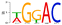Decomposition of RNA methylome reveals co-methylation patterns induced by latent enzymatic regulators of the epitranscriptome†
Lian
Liu
a,
Shao-Wu
Zhang
*a,
Yu-Chen
Zhang
a,
Hui
Liu
b,
Lin
Zhang
b,
Runsheng
Chen
ac,
Yufei
Huang
d and
Jia
Meng
*e
aKey Laboratory of Information Fusion Technology of Ministry of Education, School of Automation, Northwestern Polytechnical University, Xi'an, Shaanxi 710027, China. E-mail: zhangsw@nwpu.edu.cn
bSchool of Information and Electrical Engineering, China University of Mining and Technology, Xuzhou, 221116, China
cInstitute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China
dDepartment of Electrical and Computer Engineering, University of Texas at San Antonio, TX 78249, USA
eDepartment of Biological Sciences, Xi'an Jiaotong-Liverpool University, Suzhou, Jiangsu 215123, China. E-mail: jia.meng@xjtlu.edu.cn
First published on 29th October 2014
Abstract
Biochemical modifications to mRNA, especially N6-methyladenosine (m6A) and 5-methylcytosine (m5C), have been recently shown to be associated with crucial biological functions. Despite the intriguing advancements, little is known so far about the dynamic landscape of RNA methylome across different cell types and how the epitranscriptome is regulated at the system level by enzymes, i.e., RNA methyltransferases and demethylases. To investigate this issue, a meta-analysis of m6A MeRIP-Seq datasets collected from 10 different experimental conditions (cell type/tissue or treatment) is performed, and the combinatorial epitranscriptome, which consists of 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 758 m6A sites, is extracted and divided into 3 clusters, in which the methylation sites are likely to be hyper- or hypo-methylated simultaneously (or co-methylated), indicating the sharing of a common methylation regulator. Four different clustering approaches are used, including K-means, hierarchical clustering (HC), Bayesian factor regression model (BFRM) and nonnegative matrix factorization (NMF) to unveil the co-methylation patterns. To validate whether the patterns are corresponding to enzymatic regulators, i.e., RNA methyltransferases or demethylases, the target sites of a known m6A regulator, fat mass and obesity-associated protein (FTO), are identified from an independent mouse MeRIP-Seq dataset and lifted to human. Our study shows that 3 out of the 4 clustering approaches used can successfully identify a group of methylation sites overlapping with FTO target sites at a significance level of 0.05 (after multiple hypothesis adjustment), among which, the result of NMF is the most significant (p-value 2.81 × 10−06). We defined a new approach evaluating the consistency between two clustering results which shows that clustering results of different methods are highly correlated strongly indicating the existence of co-methylation patterns. Consistent with recent studies, a number of cancer and neuronal disease-related bimolecular functions are enriched in the identified clusters, which are biological functions that can be regulated at the epitranscriptional level, indicating the pharmaceutical prospect of RNA N6-methyladenosine-related studies. This result successfully reveals the linkage between the global RNA co-methylation patterns embedded in the epitranscriptomic data under multiple experimental conditions and the latent enzymatic regulators, suggesting a promising direction towards a more comprehensive understanding of the epitranscriptome.
758 m6A sites, is extracted and divided into 3 clusters, in which the methylation sites are likely to be hyper- or hypo-methylated simultaneously (or co-methylated), indicating the sharing of a common methylation regulator. Four different clustering approaches are used, including K-means, hierarchical clustering (HC), Bayesian factor regression model (BFRM) and nonnegative matrix factorization (NMF) to unveil the co-methylation patterns. To validate whether the patterns are corresponding to enzymatic regulators, i.e., RNA methyltransferases or demethylases, the target sites of a known m6A regulator, fat mass and obesity-associated protein (FTO), are identified from an independent mouse MeRIP-Seq dataset and lifted to human. Our study shows that 3 out of the 4 clustering approaches used can successfully identify a group of methylation sites overlapping with FTO target sites at a significance level of 0.05 (after multiple hypothesis adjustment), among which, the result of NMF is the most significant (p-value 2.81 × 10−06). We defined a new approach evaluating the consistency between two clustering results which shows that clustering results of different methods are highly correlated strongly indicating the existence of co-methylation patterns. Consistent with recent studies, a number of cancer and neuronal disease-related bimolecular functions are enriched in the identified clusters, which are biological functions that can be regulated at the epitranscriptional level, indicating the pharmaceutical prospect of RNA N6-methyladenosine-related studies. This result successfully reveals the linkage between the global RNA co-methylation patterns embedded in the epitranscriptomic data under multiple experimental conditions and the latent enzymatic regulators, suggesting a promising direction towards a more comprehensive understanding of the epitranscriptome.
