Evolution of the CRISPR-Cas adaptive immunity systems in prokaryotes: models and observations on virus–host coevolution
Eugene V.
Koonin
* and
Yuri I.
Wolf
National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA. E-mail: koonin@ncbi.nlm.nih.gov
First published on 12th September 2014
Abstract
CRISPR-Cas is an adaptive immunity system in prokaryotes that functions via a unique mechanism which involves incorporation of foreign DNA fragments into CRISPR arrays and subsequent utilization of transcripts of these inserts (known as spacers) as guide RNAs to cleave the cognate selfish element genome. Multiple attempts have been undertaken to explore the coevolution of viruses and microbial hosts carrying CRISPR-Cas using mathematical models that employ either systems of differential equations or an agent-based approach, or combinations thereof. Analysis of these models reveals highly complex co-evolutionary dynamics that ensues from the combination of the heritability of the CRISPR-mediated adaptive immunity with the existence of different degrees of immunity depending on the number of cognate spacers and the cost of carrying a CRISPR-Cas locus. Depending on the details of the models, a variety of testable, sometimes conflicting predictions have been made on the dependence of the degree of immunity and the benefit of maintaining CRISPR-Cas on the abundance and diversity of hosts and viruses. Some of these predictions have already been directly validated experimentally. In particular, both the reality of the virus–host arms race, with viruses escaping resistance and hosts reacquiring it through the capture of new spacers, and the fitness cost of CRISPR-Cas due to the curtailment of beneficial HGT have been reproduced in the laboratory. However, to test the predictions of the models more specifically, detailed studies of coevolving populations of microbes and viruses both in nature and in the laboratory are essential. Such analyses are expected to yield disagreements with the predictions of the current, oversimplified models and to trigger a new round of theoretical developments.
CRISPR-Cas: heritable adaptive immunity in archaea and bacteria
All cellular life forms on earth evolve under an incessant assault from viruses and other selfish genetic elements which translates into a perennial arms race.1–3 In the course of this arms race, cellular organisms have evolved elaborate networks of diverse, interconnected defense systems.4–6 Historically, the best known of such defense mechanisms is the adaptive immunity in vertebrates that over the decades had been characterized in exquisite detail.7–9 The key feature of adaptive immunity is immunological memory that often lasts through the lifetime of an individual, providing extremely efficient resistance against a specific, previously encountered, pathogen, but is not transmitted across generations. More recently, it has become clear that all organisms possess multiple mechanisms of innate immunity that do not involve immunological memory but instead provide non-specific protection against entire classes of pathogens that is often less efficient than the protection attained via adaptive immunity.10,11 Bacteria and archaea (collectively, prokaryotes) have been long known to possess innate immunity that includes in particular the thoroughly characterized restriction-modification systems; moreover, recent advances in comparative genomics and experimental study of virus–host interaction have revealed a variety of new defense mechanisms.6,12–15 Until recently, prokaryotes have not been thought to possess adaptive immunity but this belief was dramatically overturned with the discovery of the defense system that became known as CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas (CRISPR-associated genes).16–20The CRISPR-Cas system functions via a unique mechanism that involves incorporation of foreign DNA fragments into CRISPR repeat arrays and subsequent utilization of transcripts of these inserts (known as spacers) as guide RNAs to cleave the cognate selfish element genome.21–25 These processes are catalyzed or facilitated by complexes of multiple Cas proteins that are typically encoded in close proximity of the CRISPR cassettes. The mechanism of CRISPR-Cas is usually divided into three stages: (1) adaptation, when new 30–84 base pair long, unique spacers homologous to proto-spacer sequences in viral genomes or other alien DNA molecules are integrated into the CRISPR repeat cassettes; (2) expression and processing of pre-crRNA into short guide crRNAs; and (3) interference, when the alien DNA or RNA is targeted by a complex of Cas proteins containing a crRNA guide and cleaved within the unique target site.5,20,26,27
The defense provided by CRISPR-Cas is highly specific: a single nucleotide substitution in a “seed” region of a spacer often abrogates immunity.28 The protection against infectious elements is also extremely efficient, with the yield of the cognate virus dropping by up to 5 orders of magnitude which effectively amounts to complete resistance.19,29 The CRISPR-Cas systems are endowed with immunological memory that is ensured by the persistence of the spacers that can be transmitted across many thousands of prokaryotic cell generations. Thus, the CRISPR-Cas systems meet all the criteria for adaptive (acquired) immunity.30
In the few years that elapsed since the seminal discovery of the CRISPR mechanism, the CRISPR research evolved into a highly dynamic field of microbiology with major potential for applications in epidemiology, biotechnology and genome engineering.27,31,32 Multiple applications of CRIPSR-Cas for genome manipulation and gene expression programming already have been developed.33–36
The CRISPR-Cas systems display an enormous diversity of Cas protein repertoires and the architectures of the respective genomic loci. Comparative analysis of the sequences and structures of Cas proteins, combined with the analysis of genomic architectures, led to the classification of the CRISPR-Cas systems into three distinct types (I, II and III) that include 10 subtypes and a growing number of still unclassified, less common variants.5
