobial activity by the agar cup diffusion technique using a 1 mg mL−1 solution in DMSO.26 The test organisms were Staphylococcus aureus (DSM 1104) and Bacillus subtilis (ATCC 6633) as Gram-positive bacteria; Escherichia coli (ATCC 11775) and Pseudomonas aeruginosa (ATCC 10145) as Gram-negative bacteria. Candida albicans (DSM 70014) was also used as a representative for fungi. Each 100 mL of sterile molten agar (at 45 °C) received 1 mL of 6 h-broth culture and then the seeded agar was poured into sterile Petri dishes. Cups (8 mm in diameter) were cut in the agar. Each cup received 0.1 mL of the 1 mg mL−1 solution of the test compounds. The plates were then incubated at 37 °C for 24 h or, in case of C. albicans, for 48 h. A control using DMSO without the test compound was included for each organism. Ampicillin was used as the standard antibacterial, while clotrimazole was used as the antifungal reference. The resulting inhibition zones are recorded (Table 4).
4.2.2.2. Minimal inhibitory concentration (MIC) measurement. The minimal inhibitory concentrations (MIC) of the most active compounds were measured using the twofold serial broth dilution method.27 The test organisms were grown in their suitable broth: 24 h for bacteria and 48 h for fungi at 37 °C. Two fold serial dilutions of solutions of the test compounds were prepared using 200, 100, 50, 25, and 12.5 μg mL−1. The tubes were then inoculated with the test organisms; each 5 mL received 0.1 mL of the above inoculum and were incubated at 37 °C for 48 h. Then, the tubes were observed for the presence or absence of microbial growth. The MIC values of the prepared compounds are listed in Table 5.
4.2.2.3. Minimal bactericidal concentration (MBC) measurement. MIC tests were always extended to measure the MBC as follows: A loop-full from the tube not showing visible growth (MIC) was spread over a quarter of Müller–Hinton agar plate. After 18 h of incubation, the plates were examined for growth. Again, the tube containing the lowest concentration of the test compound that failed to yield growth on subculture plates was judged to contain the MBC of that compound for the respective test organism (Table 5).
Acknowledgements
The authors are grateful to the staff of the department of Health and Human Services, National Cancer Institute, Bethesda, Maryland, USA for carrying out the anticancer screening of the selected synthesized compounds.Notes and references
- Y. B. Lee, Y. D. Gong, D. J. Kim, C. H. Ahn, J. Y. Kong and N. S. Kang, Bioorg. Med. Chem., 2012, 20, 1303–1309 CrossRef CAS PubMed.
- J. B. Chairs, Curr. Opin. Struct. Biol., 1998, 8, 314–320 CrossRef.
- H. Gao, E. F. Yamasaki, K. K. Chan, L. L. Shen and R. M. Snapka, Mol. Pharm., 2003, 63, 1382–1388 CrossRef CAS PubMed.
- Z. Ding, J. Y. Zhou, W. Z. Wei, V. V. Baker and G. S. Wu, Oncogene, 2002, 21, 4530–4538 CrossRef CAS PubMed.
- B. Zarranz, A. Jaso, I. Aldana and A. Monge, Bioorg. Med. Chem., 2004, 12, 3711–3721 CrossRef CAS PubMed.
- P. Corona, A. Carta, M. Loriga, G. Vitale and G. Paglietti, Eur. J. Med. Chem., 2009, 44, 1579–1591 CrossRef CAS PubMed.
- P. Corona, G. Vitale, M. Loriga, G. Paglietti, P. L. Colla, G. Collu, G. Sanna and R. Loddo, Eur. J. Med. Chem., 2006, 41, 1102–1107 CrossRef CAS PubMed.
- S. Piras, M. Loriga, A. Carta, G. Paglietti, M. P. Costi and S. Ferrari, J. Heterocycl. Chem., 2006, 43, 541–548 CrossRef CAS.
- S. A. Galal, A. S. Abdelsamie, S. M. Soliman, J. Mortier, G. Wolber, M. M. Ali, H. Tokuda, N. Suzuki, A. Lida, R. A. Ramadan and H. I. El Diwani, Eur. J. Med. Chem., 2013, 69, 115–124 CrossRef CAS PubMed.
- N. S. Habib and S. A. El-Hawash, Pharmazie, 1997, 52, 594–598 CAS.
- S. A. El-Hawash, N. S. Habib and N. H. Fanaki, Pharmazie, 1999, 54, 808–813 CAS.
- N. S. Habib and S. A. El-Hawash, Boll. Chim. Farm., 2005, 144, 1–16 Search PubMed.
- S. A. El-Hawash, N. S. Habib and M. A. Kassem, Arch. Pharm. Chem. Life Sci., 2006, 339, 564–571 CrossRef CAS PubMed.
- S. A. M. El-Hawash and A. E. Abdel-Wahab, Arch. Pharm. Chem. Life Sci., 2006, 339, 437–447 CrossRef CAS PubMed.
- A. M. El-Massry, Heterocyclic Communications, 1999, 5, 555–563 CrossRef CAS.
- X. M. Wang, J. Xu, Y. P. Li, H. Li, C. S. Jiang, G. D. Yang, S. M. Lu and S. Q. Zhang, Eur. J. Med. Chem., 2013, 67, 243–251 CrossRef CAS PubMed.
- Y. Luo, S. Zhang, Z. J. Liu, W. Chen, J. Fu, Q. F. Zeng and H. L. Zhu, Eur. J. Med. Chem., 2013, 64, 54–61 CrossRef CAS PubMed.
- R. M. Herbst and D. Shemin, Org. Synth., 1939, 19, 4 CrossRef CAS.
- R. M. Herbst and D. Shemin, Org. Synth., 1939, 19, 1 CrossRef CAS.
- M. Moffitt and H. P. Schultz, J. Org. Chem., 1977, 42, 2504–2505 CrossRef CAS.
- E. A. M. Badawey, J. Heterocycl. Chem., 1996, 33, 229–233 CrossRef CAS.
- S. N. Khattab, S. Y. Hassan, A. A. Bekhit, A. M. E. Massry, V. Langer and A. Amer, Eur. J. Med. Chem., 2010, 45, 4479–4489 CrossRef CAS PubMed.
- M. R. Boyd and K. D. Paull, Drug Dev. Res., 1995, 34, 91–109 CrossRef CAS.
- A. Monks, D. Scudiero, P. Skehan, R. Shoemaker, K. Paull, D. Vistica, C. Hose, J. Langley, P. Cronise, A. Vaigro-Wolff, M. Gray-Goodrich, H. Campbell and M. Boyd, J. Natl. Cancer Inst., 1991, 83, 757–766 CrossRef CAS PubMed.
- O. S. Weislow, R. Kiser, D. L. Fine, J. Bader, R. H. Shoemaker and M. R. Boyd, J. Natl. Cancer Inst., 1989, 81, 577–586 CrossRef CAS PubMed.
- S. R. Jain and A. Kar, Planta Med., 1971, 20, 118–123 CrossRef CAS PubMed.
- A. C. Scott, in Mackie and McCartney Practical Medical Microbiology, ed. J. G. Collee, J. P. Duguid, A. G. Fraser and B. P. Marmion, Churchill Livingstone, New York, USA, 13th edn, 1989, vol. 2, pp. 161–181 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4md00257a |
| This journal is © The Royal Society of Chemistry 2015 |