Introduction
The dynamic chemical modifications of DNA together with their functions have been well established through intensive studies ranging from simple model organisms to humans in the past decade.1–3 Compared with DNA modifications, RNA modifications are largely neglected, and have yet drawn extensive attention until very recently.4–7 The presence of chemical modifications to RNA was established as early as the 1970s.8–10 Until today, people have identified over 100 post-transcriptional RNA modifications in all 3 kingdoms of life;11 however, not much is known so far about their biological, physiological and pathological functions due to technical limitations. Recently, a new technique (differently named as “MeRIP-Seq”12 or “m6A-Seq”13) proposed in 2012 by two independent groups, for the first time, enabled the global unbiased profiling of mRNA N6-methyladenosine methylome, one component of the epitranscriptome layer of gene regulation.14 Currently, the only practically feasible unbiased approach for measuring mRNA m6A is the MeRIP-Seq technique.12,13,15 MeRIP-Seq pulls down and sequences the methylated mRNA fragments with anti-m6A antibody, and an input control sample is also generated to measure the basal abundance of all genes (see Fig. 1).Similar to ChIP-Seq data analysis,16–18 since MeRIP-Seq cannot provide base-resolution, the detection of RNA methylation sites from MeRIP-Seq has been mainly formulated as the “peak detection” problem;19,20 however, as pointed out previously, a single RNA methylation site may be split into 2 sections due to the existence of introns, the peak calling of MeRIP-Seq data should ideally be splicing-aware.14,19 Nevertheless, the MeRIP-Seq technique successfully combines the essence of methylated DNA immunoprecipitation sequencing (MeDIP-Seq) and RNA sequencing (RNA-Seq) for high-resolution detection of transcriptome-wide RNA methylation modifications. Within 2 years' time, the technique has been applied to a number of important studies in humans, mouse, yeast, etc.21–25 Meanwhile, some RNA (de)methylation enzymes are identified.24,26–28 These studies together greatly enhanced our understanding of the reversible modifications to mRNA.5,29
However, one question remains to be answered is how the epitranscriptome, which consists of tens of thousands of RNA methylation sites, is regulated at the system level across multiple conditions by RNA methyltransferases and demethylases. While it is important to cumulate additional knowledge for the function of a specific RNA methylation enzyme under a particular condition to obtain all pieces of a Jigsaw puzzle, it is also necessary to integrate what we have so far for a big picture and untangle the high dimensional RNA methylome of tens of thousands of RNA methylation sites to shape an interpretable picture. The RNA methylome embraces a number of features that make a system level computational analysis necessary and feasible:
• The RNA m6A methylome consists of a large number of RNA methylation sites (ranging from 9124 to 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 293 m6A sites predicted under different conditions14). It has been even speculated that, every RNA molecule may be methylated under a specific condition. Conceivably, the dimension reduction technique is necessary to make sense of high-dimensional information. The RNA residuals are methylated or demethylated by a relatively small number of regulators (RNA methyltransferases or demethylases), each of which regulates thousands of RNA methylation sites. In this sense, high dimensional RNA methylation data can be conveniently projected into lower dimensional space of RNA methylation regulators to reveal directly the biochemical causes of the observed phenomenon.
293 m6A sites predicted under different conditions14). It has been even speculated that, every RNA molecule may be methylated under a specific condition. Conceivably, the dimension reduction technique is necessary to make sense of high-dimensional information. The RNA residuals are methylated or demethylated by a relatively small number of regulators (RNA methyltransferases or demethylases), each of which regulates thousands of RNA methylation sites. In this sense, high dimensional RNA methylation data can be conveniently projected into lower dimensional space of RNA methylation regulators to reveal directly the biochemical causes of the observed phenomenon.
• RNA methylation is non-stoichiometric, i.e., a specific residual can be methylated only on a fraction of transcripts, not necessarily all or none. This process is influenced by the “methylation potential”,30 which is the ratio of S-adenosylmethionine (SAM, the universal methyl donor cosubstrate) and S-adnosylhomocysteine (SAH, the by-product of SAM that acts as a competitive inhibitor). With the simplest approximation of an equilibrium condition, the ratio between methylated and unmodified residuals is directly proportional to the SAM/SAH ratio, and is independent of the absolute RNA abundance. The fact that the same nucleic acids are not methylated at the same level indicates the specificity of the enzymes, which is more complicatedly determined by the methylation complex.
• The RNA co-methylation pattern exists due to enzymatic regulators. Consistent with the “SAM/SAH” ratio, the group of RNA methylation sites controlled by the same RNA methylation factor will show hyper- or hypo-methylation simultaneously. This is analogous to the transcription factor (TF) network or the microRNA (miRNA) network, where the regulated target genes show a co-expression pattern consistent with their regulator. Conceivably, the hyper-methylation of a large number of RNA methylation sites may indicate the increase of methyltransferases or the decreases of demethylase. The co-regulation pattern is the key for the identification of latent regulatory factors, which may function at the protein level and cannot be directly observable from the high-throughput RNA methylation sequencing data. Due to the activities of RNA methylation enzymes and their specificity, the methylation levels of the large number of RNA methylation sites are not independent of each other but show some clustering effect. On one hand, it is likely that a single methylation factor may regulate a large number of RNA methylation sites simultaneously; on the other hand, if we consider a single RNA methylation enzyme can be a protein complex consisting of the protein products of several genes, then it is possible that the methylation status of a single site is determined by multiple proteins. Although the real regulatory relationship between RNA methylation sites and enzymes can be more complex, it is practically more convenient to start with simpler computation methods, such as K-means, and gradually increases the complexity of tools.
• RNA co-methylation patterns are likely to have specific biological functions. From an evolutionary point of view, to maximize the functionality of RNA methylation as a means of regulation, rather than aimlessly targeting a number of unrelated genes by random, natural selection should favor that an RNA methylation regulator targets functional-related genes to control specific functions so as to add the adaptability of the liver organism. It should be reasonable to assume that the targets of the same RNA methylation enzyme are likely to share functions in common; and on the other hand, for the purpose of validation, if some functions are statistically related to an RNA methylation enzyme, it is likely that identification of the enzyme and its targets is successful.
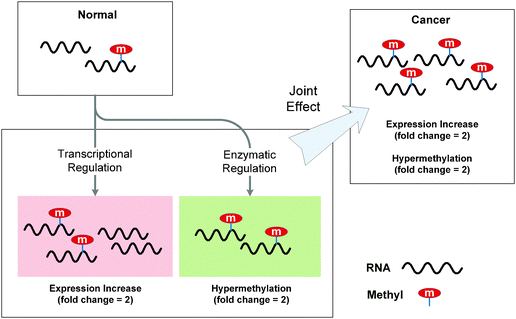
• Transcriptional regulation may indirectly affect the epitranscriptome. Although existing studies mostly focus on the changes in the absolute amount of methylation with the basal RNA expression levels ignored, it is important to notice whether the increase is triggered by transcriptional regulation or by the enzymatic regulation (RNA methyltransferases or demethylases). While the RNA “methylation potential” moderates the ratio of methylated and unmodified molecules, transcriptional regulation directly controls the absolute amount of RNA transcripts with the ratio unchanged. In practice, the changes in the absolute amount of RNA methylation can be due to a joint effect of the two (see Fig. 2).
In this study, the combinatorial RNA m6A methylome from 10 experimental conditions (different tissues, cell lines or treatments) is firstly extracted using an R package we developed to study its dynamics. Four different clustering approaches, representing different rationales, are applied to dissect the RNA methylation sites. The results confirm the existence of co-methylation patterns and their relationship with RNA methylation enzymes.