A major distinction between CRISPR-Cas and animal immune systems is that CRISPR-Cas modifies the host genome in response to infection and hence provides heritable immunity. Indeed, CRISPR-Cas is the most compelling known case for Lamarckian inheritance whereby an organism responds to an environmental cue (in this case, invasion of foreign DNA) by generating a heritable modification of the genome that provides specific adaptive response to the original challenge.37
The benefit of highly specific and efficient resistance to selfish elements enjoyed by the organisms that carry CRISPR-Cas loci appears to be countered by potential fitness cost.38,39 One likely source of the cost is autoimmunity whereby a CRISPR cassette accidentally incorporates a spacer homologous to the host DNA, with obvious deleterious consequences.40,41 This phenomenon is strikingly similar to animal autoimmunity. The other and perhaps more important source of cost does not involve any errors and appears to be inherent to the CRISPR-Cas mechanism. This potential drawback of CRISPR-Cas involves curtailment of the capture of new genes via horizontal gene transfer (HGT) that is likely to accompany the defensive action of CRISPR-Cas. In prokaryotes, HGT is a key evolutionary process that rescues clonal populations from the doom of Mueller's ratchet and often serves as the principal means of adaptation to a new environment.2,42–45 Considerable evidence has been presented that CRISPR-Cas indeed prevents adaptive HGT.46–49
The CRISPR-Cas systems show a remarkably non-uniform distribution among prokaryotes, with nearly all sequenced hyperthermophiles (mostly archaea) but less than 50% of the mesophiles (largely bacteria) encompassing CRISPR-Cas loci.5,24,39 In bacteria, the CRISPR-Cas loci demonstrate notable evolutionary volatility, with many cases reported when some of several closely related bacterial strains possessed CRISPR-Cas but the others lacked it.50,51 Numerous cases of apparent horizontal transfer (HGT) of CRISPR-Cas loci also have been reported.52,53 Furthermore, the CRISPR-Cas loci have been shown to abrogate acquisition of foreign DNA via HGT49,54 and consequently are rapidly lost under selective pressure for horizontal gene transfer as demonstrated by the propagation of antibiotic-resistant CRISPR− strains of Enterococcus faecalis derived from a CRISPR+ progenitor in a hospital environment.47 Rapid acquisition and loss of CRISPR spacers leading to intra-population heterogeneity also has been observed in experiments on both archaeal55 and bacterial56 models.
Viruses can evade CRISPR-Cas through minimal mutational or recombinational changes in proto-spacers (the sequences in the viral genome that are excised by the Cas machinery to become spacers). In some bacteria, a single proto-spacer mutation renders CRISPR-Cas ineffectual29,57,58 although other CRISPR-Cas systems show less rigid specificity.28 The hosts can regain antiviral immunity by acquiring new spacers from the same viral genome.57–60 The restoration of immunity is facilitated by an ancillary mechanism dubbed priming whereby spacers derived from a given virus or plasmid genome, even after losing their direct protective capacity, substantially enhance acquisition of new spacers from the same. Together, viral evasion of immunity and loss and regain of immune spacers and entire CRISPR-Cas loci by prokaryotes drive a co-evolutionary arms race between the mutating virus and the spacer-incorporating host. This arms race can go multiple rounds and take unexpected forms as illustrated by the finding that certain bacteriophages encode their own CRISPR-Cas system which targets host innate immunity loci, thus exploiting a host defense mechanism for counter-defense.61
The virus–host arms race combined with the horizontal mobility of the CRISPR-Cas loci results in complex co-evolutionary dynamics. It is well known from decades of studies in ecology, starting with the classic work of Lotka and Volterra, that mathematical modeling of prey–predator or host–parasite coevolution can clarify the co-evolutionary dynamics and reveal non-trivial evolutionary regimes.62–64 Therefore multiple attempts to model the coevolution of CRISPR-Cas carrying hosts and viruses have been undertaken shortly after the molecular characterization of CRISPR-Cas. Here we discuss these models, the predictions they make regarding the population dynamics of viruses and hosts, and various co-evolutionary regimes, and the first attempts on experimental validation of these predictions.
Mathematical models of CRISPR-Cas–virus coevolution and the first experimental tests
The arms race between the immune system and viruses, the common events of loss and horizontal transfer of CRISPR-Cas loci and the fitness cost that is apparently incurred by CRISPR-Cas combine to yield complex evolutionary dynamics. However, this dynamics could be simplified by the extremely high efficacy of the CRISPR-Cas-mediated immunity which decreases the virus yield by as much as 5 orders of magnitude. Given this level of immunity, the host population can be reasonably approximated by partitioning into resistant and sensitive subpopulations. This type of dynamic provides fertile ground for mathematical modeling with a potential to elucidate the interactions between different evolutionary processes and possibly discover unexpected evolutionary regimes. The mathematical models of CRISPR-Cas coevolution that have been independently developed by several research groups focused on explaining the striking features of the CRISPR-Cas systems that became apparent through comparative genomic analyses, namely their fast evolution, enormous diversity of the spacers and gene arrangements in the cas operons, and the old (distal from the leader) end uniformity.Most of the models belong to one of the two classes according to the type of abstractions that are used to represent the agents and their interactions (Table 1). The first approach employs systems of differential equations (SDE) in which the coevolving entities, i.e. host and virus strains, are represented by their abundances. The interactions of these entities with each other and with the environment are represented as rates of change. The SDE models can be analyzed numerically or analytically. The second approach, the agent-based stochastic models (ABS), represents host and virus strains or individual microbes and viruses in greater detail.