Materials and method
Multiple N6-methyladenosine MeRIP-Seq datasets from different conditions are collected for this analysis. Samples include HEK293, HepG2, U2OS cell lines and brain tissue with different treatments, some with more than 1 biological replicate. The raw data in FASTQ format (SRA) were obtained directly from Gene Expression Omnibus (GEO), and then aligned to the human reference genome assembly (hg19) with spliced aligner Tophat231 for the following analysis. The information of these datasets is summarized in Table 1.| ID | Tissue/cell | Treatment | Other info | # of replicates | # of reads (million) | Source |
|---|---|---|---|---|---|---|
| IP & input | IP & input | |||||
| h1 | HEK293T | SYSY Ab | 2 & 3 | 145 & 217 | [12] | |
| h2 | HEK293T | NEB Ab | 1 & 3 | 33 & 217 | [12] | |
| h3 | Brain | 1 & 1 | 22 & 17 | [13] | ||
| h4 | HepG2 | 4 & 3 | 68 & 85 | [13] | ||
| h5 | HepG2 | UV | 1 & 1 | 21 & 7 | [13] | |
| h6 | HepG2 | HS | 1 & 1 | 34 & 52 | [13] | |
| h7 | HepG2 | HGF | 1 & 1 | 33 & 23 | [13] | |
| h8 | HepG2 | IFN | 1 & 1 | 47 & 27 | [13] | |
| h9 | U2OS | 3 & 3 | 86 & 83 | [21] | ||
| h10 | U2OS | DAA | 3 & 3 | 80 & 87 | [21] |
To dissect the RNA methylome with the clustering approach, the joint epitranscriptome must be firstly extracted. Currently, there is no convenient tool provided with this function. For this purpose, we developed an open source R package RNA Methylation Tool (RMT) for the processing of multiple MeRIP-Seq datasets and extracting the combinatorial epitranscriptome, i.e., all the RNA methylation sites detected under one or more conditions. The general work flow of RMT is shown in Fig. 3.
 | ||
| Fig. 3 Work flow of the RMT package. The RMT package is developed for convenient extraction of the combinatorial epitranscriptome from multiple MeRIP-Seq datasets obtained from different conditions. Specifically, it requires the input of multiple MeRIP-Seq datasets in the form of aligned BAM files, and outputs all the RNA methylation sites together with their estimated methylation levels. The RMT package is available in the ESI,† S1. | ||
We will in the next detail each step of RMT work flow.
• Extract the individual RNA methylome using the exomePeak R/Bioconductor package: the very first step of the analysis is to extract all the RNA methylation sites (or the “epitranscriptome”) from each individual MeRIP-Seq experiment. As aforementioned, there are two types of samples in MeRIP-Seq, i.e., IP and control samples. As essentially an enrichment-based approach, since the pull-down reads in the IP sample are enriched with methylated fragments, there is likely a higher number of reads (or a “peak”) appearing near the methylation sites in the IP sample compared with the input control sample, thus the methylation sites may be detected with the “peak calling” method. We previously developed the exomePeak R/Bioconductor package19 for this purpose. The exomePeak package is based on the C-test for comparison of two Poisson means32 to detect the methylation sites on RNA molecule. As a splicing-aware peak caller focus on the exons only, its effectiveness on MeRIP-Seq peak calling has been demonstrated previously.19
• Merge all detected methylation sites for a combinatorial methylome: RNA methylation is reversible and dynamic under different conditions. The RNA methylome with tens of thousands of RNA methylation sites identified under different conditions in the previous step are further merged into a combined set. It is worth mentioning that the difference between the RNA methylome under different conditions is not only due to the context-specificity but also related to noise and the detectability of MeRIP-Seq, i.e., when the expression level of a gene is low, its methylation site can be difficult to detect. The combination of multiple RNA methylomes is conducted in the following way: (1) RNA methylation sites that do not overlap with those detected under a different condition are context-specific and unique, thus all are kept. (2) RNA methylation sites that overlap with those detected under a different condition are not context specific and may appear multiple times. Under this scenario, only the widest methylation sites are kept.
• Quantification of the RNA methylation level: a natural way to quantify the RNA methylation level (percentage of methylated RNA molecules) based on MeRIP-Seq data is the “IP/Input ratio”, which is defined as the ratio of the number of reads in IP and input control sample after compensating for the sequencing depth (or total number of reads). However, infinite “IP/Input ratio” might be generated when there are no reads detected in the input sample, which is not rare. Here, we adopt the way of computing gene expression in RNA-Seq with “RPKM” for a specific methylation site. The RNA methylation level is then quantified using:
 | (1) |
After extracting the combinatorial RNA methylome and the methylation levels, feature selection was conducted to select the most informatics features (RNA methylation sites) for clustering purpose. For each feature, the methylation level defined in (1) can be determined on every single biological replicate provided that the corresponding paired IP and Input control MeRIP-Seq sample is available. For the purpose of detecting co-methylated RNA methylation sites, of interests are those varying significantly across different conditions/replicates. For best clustering effect, it is important to exclude those with small variance in the methylation level. We will select features that have a larger variance in the methylation level across different conditions, then the methylation levels are standardized prior to clustering analysis using:
 | (2) |
![[x with combining circumflex]](https://www.rsc.org/images/entities/i_char_0078_0302.gif) m,j, the co-methylation pattern embedded may be extracted. Each co-methylation pattern consists of a number of methylation sites are hyper- and hypo-methylated simultaneously, indicating a common latent regulatory factor at the epitranscriptomic level. For this purpose, 4 widely used clustering approaches are adopted, including K-means, hierarchical clustering, Bayesian factor model33 and nonnegative matrix factorization,34 each reflects a different underlying assumption. The clustering results are then passed for gene ontology analysis with the topGO R/Bioconductor package35 for functional analysis.
m,j, the co-methylation pattern embedded may be extracted. Each co-methylation pattern consists of a number of methylation sites are hyper- and hypo-methylated simultaneously, indicating a common latent regulatory factor at the epitranscriptomic level. For this purpose, 4 widely used clustering approaches are adopted, including K-means, hierarchical clustering, Bayesian factor model33 and nonnegative matrix factorization,34 each reflects a different underlying assumption. The clustering results are then passed for gene ontology analysis with the topGO R/Bioconductor package35 for functional analysis.
Result and discussion
RNA methylome of individual samples
The newly developed RMT package internally adapts the exomePeak package for peak calling on every individual IP sample. Specifically, the RNA methylation sites under each condition are shown in Table 2, the number of peaks (RNA methylation sites) under a specific condition ranges from 1200 (brain) to 21127 (HepG2). The strand-specific consensus sequences for each condition are then obtained at MEME-ChIP webserver36 by extracting all the peak regions from hg19 whole genome FASTA file provided from Illumina iGenome. Despite the large variation in the numbers of detected RNA methylation sites, the enriched consensus sequences are similar to the known RRACH motif of m6A,12,13 indicating that the determined RNA methylation sites are accurate and consistent with previous studies.Combinatorial RNA methylome
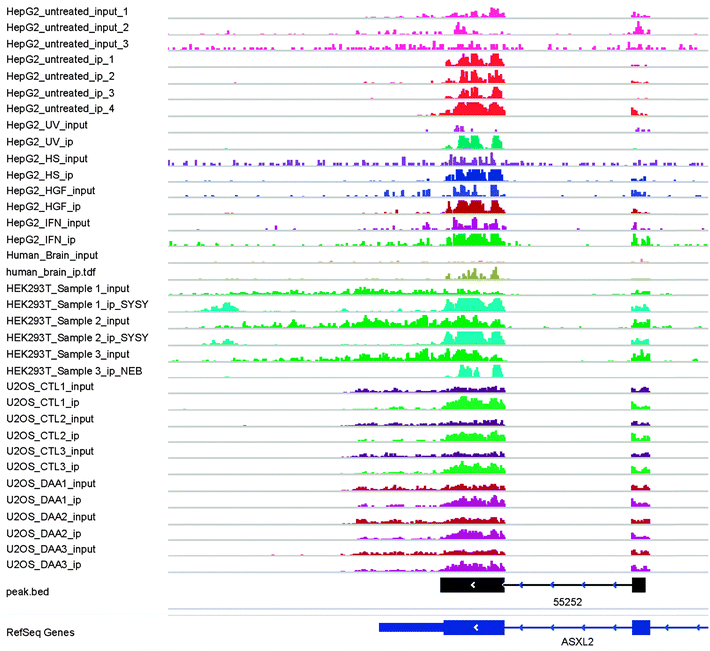
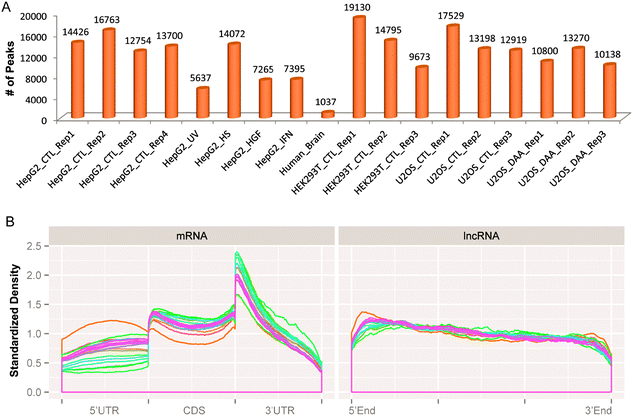
The combinatorial RNA methylome of 10 different datasets (Table 1) is then extracted by the newly developed RMT package. Thanks to the GenomicFeatures R/Bioconductor package,37 the peak merging stage takes only seconds to complete, the most time consuming step is peak calling by the exomePeak package. The 258465 RNA methylation sites detected on 18 replicates from 10 different conditions are further merged into 42758 unique RNA methylation sites using the rules previously specified (see Fig. 4 and ESI,† S2).Under individual conditions, different numbers of merged methylation sites can be observed (see Fig. 5A), and consistent with previous studies, the RNA methylation sites are more enriched near the stop codon of mRNA and on the 3′UTR region (see Fig. 5B). One interesting new observation is that the RNA methylation sites on lncRNA are consistently more enriched on the 5′end compared with the 3′end, whose cause is yet clear (see Fig. 5B).
For the merged 42758 unique RNA methylation sites, each on average appears on 1/3 (6 of 18) of biological replicates obtained under 10 different conditions. Specifically, 35.4% (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 126) appears only once, and only 33.1% (14
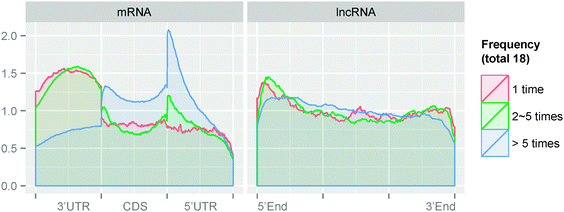
126) appears only once, and only 33.1% (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 415) appears on more than 5 biological replicates, indicating a highly dynamic landscape of RNA N6-methyladnosine. Please note that this highly specific behavior may partially be attributed to detection noise and the dynamics of transcription, i.e., given the current protocol of MeRIP-Seq, it is impossible to detect an RNA methylation site on lowly expressed genes, so dynamics of transcription will also be cumulated in MeRIP-Seq peak calling. We then compared the genomic distribution of the most frequent (appear more than 5 times) and most rare methylation sites (appear only once). As shown in Fig. 6, compared with the most frequent methylation sites, the highly specific sites are highly enriched in the 5′UTR region of mRNA. On lncRNA, however, they do not show distinct differences.
415) appears on more than 5 biological replicates, indicating a highly dynamic landscape of RNA N6-methyladnosine. Please note that this highly specific behavior may partially be attributed to detection noise and the dynamics of transcription, i.e., given the current protocol of MeRIP-Seq, it is impossible to detect an RNA methylation site on lowly expressed genes, so dynamics of transcription will also be cumulated in MeRIP-Seq peak calling. We then compared the genomic distribution of the most frequent (appear more than 5 times) and most rare methylation sites (appear only once). As shown in Fig. 6, compared with the most frequent methylation sites, the highly specific sites are highly enriched in the 5′UTR region of mRNA. On lncRNA, however, they do not show distinct differences.