| Type | Features | Major results | Ref. |
|---|---|---|---|
| SDE | Single type of virus/plasmid, 5 types of hosts | Strong defence against viruses, weak defence against plasmids | 67 |
| SDE | 3 Types of hosts, 3 types of viruses | Importance of multiple spacer–protospacer matches | 65 |
| SDE | 6 Types of hosts with and without plasmids | CRISPR-Cas loci might be selected to be labile | 82 |
| ABS | Lattice population model, CRISPR arrays in host, protospacer arrays in virus | Long-term virus–host and virus strain coexistence in spatially structured population | 69, 70 |
| ABS, SDE | Strain-level host and virus representation; CRISPR arrays in host, protospacer arrays in virus | Diversification of host and virus, incomplete selective sweeps | 72, 73 |
| ABS, SDE | Strain-level host and virus representation; CRISPR arrays in host, protospacer arrays in virus | Rapid fluctuations of host population distributed immunity (PDI), limiting effect of high PDI on virus diversity | 74 |
| ABS | Strain-level host and virus representation; CRISPR arrays in host, protospacer arrays in virus | Rapid selection sweeps, distal end conservation of CRISPR arrays | 76 |
| ABS | Strain-level host and virus representation; CRISPR arrays in host, protospacer arrays in virus | Virus diversity threshold for CRISPR-Cas maintenance | 54 |
| ABS | Individual-level host and virus representation; CRISPR arrays in host, protospacer arrays in virus | Direct correlation of virus diversity with population size, importance of multiple spacer–protospacer matches | 71 |
| SDE | 2 Types of hosts, single type of virus | Quasi-chaotic oscillations at high virus reproduction rates | 66 |
| ABS, SDE | Strain-level host and virus representation; virus mutation and recombination, CRISPR arrays in host, protospacer arrays in virus | Proximal end heterogeneity with distal end conservation of CRISPR arrays, effects of virus mutation and recombination depend on multiplicity of spacer–protospacer matches | 78, 79 |
| ABS | Evolution of CRISPR arrays via acquisition and deletion | Clock-like divergence of arrays, estimation of host phylogeny and divergence times by CRISPR array comparison | 77 |
| SDE | Levels of processed and unprocessed CRISPR locus transcripts under expression and degradation | Rapid upregulation of processed transcript abundance, phase transition-like behavior | 80, 81 |
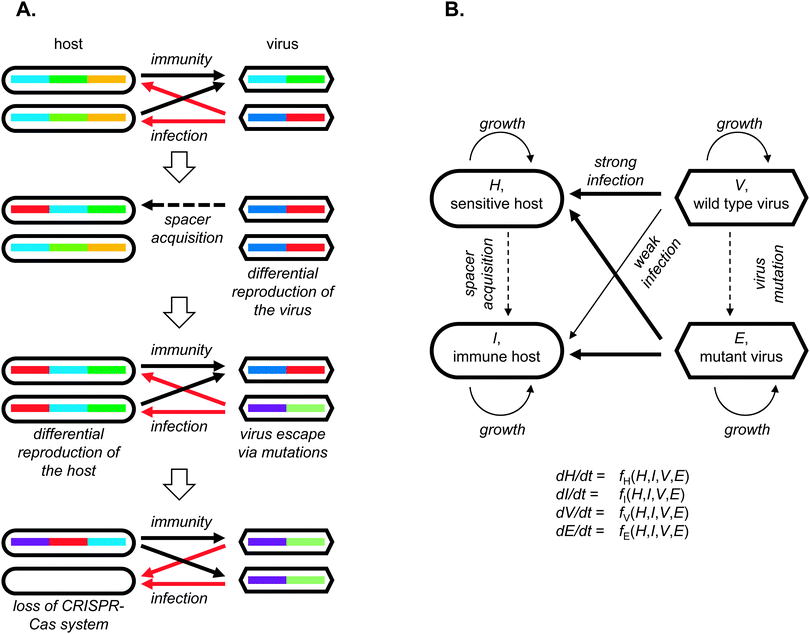
Most of the ABS-type models explicitly use CRISPR-inspired representations of the host and the virus individuals or populations. Thus, hosts are represented by arrays or sets of spacers and viruses as arrays or sets of (potential) protospacers (Fig. 1). Spacer acquisition is triggered by a contact with a protospacer-carrying virus and virus mutations replace old protospacers by novel versions. In principle, such a representation can be generalized to other hypothetical types of adaptive immune system whereby the virus is represented by a set of “anti-idiotypes” recognized in a binary manner whereas the immune memory stores “idiotypes” and is subject to decay. The adaptive nature of such systems is manifest in the interaction-dependent acquisition of an idiotype specific to the particular invading virus.