After extracting the combinatorial epitranscriptome of more than 40k RNA m6A sites, their RPKM values are determined for each individual sample, and the methylation levels are calculated and standardized based on equation (1) and (2). Feature selection was conducted to select 3274 methylation sites with sample variance larger than 30. The selected methylation sites having methylation levels changing significantly across different conditions are then passed to clustering algorithm for discovery of the co-methylation patterns. Specifically, we applied four clustering algorithms to cluster the 3274 RNA methylation sites, including K-means, hierarchical clustering (HC), non-negative matrix factorization (NMF) and Bayesian factor regression model (BFRM). An important predefined parameter for clustering analysis is the optimal number of clusters, which is pre-determined with cluster silhouettes of K-means method in a model selection procedure (see Fig. 7). In our analysis, the other 3 clustering approaches adapt the same number of clusters for easy comparison purpose. It is worth mentioning that the optimal number of clusters 3 is determined with a small subset of the data available (10 different conditions), and the actual number of RNA methylation regulatory factors can be much larger with a lot more different cell types.
After applying all 4 clustering approaches to the selected 3274 unique RNA methylation sites, as can be seen from Fig. 8, the methylation sites were divided into 3 clusters. Considering the 4 clustering methods each embraces a different rationale, it is not surprising to see that there exist distinct differences among them. While the BFRM clusters are of approximately similar size, the other 3 algorithms, especially for K-means and HC, generated clusters with quite uneven sizes.
Given the aforementioned clustering results, it is important to check whether different clustering approaches capture a relatively consistent co-methylation pattern. For this purpose, we define a new method for evaluating the consistency between two clustering approaches. Given the clustering results of N elements by two methods c = [c1,c2,…,cN] and r = [r1,r2,…,rN], where ci,ri∈{1,2,…,K} represents the cluster ID of the ith element, and K represents the total number of clusters. We first convert the clustering IDs into a pair-wise resembling matrix (PRM) C and R, specifically, for an element in pair-wise resembling matrix ci,j generated from c = [c1,c2,…,cN], we define,
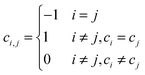 | (3) |
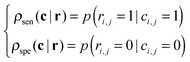 | (4) |
| ρ(c,r) = ρ(r,c) |
| = ¼[ρsen(c|r) + ρspe(c|r)] + ¼[ρsen(r|c) + ρsen(r|c)] | (5) |
| K-means | HC | NMF | BFRM | |
|---|---|---|---|---|
| K-means | NA | 0.772 | 0.805 | 0.594 |
| HC | 0.772 | NA | 0.701 | 0.574 |
| NMF | 0.805 | 0.701 | NA | 0.584 |
| BFRM | 0.594 | 0.574 | 0.584 | NA |
The high consistency between different clustering approaches indicates that co-methylation patterns are successfully captured. To find out whether the identified co-methylation patterns are corresponding to actual RNA methylation regulators, we compared them with the target genes of a known RNA m6A demethylase FTO.26 Specifically, the FTO target sites in mouse midbrain are firstly identified using the exomePeak package based on the m6A MeRIP-Seq data obtained from wild type mouse midbrain and FTO knockout condition.22 Since it has been shown previously that m6A are conserved between human and mouse, these FTO target sites are lifted to human genome hg19 assembly using the UCSC LiftOver tool.38 Not surprisingly, more than 90% of the lifted FTO target sites overlap with our combinatorial RNA methylome. We then used Fisher's exact test (FET) to compare whether the identified co-methylation patterns are significantly overlapping with the FTO target genes. Multiple hypothesis correction was also conducted to calculate the false discovery rate (FDR) using the BH method.39 As shown in Table 4, 3 out of 4 clustering approaches can successfully identify a co-methylation pattern significantly overlapping with FTO targets at a significance level of 0.05 after multiple hypothesis correction, showing that clustering methods can indeed identify a biologically meaningful co-methylation pattern corresponding to the latent regulator of the epitranscriptome. The most significant co-methylation pattern overlapping with FTO targets is from NMF with a p-value of 2.81 × 10−06 and a FDR of 3.372 × 10−05, suggesting the superior performance of the NMF clustering approach over the other 3 methods at the current setting in finding the co-methylation pattern in the epitranscriptome. Interestingly, for all 4 clustering approaches, FTO target sites are enriched in the largest of the 3 clusters. Indeed, FTO is the first mRNA demethylase discovered and plays a crucial role in human metabolism.5,6,29 It is reasonable to assume FTO is one of the most influential regulators at the epitranscriptomic layer. Please note that the captured RNA co-methylation pattern is not due to any of the transcriptional regulations. Under the ideal case of a reversible chemical reaction, the methylation potential is independent of RNA abundance and directly determines the ratio of methylated and un-modified RNA molecules.