Some SDE-type models are highly specific toward the CRISPR-Cas mechanism, limiting the scope of the model to a relatively narrow class of situations. For example, Levin et al.65 consider the interactions between three types of virus (wild type, single escape mutant and double escape mutant) and three types of host (carrying zero, one and two virus-matching spacers). Other models are more abstract and potentially are conducive to wider generalizations. Thus, Berezovskaya et al.66 use a mean field approximation to model CRISPR-mediated immunity whereas the average efficacy of the immune response depends on the virus diversity which is assumed to increase on average with the population size. Acquisition of immune memory is modeled as a constant-rate process dependent on virus–host interactions; virus mutations are approximated by a constant-rate immunity decay. Such models, in principle, can apply to a wide range of hypothetical adaptive immune systems although so far CRISPR-Cas remains the only established case of adaptive immunity in prokaryotes.
The very first model developed by Levin67 employed SDE to assess the efficacy of the CRISPR-Cas defense against lytic viruses and conjugative plasmids. The virus–host coevolution was modeled explicitly via decay of immunity caused by the accumulation of escape mutations in the phage population. Analysis of the model indicated that within an expansive domain of the parameter space, in the presence of lytic (“nasty”) viruses, CRISPR-Cas would provide a fitness advantage to microbes carrying this system over CRISPR-Cas–microbes. The range of parameters where this is the case narrows considerably in the presence of innate immunity provided by accumulating resistance mutations. In contrast, CRISPR-Cas was found not to be efficient in protection against (weakly) deleterious conjugative plasmids.
In a follow-up study Levin et al.65 combined the SDE-type model analysis with direct experiments on virus–host interaction to estimate realistic model parameters and explore evolutionary regimes. The growth rates of different virus and host strains in various combinations was predicted by the model, and the predictions were directly tested by experimental measurement of the respective rates. In particular, Levin et al. conclude that experimental observations are consistent with the evolutionary advantage conferred by CRISPR-Cas system to the host in the presence of the virus and rapid virus evasion from the CRISPR-mediated immunity, as predicted by the model. However, beyond confirming the most general predictions of the model, several unexpected observations were made through the analysis of the experimental coevolutionary system. Specifically, it has been observed that phages could invade populations of bacteria that acquired resistance through a single spacer but not through two spacers from the given phage genome, some sensitive bacteria survived even at high phage density, and even the two-spacer-carrying resistant variants were not quickly fixed in the population. These findings reveal a highly complex coevolutionary system in which resistance is not necessarily an all or none phenomenon, with single-spacer bacteria being only partially resistant, and spacer-less variants developing the resistant phenotype mechanisms that do not involve CRISPR-Cas and competing with the spacer-carrying variants.
The same group then extended the mathematical and experimental investigation of the CRISPR-Cas evolution to incorporate strongly beneficial or even essential plasmids, i.e. explicitly modeling the fitness cost of CRISPR-Cas.68 Both experiments and computer simulations consistently show that bacterial populations that carry spacers preventing the replication of an essential plasmid nevertheless can support replication of the plasmid and hence survive as a result of spontaneous loss of either the cognate spacer or the entire CRISPR-Cas locus. Fluctuation experiments show that the CRISPR deletion mutants are not induced by the plasmids but rather pre-exist, being produced at high rates (>10−4 per cell per generation). These findings confirm the extreme evolutionary lability of the CRISPR-Cas loci and suggest that the ensuing flexibility of microbial defense against foreign DNA is a beneficial condition. Put another way, there is no selection for increased stability of CRISPR-Cas loci because stable inheritance of CRISPR-Cas systems would be deleterious due to the curtailment of beneficial gene acquisition via HGT. Notably, independent experiments have shown, perhaps somewhat paradoxically, that bacterial strains that lose natural competence, with time, also tend to lose their CRISPR-Cas loci.50 Thus, it appears that in order to persist, the CRISPR-Cas systems have to be perpetually replenished via HGT.
Haerter et al. developed an ABS model to explore the impact of the CRISPR-Cas immunity on the virus–host coexistence.69,70 The simulations of virus–CRISPR interaction under this model show that long-term coexistence of viruses and hosts is readily achieved in spatially structured communities but is unstable in well-mixed ones. Furthermore, CRISPR-Cas-system provides the hosts with resistance to a wide range of virus strains even when the actual number of strains a particular host is immune to is small relative to the overall virus diversity. The other side of virus–host coevolution revealed by this model is that under conditions conducive to coevolution, CRISPR-mediated immunity promotes the coexistence of diverse virus strains, i.e. buffers the diversity of the virus population.
Weinberger et al. used a strain-level ABS model of virus–host coevolution to explore the conditions that favor the CRISPR-Cas system maintenance or loss.39 It was shown that the efficacy of CRISPR-mediated immunity sharply decreases when virus diversity reaches some critical level, whereas low virus diversity leads to a low penetrance of CRISPR-Cas system in the host population. Thus, the maximum intensity of coevolution and accordingly the maximum length of the CRISPR cassette is observed at some intermediate level of virus diversity beyond which further diversification of the viruses leads to the collapse of CRISPR-Cas. This analysis might explain the key observation in the global ecology of CRISPR-Cas, namely the presence of CRISPR-Cas in nearly all thermophilic archaea and most thermophilic bacteria but less than a half of the mesophilic microbes. The limits on mutation rates and hence on viral diversity in high-temperature environments might be responsible for the high prevalence of CRISPR-positive hosts among thermophiles. This modeling study sheds light on the more general aspect of Lamarckian adaptive evolution: this mode of evolution is advantageous under conditions of intense environmental pressure that, however, does not exceed a threshold beyond which organisms lose the ability to track the changes in the environment.