| Algorithms | Cluster ID | # of sites | p-value | FDR | Odds ratio |
|---|---|---|---|---|---|
| Note: three out of 4 clustering approaches can successfully identify a co-methylation pattern corresponding to FTO target sites at a significance level of 0.05 after multiple hypothesis correction. Interestingly, for all 4 clustering approaches, FTO target sites are enriched in the largest of the 3 clusters. Indeed, FTO is the first mRNA demethylase discovered and plays a crucial role in human metabolism.5,6,29 It is reasonable to assume FTO is one of the most influential regulators at the epitranscriptomic layer. | |||||
| K-means | Cluster 1 | 1780 | 0.012 | 0.048 | 1.651 |
| Cluster 2 | 595 | 0.977 | 0.999 | 0.568 | |
| Cluster 3 | 899 | 0.884 | 0.999 | 0.769 | |
| HC | Cluster 1 | 316 | 0.734 | 0.999 | 0.837 |
| Cluster 2 | 2272 | 0.051 | 0.153 | 1.523 | |
| Cluster 3 | 686 | 0.963 | 0.999 | 0.629 | |
| NMF | Cluster 1 | 998 | 0.999 | 0.999 | 0.408 |
| Cluster 2 | 1597 | 2.81 × 10 −06 | 3.372 × 10 −05 | 2.793 | |
| Cluster 3 | 679 | 0.995 | 0.999 | 0.481 | |
| BFRM | Cluster 1 | 1249 | 0.003 | 0.018 | 1.791 |
| Cluster 2 | 990 | 0.655 | 0.999 | 0.934 | |
| Cluster 3 | 1035 | 0.999 | 0.999 | 0.484 | |
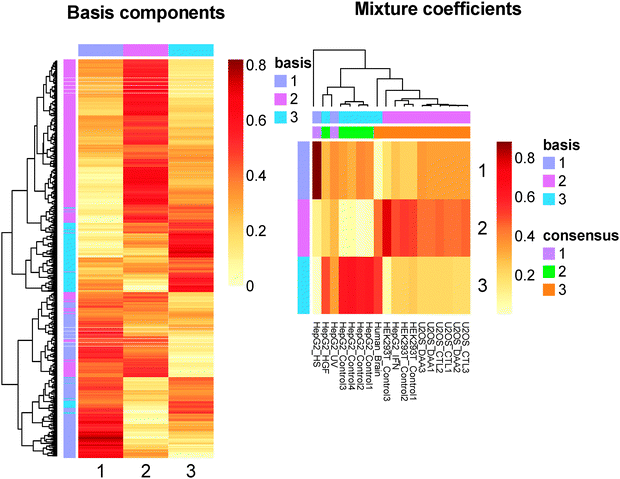
Among the 4 clustering approaches, NMF gives the most biologically significant result (see Fig. 9). NMF decomposes the RNA methylome into the product of two non-negative matrices, which essentially assume a regression-like model. Under the framework of NMF, a single methylation site may be regulated by multiple RNA methylation regulators, which is substantially different from Kmeans and HC. The recent identification of the METTL3-WTAP-METTL14 RNA methylation complex24,27 provides a good example of complex dependence relationship. All 3 genes are required to form an RNA methylation complex. It is worth mentioning that although the actual regulatory relationship at the epitranscriptomic layer is more complex, it does not prohibit using a simpler and more straightforward approach.
Conceivably, the 3 clusters of RNA methylation sites identified from NMF are roughly corresponding to 3 regulators of the epitranscriptome, through which different biomolecular functions can be regulated at the epitranscriptomic level. To reveal these functions, gene ontology enrichment analysis was conducted using the topGO R/Bioconductor package35 against the Biological processes category of gene ontology database. As shown in Fig. 10, relative distinct biological functions are enriched in different clusters. There is essentially no overlap in function between different clusters when keeping the top 20 mostly enriched functions for each cluster, indicating that the regulatory mechanism at the epitranscriptomic level is potentially highly specific.
 | ||
| Fig. 10 Gene ontology enrichment result of K-means. Relative distinct biological functions are enriched in different clusters. There is essentially no overlap in function between different clusters when keeping the top 20 mostly enriched functions for each cluster. This analysis was conducted using the topGO R/Bioconductor package.35 | ||
Specifically, the functions enriched in cluster 2, which is corresponding to FTO target sites, are shown in Fig. 11. There are a number of important biological functions enriched in this cluster, including neuron differentiation (p-value 0.0014), neurogenesis (0.0044), etc. Consistent with previous studies on FTO, these functions are highly related to neuronal diseases22 and cancer, indicating the prospect of RNA m6A and FTO studies.
Conclusion and discussion
In summary, with a newly developed R package RMT, we extracted the combinatorial epitranscriptome of more than 40000 RNA methylation sites from 18 biological replicates obtained under 10 different conditions, and then partially clustered them into 3 groups using 4 different clustering approaches after feature and model selection. The methylation sites belonging to the same cluster are likely to be hyper- and hypo-methylated simultaneously (or co-methylated), indicating that they are regulated by a common RNA methylation factor (methyltransferase or demethylase). We defined a new approach evaluating the consistency between two clustering results and show that, despite the discrepancy between different clustering approaches, the 4 clustering results are still highly correlated, capturing consistent patterns embedded in the epitranscriptome. To examine whether the co-methylation patterns are biologically meaningful, the target RNA methylation site of a known RNA demethylase (FTO) is identified from mouse and lifted to human, and we show that FTO target sites are significantly overlapping with the identified co-methylation pattern. Many important biological functions are significantly enriched and thus may be regulated by different RNA methylation factors at the epitranscriptomic level, a layer of gene regulation that has been missing for decades.14Computational reconstruction of the entire RNA methylation network is especially difficult due to the following reasons. Firstly, till this day, only a small number of genes related to RNA methylation are identified.11 Although it is possible to predict enzyme targets from enzyme knockout dataset, to the best of our knowledge, no effort has been made so far to computationally predict the target sites of a specific RNA methyltransferase or demethylase. Secondly, so far there are only a very limited amount of MeRIP-Seq dataset available. And the methylation levels for lowly expressed genes are difficult to estimate. It is clear that precision achieved on lowly and highly expressed transcripts are different. From a computational perspective, existing methods are not yet optimized for the intrinsic features of MeRIP-Seq, and advanced computational approaches that can take care of this discrepancy are still needed. With more data cumulated and revised mathematical models, it is promising that future work may more explicitly and specifically associate methylation sites with RNA methylation factors.
Limited by the available data and knowledge, this study provides only the RNA methylation sites that are likely to share a common regulator, but fails to specify what actually those regulators (gene or protein) are. Nevertheless, it demonstrated, for the first time, the feasibility of dissecting the RNA methylome based on the RNA co-methylation patterns induced by RNA methyltransferases and demethylases, implying a promising direction in untangling the RNA methylome based on its regulators, through which the biological meaning and underlying mechanism can be revealed in a deeper and more concise manner. Conceivably, with the eliminated regulatory relationship, the manipulation of the entire RNA methylome can be achieved through a much less number of enzymatic regulators. This work can also inspire RNA methylation is an open question, many methods are not implanted. This work may also potentially be integrated with other related studies, e.g., a joint analysis of RNA methylation sites and transcription starting sites,40–42 or suggests the combinatorial patterns of different post-transcriptional RNA modifications, like in chromatin modifications and transcription factor binding.43,44
Acknowledgements
We thank the support from the National Natural Science Foundation of China (61473232, 61401370, 91430111, 61170134 and 61201408) to SWZ, JM and HL; the National Science Foundation of USA (CCF-1246073) to YH; The graduate starting seed fund of Northwestern Polytechnical University (Z2014145) to YCZ and LL; Fundamental Research Funds for the Central Universities (2014QNB47, 2014QNA84) to LZ and HL. We also thank computational support from the UTSA Computational Systems Biology Core, funded by the National Institute on Minority Health and Health Disparities (G12MD007591) from the National Institutes of Health.References
- B. E. Bernstein, A. Meissner and E. S. Lander, The mammalian epigenome, Cell, 2007, 128, 669–681 CrossRef CAS PubMed.
- C. Bock, Analysing and interpreting DNA methylation data, Nat. Rev. Genet., 2012, 13, 705–719 CrossRef CAS PubMed.
- P. W. Laird, Principles and challenges of genomewide DNA methylation analysis, Nat. Rev. Genet., 2010, 11, 191–203 CrossRef CAS PubMed.
- C. He, Grand challenge commentary: RNA epigenetics?, Nat. Chem. Biol., 2010, 6, 863–865 CrossRef CAS PubMed.
- Y. Fu, D. Dominissini, G. Rechavi and C. He, Gene expression regulation mediated through reversible m6A RNA methylation, Nat. Rev. Genet., 2014, 15, 293–306 CrossRef CAS PubMed.
- K. D. Meyer and S. R. Jaffrey, The dynamic epitranscriptome: N6-methyladenosine and gene expression control, Nat. Rev. Mol. Cell Biol., 2014, 15, 313–326 CrossRef CAS PubMed.
- R. Liebers, M. Rassoulzadegan and F. Lyko, Epigenetic Regulation by Heritable RNA, PLoS Genet., 2014, 10, e1004296 Search PubMed.
- R. Desrosiers, K. Friderici and F. Rottman, Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 3971–3975 CrossRef CAS.
- U. Schibler, D. E. Kelley and R. P. Perry, Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells, J. Mol. Biol., 1977, 115, 695–714 CrossRef CAS PubMed.
- D. T. Dubin and R. H. Taylor, The methylation state of poly A-containing-messenger RNA from cultured hamster cells, Nucleic Acids Res., 1975, 2, 1653–1668 CrossRef CAS PubMed.
- M. A. Machnicka, K. Milanowska, O. Osman Oglou, E. Purta, M. Kurkowska, A. Olchowik, W. Januszewski, S. Kalinowski, S. Dunin-Horkawicz, K. M. Rother, M. Helm, J. M. Bujnicki and H. Grosjean, MODOMICS: a database of RNA modification pathways–2013 update, Nucleic Acids Res., 2013, 41, D262–D267 CrossRef CAS PubMed.
- K. D. Meyer, Y. Saletore, P. Zumbo, O. Elemento, C. E. Mason and S. R. Jaffrey, Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons, Cell, 2012, 149, 1635–1646 CrossRef CAS PubMed.
- D. Dominissini, S. Moshitch-Moshkovitz, S. Schwartz, M. Salmon-Divon, L. Ungar, S. Osenberg, K. Cesarkas, J. Jacob-Hirsch, N. Amariglio, M. Kupiec, R. Sorek and G. Rechavi, Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq, Nature, 2012, 485, 201–206 CrossRef CAS PubMed.
- Y. Saletore, K. Meyer, J. Korlach, I. D. Vilfan, S. Jaffrey and C. E. Mason, The birth of the Epitranscriptome: deciphering the function of RNA modifications, Genome Biol., 2012, 13, 175 CrossRef CAS PubMed.
- D. Dominissini, S. Moshitch-Moshkovitz, M. Salmon-Divon, N. Amariglio and G. Rechavi, Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing, Nat. Protoc., 2013, 8, 176–189 CrossRef CAS PubMed.
- Y. Zhang, T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson, B. E. Bernstein, C. Nusbaum, R. M. Myers, M. Brown and W. Li, Model-based analysis of ChIP-Seq (MACS), Genome Biol., 2008, 9, R137 CrossRef PubMed.
- P. J. Park, ChIP–seq: advantages and challenges of a maturing technology, Nat. Rev. Genet., 2009, 10, 669–680 CrossRef CAS PubMed.
- R. A. Harris, T. Wang, C. Coarfa, R. P. Nagarajan, C. Hong, S. L. Downey, B. E. Johnson, S. D. Fouse, A. Delaney and Y. Zhao, Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications, Nat. Biotechnol., 2010, 28, 1097–1105 CrossRef CAS PubMed.
- J. Meng, X. Cui, M. K. Rao, Y. Chen and Y. Huang, Exome-based analysis for RNA epigenome sequencing data, Bioinformatics, 2013, 29, 1565–1567 CrossRef CAS PubMed.
- Y. Li, S. Song, C. Li and J. Yu, MeRIP-PF: an easy-to-use pipeline for high-resolution peak-finding in MeRIP-Seq data, Genomics, Proteomics Bioinf., 2013, 11, 72–75 CrossRef CAS PubMed.
- J.-M. Fustin, M. Doi, Y. Yamaguchi, H. Hida, S. Nishimura, M. Yoshida, T. Isagawa, M. S. Morioka, H. Kakeya, I. Manabe and H. Okamura, RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock, Cell, 2013, 155, 793–806 CrossRef CAS PubMed.
- M. E. Hess, S. Hess, K. D. Meyer, L. A. Verhagen, L. Koch, H. S. Bronneke, M. O. Dietrich, S. D. Jordan, Y. Saletore, O. Elemento, B. F. Belgardt, T. Franz, T. L. Horvath, U. Ruther, S. R. Jaffrey, P. Kloppenburg and J. C. Bruning, The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry, Nat. Neurosci., 2013, 16, 1042–1048 CrossRef CAS PubMed.
- S. Schwartz, S. D. Agarwala, M. R. Mumbach, M. Jovanovic, P. Mertins, A. Shishkin, Y. Tabach, T. S. Mikkelsen, R. Satija, G. Ruvkun, S. A. Carr, E. S. Lander, G. R. Fink and A. Regev, High-Resolution Mapping Reveals a Conserved, Widespread, Dynamic mRNA Methylation Program in Yeast Meiosis, Cell, 2013, 155, 1409–1421 CrossRef CAS PubMed.
- J. Liu, Y. Yue, D. Han, X. Wang, Y. Fu, L. Zhang, G. Jia, M. Yu, Z. Lu, X. Deng, Q. Dai, W. Chen and C. He, A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation, Nat. Chem. Biol., 2014, 10, 93–95 CrossRef CAS PubMed.
- Y. Wang, Y. Li, J. I. Toth, M. D. Petroski, Z. Zhang and J. C. Zhao, N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells, Nat. Cell Biol., 2014, 16, 191–198 CrossRef CAS PubMed.
- G. Jia, Y. Fu, X. Zhao, Q. Dai, G. Zheng, Y. Yang, C. Yi, T. Lindahl, T. Pan and Y.-G. Yang, N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO, Nat. Chem. Biol., 2011, 7, 885–887 CrossRef CAS PubMed.
- X.-L. Ping, B.-F. Sun, L. Wang, W. Xiao, X. Yang, W.-J. Wang, S. Adhikari, Y. Shi, Y. Lv, Y.-S. Chen, X. Zhao, A. Li, Y. Yang, U. Dahal, X.-M. Lou, X. Liu, J. Huang, W.-P. Yuan, X.-F. Zhu, T. Cheng, Y.-L. Zhao, X. Wang, J. M. R. Danielsen, F. Liu and Y.-G. Yang, Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase, Cell Res., 2014, 24, 177–189 CrossRef CAS PubMed.
- V. Khoddami and B. R. Cairns, Identification of direct targets and modified bases of RNA cytosine methyltransferases, Nat. Biotechnol., 2013, 31, 458–464 CrossRef CAS PubMed.
- M. Lee, B. Kim and V. N. Kim, Emerging Roles of RNA Modification: m6A and U-Tail, Cell, 2014, 158, 980–987 CrossRef CAS PubMed.
- R. Carmel and D. W. Jacobsen, Homocysteine in health and disease, Cambridge University Press, 2001 Search PubMed.
- D. Kim, G. Pertea, C. Trapnell, H. Pimentel, R. Kelley and S. L. Salzberg, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions, Genome Biol., 2013, 14, R36 CrossRef PubMed.
- J. Przyborowski and H. Wilenski, Homogeneity of results in testing samples from Poisson series: With an application to testing clover seed for dodder, Biometrika, 1940, 31, 313–323 Search PubMed.
- C. M. Carvalho, J. Chang, J. E. Lucas, J. R. Nevins, Q. Wang and M. West, High-Dimensional Sparse Factor Modeling: Applications in Gene Expression Genomics, J. Am. Stat. Assoc., 2008, 103, 1438–1456 CrossRef CAS PubMed.
- D. D. Lee and H. S. Seung, Algorithms for non-negative matrix factorization, Advances in neural information processing systems, 2001, pp. 556–562 Search PubMed.
- A. Alexa and J. Rahnenfuhrer, topGO: enrichment analysis for gene ontology, R package version 2.8, 2010 Search PubMed.
- P. Machanick and T. L. Bailey, MEME-ChIP: motif analysis of large DNA datasets, Bioinformatics, 2011, 27, 1696–1697 CrossRef CAS PubMed.
- M. Lawrence, W. Huber, H. Pages, P. Aboyoun, M. Carlson, R. Gentleman, M. T. Morgan and V. J. Carey, Software for computing and annotating genomic ranges, PLoS Comput. Biol., 2013, 9, e1003118 CAS.
- M. Lawrence, V. Carey, R. Gentleman, I. XML, L. IRanges and M. M. Lawrence, Package ‘rtracklayer’, 2013 Search PubMed.
- Y. Benjamini and Y. Hochberg, Controlling the false discovery rate: a practical and powerful approach to multiple testing, J. R. Stat. Soc., Ser. B: Methodol., 1995, 289–300 Search PubMed.
- W. Chen, T.-Y. Lei, D.-C. Jin, H. Lin and K.-C. Chou, PseKNC: A flexible web server for generating pseudo K-tuple nucleotide composition, Anal. Biochem., 2014, 456, 53–60 CrossRef CAS PubMed.
- S.-H. Guo, E.-Z. Deng, L.-Q. Xu, H. Ding, H. Lin, W. Chen and K.-C. Chou, iNuc-PseKNC: a sequence-based predictor for predicting nucleosome positioning in genomes with pseudo k-tuple nucleotide composition, Bioinformatics, 2014, 30, 1522–1529 CrossRef CAS PubMed.
- W. Chen, P.-M. Feng, E.-Z. Deng, H. Lin and K.-C. Chou, iTIS-PseTNC: A sequence-based predictor for identifying translation initiation site in human genes using pseudo trinucleotide composition, Anal. Biochem., 2014, 462, 76–83 CrossRef CAS PubMed.
- L. Ferraris, A. P. Stewart, J. Kang, A. M. DeSimone, M. Gemberling, D. Tantin and W. G. Fairbrother, Combinatorial binding of transcription factors in the pluripotency control regions of the genome, Genome Res., 2011, 21, 1055–1064 CrossRef CAS PubMed.
- M. Kato, N. Hata, N. Banerjee, B. Futcher and M. Q. Zhang, Identifying combinatorial regulation of transcription factors and binding motifs, Genome Biol., 2004, 5, R56 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Software availability: the open source R package “RMT” for extracting a combinatorial epitranscriptome from multiple MeRIP-Seq experiments. See DOI: 10.1039/c4mb00604f |
| This journal is © The Royal Society of Chemistry 2015 |