In a subsequent refinement of the strain-based ABS approach, Iranzo et al. explored the population dynamics of viruses and hosts in the course of coevolution.71 The results indicate that generally, virus diversity dramatically increases with the virus population size which itself grows with the host population size. Therefore, even at equal mutation rates, the efficacy of CRISPR-based immunity is predicted to be limited in environments that support larger host and virus populations. It has been found that CRISPR-Cas stabilizes the stochastic virus–host system in the intermediate range of viral mutation rate, resulting in an extended coexistence of viruses and the microbial hosts. Analysis of this model additionally suggests that the highly efficient spacer acquisition mechanism and its ability to utilize a wide range of viral sequences as protospacers are beneficial primarily not due to the more rapid response to emerging infections but rather because multiple spacer–protospacer matches limit the rate of generation of immunity escape mutants in virus populations. These findings are compatible with the observation of Levin et al. that multiple spacer matches yield a higher efficacy of immunity than single matches.65
Berezovskaya et al. incorporated the inferred trend of the virus diversity to increase and conversely the efficacy of adaptive immunity to decrease with populations size in an SDE-based model which was exhaustively explored analytically.66 This analysis of the model has shown that the combination of adaptive immunity that is inversely dependent on the virus abundance with rapid virus multiplication leads to emergence of high-amplitude quasi-chaotic oscillations in both host and virus populations. Under this regime, the relative abundance of the immune and non-immune hosts can fluctuate rapidly. Such unpredictable behavior might explain the observed inconsistency in CRISPR-Cas status and spacer composition in relatively closely related microbial lineages.
Childs et al. employed a combined ABS/SDE model in which the spacers and protospacers in the host and virus respectively evolve stochastically in an ABS-type settings, with SDE-determined behavior between the random events.72 This model displayed a rich behavior with diversification of host and virus into multiple strains. Incomplete selective sweeps and waves of recurrence of older virus lineages are predicted to be common. The model suggests a long-term maintenance of host and virus diversity which appears to be consistent with observations in various natural habitats.73
This model has been further advanced to introduce the concept of Population Distributed Immunity (PDI) whereby multiple alleles providing resistance to the same virus (i.e. multiple spacers from the same genome) are distributed across the host population.74 Computer simulations under the model revealed broad fluctuations in the PDI values, with PDI reaching it maximum immediately before the host population size peaks and dropping almost to zero between peaks. It has been found that elevated PDI stabilizes the host population and conversely leads to decreased virus diversity and density. The high PDI values observed in the simulations were found to be compatible with the findings from coevolutionary experiments.75
Several modeling studies have addressed more specific aspects of the coevolution of viruses and CRISPR-Cas loci. In particular, Weinberger et al. employed an ABS model to explore the evolution of the spacer pattern in the CRISPR cassettes of an evolving population.76 The model predicts rapid selection sweeps in the host population leading to the apparent conservation of the trailer end of the CRISPR arrays. Conversely, the ongoing loss of spacers is predicted to result in polyclonal virus blooms that can be prevented by mechanisms preserving the spacers across the entire cassette.
Kupczok and Bollback explored an ABS model that was tuned to the analysis of the tempo and mode of the CRISPR array evolution.77 It has been found that the divergence of the spacer content of CRISPR arrays with time could be predicted using the model parameters estimated from empirical data and that comparison of CRISPR cassettes can be used for the host strain classification and phylogeny.
The model of He and Deem,78 one of the first theoretical analyses of CRISPR-Cas, combined the ABS and SDE approaches to explore the coevolution of phages and microbial host and the evolution of CRISPR arrays. This model recapitulates, under a broad range of parameters, the well-known heterogeneity of the spacer content in CRISPR arrays whereby the old, leader-distal end of the cassette is more homogeneous than the leader-proximal end. In particular, this distribution of spacers can obtain even under a uniform spacer deletion rate. The model additionally incorporated the effects of viral recombination on the coevolutionary arms race, showing that in cases when effective protection by CRISPR can be achieved in the presence of a mismatch between the spacer and the protospacer, recombination became the most efficient route of viral escape. In a subsequent development of this approach, Han et al.79 explored the effects of different mechanisms of spacer deletion. This model predicted that the diversity of the spacers increases with the diversity of the phage population, in agreement with experimental data.59
Most of the CRISPR-Cas models explore the relatively long-term host–virus interaction and coevolution at the time scales measured in many generations of microorganisms. In contrast, the SDE model developed and explored by Djordjevic et al. addresses the function and regulation of the CRISPR-Cas system in the course of a single infection.80 Analysis of this model shows how a relatively small increase of a CRISPR locus transcription can lead to a disproportionally large and rapid increase the concentration of the crRNA in the host cell. A follow-up analysis suggests that such phase transition-like behavior is an essential feature of diverse prokaryotic defense systems that include potentially toxic proteins.81
Conclusions
The efforts on mathematical modeling of CRISPR-mediated immunity are only 4 years old but by capitalizing on the extensive earlier work on predator–prey dynamics and parasite–host coevolution, a lot of ground has already been covered. The models converge on several key findings that broadly agree with qualitative observations from comparative genomic analyses. In particular, it has become clear that the combination of the heritability of the CRISPR-mediated adaptive immunity with the existence of different degrees of immunity depending on the number of cognate spacers and the cost of carrying a CRISPR-Cas locus translates into a highly complex co-evolutionary dynamics. This dynamics includes oscillations of the host and virus population size that might be intrinsically unpredictable (quasi-chaotic).66 Depending on the details of the models, a variety of testable, sometimes conflicting predictions have been made on the dependence of the degree of immunity and the benefit of maintaining CRISPR-Cas on the abundance and diversity of hosts and viruses. In particular, contrary to the naïve expectation that the benefit of CRISPR-Cas would increase proportional to the virus diversity, model analysis has suggested that the maximum activity and efficacy of CRISPR-Cas are expected at intermediate virus diversity,39 a prediction that remains to be tested experimentally.Some of the key results of mathematical modeling have already been directly validated experimentally. Thus, it has been shown that the virus–host arms race, with viruses escaping resistance and hosts reacquiring it through the capture of new spacers is a tangible reality that can be reproduced in the laboratory.65 Moreover, the fitness cost incurred by CRISPR-Cas due to the curtailment of beneficial HGT also has been observed experimentally and shown to result in frequent loss of the CRISPR-Cas loci and competition between CRISPR-Cas and other, in particular phenotypic, resistance mechanisms.68 These findings are broadly compatible with the extremely patchy distribution of CRISPR-Cas among the available archaeal and bacterial genomes.6 However, comparative genomic analysis at the level of species or even strains of bacteria and archaea might not be particularly informative for understanding the microevolutionary dynamics predicted by the mathematical models of CRISPR-Cas because the key events occur at the population level.77 Therefore, in order to test the predictions of the models, detailed studies of coevolving microbes and viruses both in nature and in the laboratory are essential. Beyond doubt, such analyses will yield disagreements with the predictions of the current, oversimplified models, stimulating a new round of theoretical developments.
Acknowledgements
We thank Alexander Lobkovsky for helpful discussions. The authors' research is supported by intramural funds of the US Department of Health and Human Research (to the National Library of Medicine).References
- A. Stern and R. Sorek, BioEssays, 2011, 33, 43–51 CrossRef CAS PubMed.
- E. V. Koonin and Y. I. Wolf, Front. Cell. Infect. Microbiol., 2012, 2, 119 Search PubMed.
- P. Forterre and D. Prangishvili, Ann. N. Y. Acad. Sci., 2009, 1178, 65–77 CrossRef CAS PubMed.
- J. Haaber, J. E. Samson, S. J. Labrie, V. Campanacci, C. Cambillau, S. Moineau and K. Hammer, Appl. Environ. Microbiol., 2010, 76, 7085–7092 CrossRef CAS PubMed.
- K. S. Makarova, D. H. Haft, R. Barrangou, S. J. Brouns, E. Charpentier, P. Horvath, S. Moineau, F. J. Mojica, Y. I. Wolf, A. F. Yakunin, J. van der Oost and E. V. Koonin, Nat. Rev. Microbiol., 2011, 9, 467–477 CrossRef CAS PubMed.
- K. S. Makarova, Y. I. Wolf and E. V. Koonin, Nucleic Acids Res., 2013, 41, 4360–4377 CrossRef CAS PubMed.
- M. D. Cooper and M. N. Alder, Cell, 2006, 124, 815–822 CrossRef CAS PubMed.
- T. Boehm, Nat. Rev. Immunol., 2011, 11, 307–317 CrossRef CAS PubMed.
- T. Boehm, Curr. Biol., 2012, 22, R722–R732 CrossRef CAS PubMed.
- S. Akira, S. Uematsu and O. Takeuchi, Cell, 2006, 124, 783–801 CrossRef CAS PubMed.
- R. Medzhitov, Immunity, 2009, 30, 766–775 CrossRef CAS PubMed.
- T. R. Blower, G. P. Salmond and B. F. Luisi, Curr. Opin. Struct. Biol., 2011, 21, 109–118 CrossRef CAS PubMed.
- T. R. Blower, T. J. Evans, R. Przybilski, P. C. Fineran and G. P. Salmond, PLoS Genet., 2012, 8, e1003023 CAS.
- R. Leplae, D. Geeraerts, R. Hallez, J. Guglielmini, P. Dreze and L. Van Melderen, Nucleic Acids Res., 2011, 39, 5513–5525 CrossRef CAS PubMed.
- K. Vasu and V. Nagaraja, Microbiol. Mol. Biol. Rev., 2013, 77, 53–72 CrossRef CAS PubMed.
- E. V. Koonin and K. S. Makarova, RNA Biol., 2013, 10, 679–686 CrossRef CAS PubMed.
- D. Bhaya, M. Davison and R. Barrangou, Annu. Rev. Genet., 2011, 45, 273–297 CrossRef CAS PubMed.
- H. Deveau, J. E. Garneau and S. Moineau, Annu. Rev. Microbiol., 2010, 64, 475–493 CrossRef CAS PubMed.
- L. A. Marraffini and E. J. Sontheimer, Nat. Rev. Genet., 2010, 11, 181–190 CrossRef CAS PubMed.
- B. Wiedenheft, S. H. Sternberg and J. A. Doudna, Nature, 2012, 482, 331–338 CrossRef CAS PubMed.
- D. H. Haft, J. Selengut, E. F. Mongodin and K. E. Nelson, PLoS Comput. Biol., 2005, 1, e60 CrossRef PubMed.
- R. Jansen, J. D. Embden, W. Gaastra and L. M. Schouls, Mol. Microbiol., 2002, 43, 1565–1575 CrossRef CAS.
- E. V. Koonin and K. S. Makarova, F1000 Biol. Rep., 2009, 1, 5 Search PubMed.
- K. S. Makarova, N. V. Grishin, S. A. Shabalina, Y. I. Wolf and E. V. Koonin, Biol. Direct, 2006, 1, 7 CrossRef PubMed.
- F. J. Mojica, C. Diez-Villasenor, J. Garcia-Martinez and E. Soria, J. Mol. Evol., 2005, 60, 174–182 CrossRef CAS PubMed.
- J. van der Oost, M. M. Jore, E. R. Westra, M. Lundgren and S. J. Brouns, Trends Biochem. Sci., 2009, 34, 401–407 CrossRef CAS PubMed.
- D. Bikard and L. A. Marraffini, Curr. Opin. Immunol., 2012, 24, 15–20 CrossRef CAS PubMed.
- E. Semenova, M. M. Jore, K. A. Datsenko, A. Semenova, E. R. Westra, B. Wanner, J. van der Oost, S. J. Brouns and K. Severinov, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 10098–10103 CrossRef CAS PubMed.
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero and P. Horvath, Science, 2007, 315, 1709–1712 CrossRef CAS PubMed.
- J. Rimer, I. R. Cohen and N. Friedman, BioEssays, 2014, 36, 273–281 CrossRef CAS PubMed.
- L. Fabre, J. Zhang, G. Guigon, S. Le Hello, V. Guibert, M. Accou-Demartin, S. de Romans, C. Lim, C. Roux, V. Passet, L. Diancourt, M. Guibourdenche, S. Issenhuth-Jeanjean, M. Achtman, S. Brisse, C. Sola and F. X. Weill, PLoS One, 2012, 7, e36995 CAS.
- D. Carroll, Mol. Ther., 2012, 20, 1658–1660 CrossRef CAS PubMed.
- L. Qi, R. E. Haurwitz, W. Shao, J. A. Doudna and A. P. Arkin, Nat. Biotechnol., 2012, 30, 1002–1006 CrossRef CAS PubMed.
- P. Mali, L. Yang, K. M. Esvelt, J. Aach, M. Guell, J. E. Dicarlo, J. E. Norville and G. M. Church, Science, 2013, 339, 823–826 CrossRef CAS PubMed.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS PubMed.
- A. K. Cain and C. J. Boinett, Nat. Rev. Microbiol., 2013, 11, 226 CrossRef CAS PubMed.
- E. V. Koonin and Y. I. Wolf, Biol. Direct, 2009, 4, 42 CrossRef PubMed.
- E. R. Westra, D. C. Swarts, R. H. Staals, M. M. Jore, S. J. Brouns and J. van der Oost, Annu. Rev. Genet., 2012, 46, 311–339 CrossRef CAS PubMed.
- A. D. Weinberger, Y. I. Wolf, A. E. Lobkovsky, M. S. Gilmore and E. V. Koonin, mBio, 2012, 3, e00456 CrossRef CAS PubMed.
- A. Stern, L. Keren, O. Wurtzel, G. Amitai and R. Sorek, Trends Genet., 2010, 26, 335–340 CrossRef CAS PubMed.
- D. Paez-Espino, W. Morovic, C. L. Sun, B. C. Thomas, K. Ueda, B. Stahl, R. Barrangou and J. F. Banfield, Nat. Commun., 2013, 4, 1430 CrossRef PubMed.
- W. F. Doolittle, Trends Cell Biol., 1999, 9, M5–M8 CrossRef CAS.
- W. F. Doolittle and O. Zhaxybayeva, Genome Res., 2009, 19, 744–756 CrossRef CAS PubMed.
- E. V. Koonin, K. S. Makarova and L. Aravind, Annu. Rev. Microbiol., 2001, 55, 709–742 CrossRef CAS PubMed.
- N. Takeuchi, K. Kaneko and E. V. Koonin, G3 (Bethesda), 2014, pp. 325–339 Search PubMed.
- L. A. Marraffini and E. J. Sontheimer, Science, 2008, 322, 1843–1845 CrossRef CAS PubMed.
- K. L. Palmer and M. S. Gilmore, mBio, 2010, 1, e00227 CrossRef PubMed.
- K. L. Palmer, V. N. Kos and M. S. Gilmore, Curr. Opin. Microbiol., 2010, 13, 632–639 CrossRef CAS PubMed.
- D. Bikard, A. Hatoum-Aslan, D. Mucida and L. A. Marraffini, Cell Host Microbe, 2012, 12, 177–186 CAS.
- P. Jorth and M. Whiteley, mBio, 2012, 3, e00309 CrossRef CAS PubMed.
- M. Pleckaityte, M. Zilnyte and A. Zvirbliene, BMC Microbiol., 2012, 12, 301 CrossRef CAS PubMed.
- P. Horvath, A. C. Coute-Monvoisin, D. A. Romero, P. Boyaval, C. Fremaux and R. Barrangou, Int. J. Food Microbiol., 2009, 131, 62–70 CrossRef CAS PubMed.
- S. Chakraborty, A. P. Snijders, R. Chakravorty, M. Ahmed, A. M. Tarek and M. A. Hossain, Mol. Phylogenet. Evol., 2010, 56, 878–887 CrossRef CAS PubMed.
- A. D. Weinberger and M. S. Gilmore, Cell Host Microbe, 2012, 12, 125–126 CAS.
- S. Erdmann and R. A. Garrett, Mol. Microbiol., 2012, 85, 1044–1056 CrossRef CAS PubMed.
- M. J. Lopez-Sanchez, E. Sauvage, V. Da Cunha, D. Clermont, E. Ratsima Hariniaina, B. Gonzalez-Zorn, C. Poyart, I. Rosinski-Chupin and P. Glaser, Mol. Microbiol., 2012, 85, 1057–1071 CrossRef CAS PubMed.
- H. Deveau, R. Barrangou, J. E. Garneau, J. Labonte, C. Fremaux, P. Boyaval, D. A. Romero, P. Horvath and S. Moineau, J. Bacteriol., 2008, 190, 1390–1400 CrossRef CAS PubMed.
- A. F. Andersson and J. F. Banfield, Science, 2008, 320, 1047–1050 CrossRef CAS PubMed.
- J. F. Heidelberg, W. C. Nelson, T. Schoenfeld and D. Bhaya, PLoS One, 2009, 4, e4169 Search PubMed.
- P. Horvath, D. A. Romero, A. C. Coute-Monvoisin, M. Richards, H. Deveau, S. Moineau, P. Boyaval, C. Fremaux and R. Barrangou, J. Bacteriol., 2008, 190, 1401–1412 CrossRef CAS PubMed.
- K. D. Seed, D. W. Lazinski, S. B. Calderwood and A. Camilli, Nature, 2013, 494, 489–491 CrossRef CAS PubMed.
- A. J. Lotka, Elements of Physical Biology, Williams, Wilkins, 1925 Search PubMed.
- V. Volterra, Le cons sur la Theorie Mathematique de la Lutte popur la Vie, Gauthier, Villare, 1931 Search PubMed.
- R. M. May, Stability and Complexity in Model Ecosystems, Princeton University Press, 2001 Search PubMed.
- B. R. Levin, S. Moineau, M. Bushman and R. Barrangou, PLoS Genet., 2013, 9, e1003312 CAS.
- F. S. Berezovskaya, Y. I. Wolf, E. V. Koonin and G. P. Karev, Biol. Direct, 2014, 9, 13 CrossRef PubMed.
- B. R. Levin, PLoS Genet., 2010, 6, e1001171 Search PubMed.
- W. Jiang, I. Maniv, F. Arain, Y. Wang, B. R. Levin and L. A. Marraffini, PLoS Genet., 2013, 9, e1003844 CAS.
- J. O. Haerter and K. Sneppen, mBio, 2012, 3, e00126 CrossRef CAS PubMed.
- J. O. Haerter, A. Trusina and K. Sneppen, J. Virol., 2011, 85, 10554–10560 CrossRef CAS PubMed.
- J. Iranzo, A. E. Lobkovsky, Y. I. Wolf and E. V. Koonin, J. Bacteriol., 2013, 195, 3834–3844 CrossRef CAS PubMed.
- L. M. Childs, N. L. Held, M. J. Young, R. J. Whitaker and J. S. Weitz, Evolution, 2012, 66, 2015–2029 CrossRef PubMed.
- W. E. England and R. J. Whitaker, Biochem. Soc. Trans., 2013, 41, 1431–1436 CAS.
- L. M. Childs, W. E. England, M. J. Young, J. S. Weitz and R. J. Whitaker, PLoS One, 2014, 9, e101710 Search PubMed.
- C. L. Sun, R. Barrangou, B. C. Thomas, P. Horvath, C. Fremaux and J. F. Banfield, Environ. Microbiol., 2013, 15, 463–470 CrossRef CAS PubMed.
- A. D. Weinberger, C. L. Sun, M. M. Plucinski, V. J. Denef, B. C. Thomas, P. Horvath, R. Barrangou, M. S. Gilmore, W. M. Getz and J. F. Banfield, PLoS Comput. Biol., 2012, 8, e1002475 CAS.
- A. Kupczok and J. P. Bollback, BMC Evol. Biol., 2013, 13, 54 CrossRef PubMed.
- J. He and M. W. Deem, Phys. Rev. Lett., 2010, 105, 128102 CrossRef.
- P. Han, L. R. Niestemski, J. E. Barrick and M. W. Deem, Phys. Biol., 2013, 10, 025004 CrossRef PubMed.
- M. Djordjevic and K. Severinov, Biol. Direct, 2012, 7, 24 CrossRef CAS PubMed.
- M. Djordjevic, BioSystems, 2013, 112, 139–144 CrossRef CAS PubMed.
- W. Jiang, D. Bikard, D. Cox, F. Zhang and L. A. Marraffini, Nat. Biotechnol., 2013, 31, 233–239 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |