The yeast galactose network as a quantitative model for cellular memory
Sarah R.
Stockwell
a,
Christian R.
Landry
b and
Scott A.
Rifkin
 *a
*a
aSection of Ecology, Behavior, and Evolution, Division of Biology, University of California, San Diego, La Jolla, CA 92093-0116, USA. E-mail: sarifkin@ucsd.edu; Tel: +1 858 822 5748
bDépt. de Biologie, Institut de Biologie Intégrative et des Systèmes/PROTEO, Université Laval, Québec (Québec) G1V 0A6, Canada
First published on 14th October 2014
Abstract
Recent experiments have revealed surprising behavior in the yeast galactose (GAL) pathway, one of the preeminent systems for studying gene regulation. Under certain circumstances, yeast cells display memory of their prior nutrient environments. We distinguish two kinds of cellular memory discovered by quantitative investigations of the GAL network and present a conceptual framework for interpreting new experiments and current ideas on GAL memory. Reinduction memory occurs when cells respond transcriptionally to one environment, shut down the response during several generations in a second environment, then respond faster and with less cell-to-cell variation when returned to the first environment. Persistent memory describes a long-term, arguably stable response in which cells adopt a bimodal or unimodal distribution of induction levels depending on their preceding environment. Deep knowledge of how the yeast GAL pathway responds to different sugar environments has enabled rapid progress in uncovering the mechanisms behind GAL memory, which include cytoplasmic inheritance of inducer proteins and positive feedback loops among regulatory genes. This network of genes, long used to study gene regulation, is now emerging as a model system for cellular memory.
Introduction
In a heterogeneous and changing environment, cells benefit from storing information about past conditions in order to respond appropriately to new circumstances. Cells that are part of a larger organism record transient developmental signals during differentiation,1 and free-living microbes use stored information to follow signaling gradients2 and find and use nutrients efficiently. While animals can store information in the pattern of connections among neurons, single cells retain it in the concentrations and interactions of specific molecules. In recent years, new genetic tools and modeling approaches have allowed researchers to probe deeply into the molecular mechanisms of cellular memory. Many of these studies have capitalized on one of the primary model systems in biology to investigate how eukaryotic cells track and remember their environment: the galactose (GAL) network in budding yeast (Saccharomyces cerevisiae).The GAL network helps govern how yeast cells use and decide between some available carbon sources. Yeast cells grow best on glucose, a simple sugar that can directly enter glycolysis. If glucose is unavailable but galactose is present, cells can import galactose instead and enzymatically modify it for use as fuel. The GAL network is a small set of genes that regulates and performs galactose import and metabolism. Using galactose requires cells to devote substantial extra resources to making GAL mRNA and proteins, so the GAL enzymes are under tight control, preventing the cell from diverting resources to galactose metabolism when glucose is abundant. When galactose is the sole carbon source, the galactose-metabolizing enzymes are expressed at 1000 times their level in glucose,3 making them some of the most tightly regulated proteins in yeast.
When the concentrations of glucose and galactose change, cells alter the expression of the GAL genes in response. Unexpectedly, recent experiments have revealed that the response of yeast cells to current nutrient conditions depends on which nutrients were available several generations in the past. In this review we classify and discuss mechanisms behind this cellular memory.
Research on GAL memory has so far uncovered two kinds of cellular memory. These are generated by multiple molecular mechanisms. Reinduction memory accelerates the transition to galactose metabolism after previous experience of galactose. Yeast cells induce GAL genes more quickly and with less cell-to-cell variation during induction if they were exposed to galactose within the previous 12 hours.4Persistent memory affects the ability of naive cells to respond to new galactose. Yeast cells induce or fail to induce the GAL genes when switched to galactose depending on the media in which they had been cultured beforehand. Authors disagree on whether the cells retain this fate indefinitely.5–7
Tight control of GAL expression has made the GAL genes a canonical model system for studying gene regulation,3,8–10 as well as a workhorse tool in molecular biology for manipulating gene expression. As a result, it is one of the most thoroughly studied gene networks in eukaryotes,8 with hundreds of scientific publications ranging over more than 100 years.11 Researchers have parlayed this detailed knowledge of the GAL system into rapid progress in identifying general mechanisms behind cellular memory. Such mechanisms include interlocking regulatory feedback loops, secondary and overlapping functions of network proteins, and subtle effects of chromatin modification. We synthesize recent experimental and theoretical work on cellular memory in the GAL pathway into an overall framework for investigating GAL induction and discuss the current understanding of its mechanisms.
Overview of the GAL network
The proteins of the GAL network allow a yeast cell to sense glucose and galactose levels and allocate metabolic resources appropriately (Fig. 1) (for reviews, see ref. 3, 10, 12 and 13). The main regulatory components of the network consist of genes coding for a transcriptional activator (Gal4p), a transcriptional repressor (Gal80p), and an inducer (Gal3p). The structural genes include GAL2, which encodes a membrane-bound galactose transporter, and the genes coding for enzymes that modify galactose into glucose-6-phosphate for use in glycolysis (Gal1p, Gal7p, Gal10p). In both experimental and modeling studies, the concentration of GAL1 mRNA or protein, or a fluorescent protein driven by the GAL1 promoter, is often used as a proxy for the overall induction level of the pathway.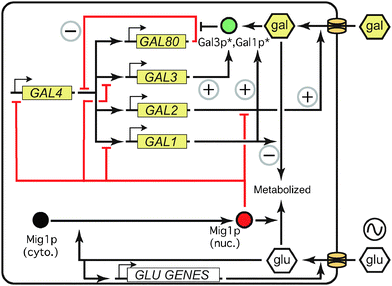 | ||
| Fig. 1 The GAL network is controlled by interlocking positive and negative feedback loops. Asterisks indicate activation by intracellular galactose. Red indicates repressive effects; green represents inducers. Positive and negative feedback loops are marked with circled + and − signs, respectively. See text for details. Adapted with permission from ref. 29. | ||
In the presence of non-repressing, non-inducing carbon sources such as raffinose or glycerol (which apparently neither activate nor repress the GAL network,14 although raffinose contains both glucose and galactose) the activator Gal4p binds specific regulatory sequences upstream of all the GAL genes except GAL4 itself.3 However, the repressor Gal80p binds Gal4p, preventing Gal4p from recruiting other proteins and initiating transcription. When galactose enters the cell, either via a passive diffusion process or facilitated by the transporter Gal2p,15 it binds and activates the inducer Gal3p in the cytoplasm. Activated Gal3p frees Gal4p from the inhibition of Gal80p,16 perhaps by reducing Gal80p dimerization.17 Gal4p then instigates transcription of the target genes, increasing their expression by several-fold (regulatory genes GAL3,80) or up to 1000-fold (structural genes GAL1,2,7,10) over the course of a few hours.3
This regulatory structure includes five feedback loops, which interact to control GAL induction (Fig. 1). GAL2 and GAL3 form positive feedback loops, inducing their own transcription via galactose signaling. Because Gal80p represses the network, its upregulation creates a negative feedback loop, limiting the extent of overall GAL induction. By metabolizing intracellular galactose, the enzymes (Gal1p, Gal7p, Gal10p) reduce the induction signal, contributing to a second, nontranscriptional negative feedback loop. Finally, in addition to its role as an enzyme, Gal1p can serve as an inducer of the GAL network, creating a third positive feedback loop. GAL1 and GAL3 are paralogs, and related species such as Kluyveromyces lactis retain the ancestral state of a single protein that acts as both inducer and enzyme.18 In S. cerevisiae, Gal1p is substantially less effective than Gal3p as an initial inducer; in gal3 mutants, GAL1 takes days rather than hours to induce the GAL pathway.19 However, Gal1p is much more abundant than Gal3p in wild-type cells once induction is underway (upregulated 1000× vs. 3× for Gal3p),3,20 and recent evidence suggests that Gal1p takes over the inducer function of Gal3p in late induction.21 Gal1p and Gal3p thus form interlocking positive feedback loops (Fig. 1). Feedback through the five loops in the GAL network produces the persistent cellular memory of past galactose concentrations.22–24
Glucose represses the GAL network in a number of ways, many of which are mediated by Mig1p (Fig. 1) and/or Gal80p.14,25,26 Glucose signals cytoplasmic Mig1p to enter the nucleus and transcriptionally repress GAL1, GAL3, and GAL4. Glucose is also known to remove the transcriptional machinery from the GAL1 promoter.27 By downregulating the activator Gal4p, glucose lowers the expression of the other GAL genes indirectly. Glucose has also been shown to trigger degradation of membrane-bound Gal2p28 and accelerate the decay of GAL1 and GAL3 transcripts,29 which affects growth rates.30
The galactose network exhibits two types of memory of past conditions
Experiments on the GAL network have uncovered two different kinds of cellular memory. In both cases, the memory was revealed only when experimental designs differed from the usual conditions of GAL experiments, which are steady-state induction levels at very high or low galactose and glucose concentrations. We define two types of memory that were discovered by departing from those conditions. Reinduction memory was revealed by tracking the dynamics of GAL induction over time,4,31 while persistent memory was discovered by culturing the yeast cells in only partially inducing media.5,6 Because cellular memory is only revealed in the aftermath of environmental changes, we detail below both the molecular mechanisms underlying these two types of memory and the experimental conditions that revealed them.Reinduction memory affects transient induction patterns
When yeast cells that have been cultured in glucose overnight are transferred to galactose, they induce the GAL genes slowly, beginning with a 3-hour lag and taking over 8 hours to reach full induction of Gal1p4,20,32 (Fig. 2, point A; Fig. 3A and D). If this induced population is then transferred to back to glucose, GAL transcription stops due to glucose repression. The GAL gene products begin to degrade and/or be diluted out (Fig. 2, point B). However, if the cells are returned once again to galactose within 12 hours, the cells reinduce Gal1p much more quickly than before and without a lag4,20,32,33 (Fig. 2, point C; Fig. 3A and D). Between galactose exposures, the cells divide up to 7 times under glucose repression, so most of the population facing the second galactose induction will have had no direct experience of the first galactose episode. Instead, they inherit a reinduction memory from their ancestors. If the interval of glucose repression lasts longer than 15 hours, the reinduction memory disappears and cells respond naively (slowly) to a new galactose exposure4 (Fig. 2, point D), suggesting that the memory mechanism lasts 12–15 hours.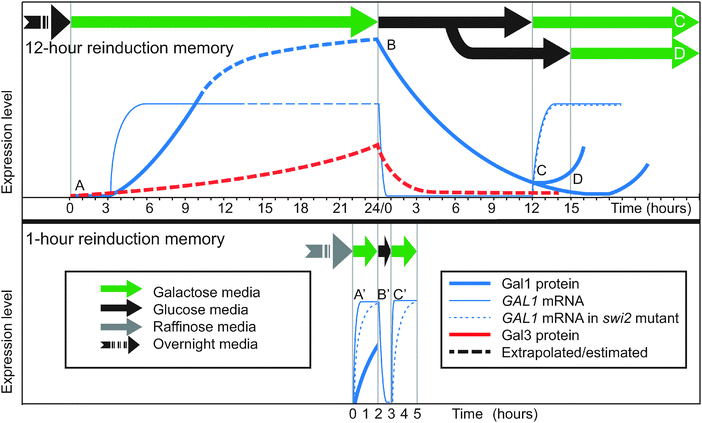 | ||
| Fig. 2 Timecourses of reinduction memory experiments. Top: 12-hour reinduction memory.4,32 Bottom: 1-hour reinduction memory.20 Timecourse curves are approximate. GAL1 and GAL3 mRNA and proteins are not on the same vertical scale. (A) Primary induction after overnight glucose. (A′) Primary induction after overnight raffinose. (B, B′) Glucose repression. (C) Secondary induction after 12 hours is faster than primary induction. (C′) Secondary induction after 1 hour is faster than primary induction for wild-type. Secondary induction is the same as primary induction (A′) for swi2Δ, indicating loss of the memory phenotype in the mutant. (D) Secondary induction after 15 hours is as slow as primary induction. Data sources (“WT” = wild-type): (A) Gal1p:4 Fig. 1c,14 Fig. 6. GAL1 WT:32 Fig. 1a. (A′) Gal1p:58 Fig. 4, in a neutral medium similar to raffinose. GAL1 WT:20 Fig. 6 (GAL1) and Fig. 2 (GAL10, similar). GAL1 swi2Δ:20 Fig. 6. (B, B′) Gal1p:4 Fig. S3A. GAL1:33 Fig. 2c. (C) Gal1p:4 Fig. 1c. GAL1 WT:32 Fig. 1c,20 Fig. 1a. GAL1 swi2Δ:20 Fig. 1b. (C′) GAL1 WT:20 Fig. 6 (GAL1), Fig. 2 (GAL10, similar). GAL1 swi2Δ:20 Fig. 6c. (D) Gal1p:4 Fig. S3B. | ||
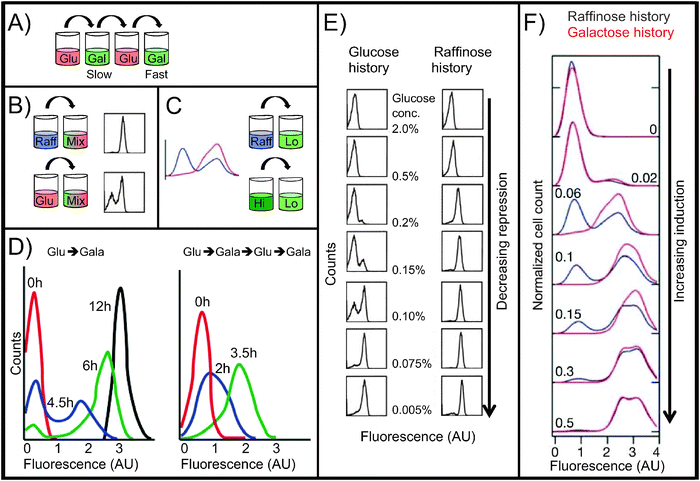 | ||
| Fig. 3 Reinduction and persistent memory. All histograms are flow cytometry measurements of single-cell fluorescence intensity of GAL gene reporters. Fluorescence intensity is on a log10 scale. All strains are wild-type except for GAL reporters. (A) Reinduction memory experiment (12-hour):4 glucose, galactose (slow GAL induction), glucose, galactose (fast GAL induction). (B) Persistent memory experiment:5 raffinose or glucose, then a mix of galactose and glucose (14 h). (C) Persistent memory experiment:6 raffinose or high galactose, then low galactose (27 h). (D) Reinduction memory data. Timecourses of Gal1p-GFP fusion protein expression in cells growing in 2% galactose media. Left: initial galactose induction after pre-growth in glucose (>24 h). Right: reinduction in galactose after pre-growth in glucose (>24 h), galactose (24 h), and glucose again (12 h). The second induction is faster and unimodal, demonstrating reinduction memory. Adapted with permission from ref. 4. (E) Persistent memory under weak repression, showing the effects of pre-growth history in glucose vs. raffinose. Following the pre-growth phase, cells were grown for 14 h in 2% galactose plus the indicated concentration of glucose. Flow cytometry measurements show the fluorescence intensity of a GFP reporter of GAL1 promoter activity. In moderately repressing medium (center images), glucose-history cells are bimodally distributed; raffinose-history cells are unimodal and the ON peak displays a graded response to glucose repression. Adapted with permission from ref. 5. (F) Persistent memory under weak induction, showing the effects of pre-growth for 12 h in raffinose (blue) vs. 2% galactose (red). After pre-growth, cells were grown for 27 h in the indicated concentration of galactose. Flow cytometry measurements show the fluorescence intensity of a YFP reporter of GAL1 promoter activity. Raffinose-history cells are bimodally distributed; galactose-history cells are unimodal and display a graded response to galactose concentration. Adapted with permission from ref. 6. | ||
It was initially unclear whether the mechanism of reinduction memory lay in the state of the GAL1 promoter (e.g., nucleosome composition and position) or in the concentration of primarily cytoplasmic signaling proteins such as Gal3p and Gal1p. To investigate this, Zacharioudakis et al.4 performed an elegant experiment to determine which cell compartment (cytoplasm or nucleus) was required for reinduction memory. Normally when yeast cells mate, the plasma membranes fuse and the two nuclei join together to create a diploid cell. In kar1-1 mutants, the cell bodies fuse but the nuclei remain separate, forming a heterokaryon cell. The researchers grew kar1-1 mutants first to full induction in galactose and then in glucose for 12 hours, giving them reinduction memory. They cultured another strain with GFP-labeled Gal1p in glucose only, making cells that were naive to galactose. When the two cell types mated, they formed a heterokaryon cell in which the naive nucleus with the GAL1-GFP gene was surrounded by the cytoplasm of a previously-induced cell. When placed in galactose, the heterokaryon cells displayed the memory phenotype by inducing Gal1p-GFP rapidly, demonstrating that cytoplasmic factors are necessary and sufficient for reinduction memory.
Additional experiments by the same team showed via mutant analysis that GAL1 is required for reinduction memory, but its paralog GAL3 is not.4 This was surprising, because of Gal3p's role as the initial inducer of the pathway in naive cells. However, the greater abundance of Gal1p coupled with its stability means that even after 6–7 dilutions due to cell division over 12 hours in glucose, its concentration suffices to rapidly reinduce the pathway.4
Chromatin remodelers are important for reinduction memory on short timescales
Although Zacharioudakis and co-workers showed that the cytoplasmic protein Gal1p is the dominant mechanism for reinduction memory, chromatin remodeling factors may play a role downstream of Gal1p signaling. Two proteins known to affect GAL1 promoter nucleosomes have been proposed to be contributors to reinduction memory: SWI/SNF, a conserved chromatin remodeling enzyme,20,31 and H2A.Z, a histone variant.33 In both cases, the proteins appear to facilitate rapid GAL1/10 induction in general,32 as does the RSC protein complex which prevents Gal4p from being blocked by nucleosomes by occupying the Gal4p binding site on the shared GAL1/10 promoter.34 SWI/SNF, in particular, contributes to a variant of reinduction memory that follows very brief glucose repression.To explore the role of SWI/SNF in reinduction memory, researchers tested GAL induction on a shorter timescale than 12–15 hours (Fig. 2, bottom).20,31 Yeast cells were pre-cultured in the non-inducing but also non-repressing sugar raffinose rather than in glucose. Upon transfer to galactose, the cells fully induced GAL1 mRNA in only 40 minutes (Fig. 2, point A′). After this primary induction, yeast were transferred to glucose for a 1-hour repression interval (Fig. 2, point B′), and then returned to galactose for a second induction (Fig. 2, point C′). The second induction was much faster than the first (10 minutes rather than 40 minutes), demonstrating reinduction memory. When the experiment was repeated in a swi2 mutant, the reinduction memory disappeared: the secondary induction (Fig. 2, point C′) was no faster than primary induction (Fig. 2, point A′). These and other experiments showed that 1-hour memory depends on different factors than the 12-hour memory documented by Zacharioudakis and co-workers.4 1-hour memory requires SWI/SNF (SWI2), but not GAL1.20 By contrast, 12-hour memory depends on GAL1,4,20 but not SWI/SNF.20
These differing requirements are related to the fact that the speed of secondary induction depends on the length of the glucose repression interval.4 Induction is very slow after >15 hours in glucose (Fig. 2, point D), moderately slow after 12 hours (Fig. 2, point C), and rapid after 1 hour (Fig. 2, point C′). Chromatin remodeling may only affect very fast inductions. SWI/SNF acts to remove nucleosomes from the GAL1 promoter when induced Gal4p signals it to do so, and nucleosome removal is thought to be a prerequisite for GAL1 transcription.27 Cells lacking SWI/SNF take longer to remove nucleosomes from GAL1,27 which limits how quickly transcription of GAL1 can begin in response to the Gal1p/Gal3p induction signal. Kundu and Peterson proposed a model for how SWI/SNF affects reinduction memory based on their results with the mutant swi2:20 in circumstances where the induction signal is weak (e.g., Gal1p after 12 or more hours of attenuation in glucose), induction is slow, so the delay caused by absent SWI/SNF is inconsequential (Fig. 2, point C). However, when inducers are abundant (e.g., Gal1p and Gal3p after only 1 hour in glucose), the induction signal is strong, and induction is very fast (Fig. 2, point C′). If SWI/SNF is impaired under these circumstances, nucleosome remodeling becomes the rate-limiting step. As a consequence, 1-hour reinduction is as slow as primary induction in swi2 mutants, and these mutants behave, at this short timescale, as if they have no memory. The observation that 12-hour memory requires GAL1 while 1-hour memory does not presumably stems from the fact that Gal3p, the primary but partially redundant inducer, persists for 1 hour in glucose but has degraded or been diluted by cell division after 12 hours.20
Another nucleosome-related protein was also proposed to affect reinduction memory, but recent evidence stands against its playing a role. H2A.Zp is a histone variant that is especially abundant at the nucleosome occupying the Gal4p binding site in the GAL1 promoter.34 When H2A.Z is absent33 or impaired,32,33GAL1 expression in galactose is slower to rise and reaches a lower steady-state concentration.32 The first 2 hours of secondary induction is also slowed in these mutants, which suggested a role for H2A.Z in reinduction memory.33 However, longer induction timecourses showed that the H2A.Z-deficient strains are slower in both primary and secondary GAL1 induction and retain reinduction memory.32 It now appears that the presence of H2A.Zp in promoter nucleosomes helps accelerate GAL1 induction in general. The mechanism of this acceleration is being actively studied.
In summary (Fig. 2 and 3A and D): reinduction memory speeds up GAL induction when fully induced cells are switched to glucose for up to 12 hours and then re-exposed to galactose. Only the transient induction states are affected—reinduction memory does not affect the equilibrium induction level of the cells. The main mechanism for encoding and inheriting this type of cellular memory is the persistence of Gal1p through the glucose repression interval and its secondary role as an inducer, although Gal3p and the SWI/SNF chromatin remodeling complex play a role in 1-hour reinduction memory.
Primary and secondary inductions of yeast cultures differ not only in how quickly they respond to galactose but also in how uniformly they respond.4 In primary induction, the population of cells induces Gal1p in a bimodal way (Fig. 3D, left): for the first several hours in galactose, some cells are OFF (low Gal1p expression) and some quickly turn ON (high Gal1p expression), with few at intermediate levels. By 12 hours, the population reaches a unimodal equilibrium: all cells are ON, and remain so as long as conditions do not change. During secondary induction, the population induces in a unimodal fashion, with all cells increasing their Gal1p expression at approximately the same rate. The two induction profiles both eventually give rise to the same distribution, with all cells ON, but the transient induction patterns depend on whether the cell was previously exposed, and remembers being exposed, to galactose. This phenomenon of different sugar histories producing different population patterns also appears in a second type of GAL memory: persistent memory.
Persistent memory affects equilibrium induction levels when the GAL network is partially induced
In constant high concentrations of galactose, a population of yeast cells eventually settles into an equilibrium in which the GAL network in all cells is highly induced. Similarly, a population in constant low galactose and/or high glucose will become homogeneously repressed. These environmental conditions describe the corners of the carbon landscape shown in Fig. 4, which indicates the long-term induction level of cells in various concentrations of glucose and galactose. The corners of this landscape have been well explored, but interesting patterns emerge in the terra incognita between them. In particular, when the GAL network is only partially induced (by weak glucose repression or low galactose activation), the cell population may be bimodally distributed, with some cells entirely ON and others firmly OFF, but few in between (Fig. 3E, left; Fig. 4, blue arrows). This bimodal induction pattern is a hallmark of persistent memory in the GAL network. This is a form of cellular memory in which some cells, after being transferred from one medium to another, remember, perhaps indefinitely, their prior induction state.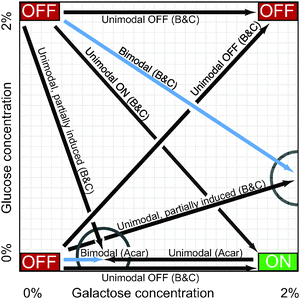 | ||
| Fig. 4 The carbon landscape. The arrows summarize persistent memory experiments described in the text, which measured GAL1 reporter expression at various concentrations of galactose (GAL inducer) and glucose (GAL repressor) in wild-type yeast. Drawing is not to scale. “OFF” and “ON” refer to the expression level of GAL1 at the extremes of glucose and galactose concentration. Cells were incubated in one carbon source (arrow bases) and then transferred to a new medium (arrowheads) for 14–27 h. Blue arrows indicate bimodal expression in the new medium, i.e., some cells are induced and others are not. Black arrows indicate a homogeneous (unimodal) cell population in the new medium. Partial circles highlight points on the carbon landscape where cellular memory of the sugar (carbon source) history affects steady-state GAL expression in the new medium (i.e., persistent memory). Acar:6 B&C.5 | ||
Persistent GAL memory was first documented by Biggar and Crabtree in 20015 when they grew cells overnight in repressing conditions (glucose) or non-inducing/non-repressing conditions (raffinose), and then transferred each population of cells to a second medium containing both galactose and glucose for 14 hours (Fig. 3B). The second medium was fully inducing (high galactose), but also partially repressing (moderate glucose). In this second environment, cells adopted different induction patterns depending on the sugar in which they had initially grown (Fig. 3E, center images). Cells from a repressing glucose history had bimodally distributed values of GAL1 promoter activity, with some cells fully ON while others remained OFF. By contrast, raffinose-history cells formed homogeneous populations in which all cells were induced to the same intermediate level, which varied by glucose concentration—a graded response. The differing induction patterns showed that cells retained memory of their earlier environments, and that they retained this memory for at least 14 hours in the new media. The researchers also tested very high or low glucose concentrations in the second medium (Fig. 3E, top and bottom images), and found that, at extreme glucose concentrations, the new environment overrode the GAL memory.
The authors did not uncover the mechanism by which cells retained a memory of glucose, but they repeated the glucose-history experiment in gal80 and mig1 mutant backgrounds and found an interesting result. Although Mig1p is involved in most of the known mechanisms of glucose repression on GAL genes, cells lacking a functional MIG1 gene preserved the wild-type bimodal induction pattern, indicating that they retained a memory of glucose repression. By contrast, the gal80 strain induced homogeneously, like wild-type cells with a raffinose history. It appears that the Gal80p-dependent mechanisms of glucose repression, such as those affecting Gal3p levels and intracellular galactose concentrations,14,35 play a larger role in this kind of persistent memory.
An experiment by Acar et al.6 found a similar memory effect, but in the absence of glucose. Here, researchers tested weakly inducing conditions instead of partial repression (Fig. 3C and F). They grew cells in raffinose or galactose, then induced them in low-galactose media. In this case, the raffinose-history cells were bimodal after 27 hours, with some cells lingering in their previous OFF state, and the galactose-history cells were unimodal. As before, persistent memory was apparent only in media that partially induced the pathway.
Table 1 summarizes these experiments. Fig. 4 provides a graphical representation, showing the two points on the carbon landscape at which the sugar history determined whether cells responded bimodally or unimodally. The experimenters measured induction after 14–27 hours in the second medium, suggesting that GAL memory in these conditions may persist for a very long time.
| Paper | History media | Induction media | Induction: wild-type | Induction: mutant |
|---|---|---|---|---|
| Acar et al.6 | 2% raffinose, 12 h | 2% raffinose + 0–0.5% galactose, 27 h | Intermediate galactose: bimodal. High/low galactose: unimodal ON/OFF [2a]. | gal2Δ: similar to WT but with lower ON peak [2b]. |
| GAL3-constitutive: unimodal, graded by galactose concentration; same as galactose history [2c]. | ||||
| GAL80-constitutive: all cells OFF in all galactose concentrations [2d]. | ||||
| 2% raffinose + 2% galactose, 12 h | 2% raffinose + 0–0.5% galactose, 27 h | Unimodal, graded by galactose concentration [2a]. | gal2Δ: similar to WT but somewhat bimodal at some galactose concentrations [2b]. | |
| GAL3-constitutive: unimodal, graded by galactose concentration; same as raffinose history [2c]. | ||||
| GAL80-constitutive: unimodal ON or OFF at most galactose concentrations [2d]. | ||||
| Biggar & Crabtree5 | 2% glucose, overnight | 2% galactose + 0–2% glucose, 14 h | Bimodal at intermediate glucose (0.1–0.2%). Unimodal at more extreme concentrations [4, 6a]. | mig1Δ: similar to WT except bimodal at slightly lower glucose (0.075%) [5a]. gal80Δ: unimodal, graded by glucose concentration [5b]. |
| 2% raffinose, overnight | 2% galactose + 0–2% glucose, 14 h | Unimodal, graded by glucose concentration [6b]. |
Model predictions and experimental tests implicate feedback loops in persistent memory
The bimodal induction patterns offered a clue to the mechanism underlying this persistent memory. A population with two widely separated expression states that persist despite changes—that are remembered—can indicate the existence of a bistable system.7,36 In such a system, cells can switch between states (ON and OFF) only when strongly perturbed. A weakly repressing or inducing medium may not be sufficient to push some cells out of the stable state they occupied in an earlier environment. This can produce the bimodal pattern observed in the experiments described above: some cells remain in their former OFF state despite the new conditions. Bistable networks are often composed of positive feedback loops, and experiments and dynamical models of the GAL network have implicated the multiple feedback loops of the GAL network as a mechanism behind history-dependent bimodal induction patterns.The induction dynamics of the GAL pathway have been a popular subject for modelers.6,7,23,24,29,37–41 Models of how positive feedback among GAL genes can lead to history-dependent bimodality have tried to establish how each of four transcriptional feedback loops (GAL1, GAL2, GAL3, GAL80) strengthens or attenuates persistent memory.6,23,24,41 These models have been informed by experiments with mutants that disable GAL regulatory genes or hold the genes' expression constant to disrupt feedback loops.
The strongest candidate for a positive feedback loop that could confer bistability and thereby memory is the loop involving GAL3, the primary inducer of the network (Fig. 1). Acar et al.6 created a constitutive mutant of this gene that retained the coding sequences, but that was driven by a doxycycline-inducible promoter. This allowed the expression of Gal3p to be tuned to a desired constant level, preserving its regulatory function but abolishing its feedback. The GAL network in these mutants lost all its dependence on history—the steady-state GAL1 expression of raffinose-grown and galactose-grown cells was identical and the populations induced gradually and unimodally. The researchers concluded that the GAL3 positive feedback loop was critical to maintaining cellular memory.
The same group tested a constitutive GAL80 mutant controlled in the same manner. By contrast, this mutation widened the range of history-dependent galactose concentrations and drove the ON and OFF peaks farther apart. The mutant intensified persistent memory, demonstrating that negative feedback of GAL80 in normal cells could work by attenuating a bistable switch that encodes memory. The authors suggested that this negative feedback in wild-type GAL80 increases the rate at which stochastic fluctuations in the cell succeed in pushing the GAL network out of its historical induction state, resulting in a cell losing its memory of that state. They used a model of stochastic switching rates to estimate the conditions under which cells would tend to stick in their original induction state and display persistent memory. Later modeling work42 demonstrated that stochasticity can narrow the parameter range in which bistability-based memory is observed, providing theoretical support for the GAL80 feedback explanation. Another group explored switching times in more detail in a system with very attenuated GAL feedback and with constitutive Gal80p concentrations low enough that the dilution of cytoplasmic Gal80p by cell division governed whether and when cells turned the GAL network on.43 Finally, a recent paper showed that the frequency and size of transcriptional bursts can play an important role in the strength of feedback and memory in cells with synthetic GAL promoters.44 It remains to be seen how much of a role stochastic effects play in GAL induction of wild-type cells.
Ramsey and colleagues24 used a constitutive double mutant of GAL80 and GAL3 to measure the joint effects of these negative and positive feedback loops. The experiment and accompanying model showed that in wild-type populations, the two feedback loops reduce cell-to-cell variability both during the induction process and at steady state. The findings agreed with Acar et al.6 in that both papers found more population heterogeneity in the GAL80 loop mutant, but Ramsey et al.'s explanation for this was different: their model suggested that negative feedback of wild-type GAL80 reduces expression noise. If this is true, then the GAL80 negative feedback may act to strengthen memory in cells where Gal3p is held constant (by reducing the stochastic fluctuations that permit cells to change state), but act to weaken memory in cells with intact Gal3p feedback (by attenuating the GAL3 positive feedback loop).
These two papers also illustrate the fact that yeast strains vary substantially in their sensitivity to galactose and thus in the conditions under which they display memory. Ramsey et al. measured a timecourse of one of the conditions that had produced bimodality in the Acar et al. paper (raffinose-grown cells induced in 0.1% galactose) and found unimodal induction at all timepoints instead. Song 2010 et al. shed light on this apparent contradiction by testing GAL induction in BY4741/BY4742 diploid cells, which are similar to Ramsey's strain but distinct from the W303 strain used by Acar et al. While Acar et al. found bimodality in W303 between 0.02–0.3% galactose, Song et al. found clear bimodality in BY4741/BY4742 only between 0.005–0.0087% galactose, suggesting that Ramsey's galactose concentration overshot the bimodal region for the sensitive BY4741/BY4742 strain.
In addition to GAL3 and GAL80, other members of the GAL network may play a role in persistent memory. A recent study has found evidence that the GAL1 feedback loop is also important in maintaining persistent memory and bimodality once the network is activated.23 This is consistent with microscopy results suggesting that Gal1p takes the place of Gal3p in blocking Gal80p repression during late induction.21
The transporter Gal2p regulates intracellular galactose and thus the level of activated inducer, forming another positive feedback loop (Fig. 1). An analogous protein (lactose permease) confers bistability on the lac operon in E. coli under some circumstances,45,46 so one might expect that Gal2p would be a source of bistability in yeast. Two groups independently measured the steady-state induction patterns of gal2 deletion mutants in the same strain background (W303), but in different environmental conditions. They interpreted their results quite differently.13 The first group6 found that gal2 mutants retained memory and raffinose-history-dependent bimodality, and concluded that the GAL2 loop was not required for persistent memory. The second group47 measured the mutants with a raffinose/sucrose history and found that the position of the ON peak depended on galactose concentrations—a graded, but still bimodal, response. They inferred a critical role for GAL2 feedback in bimodal induction. The difference between the two interpretations may lie in the fact that the groups measured different ranges of galactose concentrations (and perhaps also in the fact that sucrose is broken down into glucose and fructose). The first group found an ON peak in a consistent position for galactose from 0–0.3%, and the second group found a graded ON peak for galactose levels between 0.2–3.0%. Where the galactose concentrations overlapped, the data were similar. This confusion illustrates that the GAL network can display qualitatively different behavior over different parts of its dynamic range, demonstrating that the dynamic behavior of this molecular network is finely attuned to environmental conditions and that apparently incommensurate results with diverging implications for the mechanisms of memory can often be reconciled by careful attention to where the cells are located on the carbon landscape (Fig. 4).
It is worth noting that while feedback-mediated bistability is a powerful and popular hypothesis for explaining persistent memory, it has not been conclusively shown to be the mechanism. Most authors have measured only single timepoints in the induction timecourse.6,7,41,47 These snapshots do reveal history dependence and memory, because the cell populations display different induction patterns depending on the media in which they were pre-grown. However, the Zacharioudakis et al. reinduction memory experiments4 demonstrate that GAL induction can be transiently history-dependent without being bistable. A convincing demonstration of true bistability would be to measure the induction pattern of the population over time and show that cells switch from OFF to ON and vice versa. Acar et al.6 found this to be the case in a mutant with elevated switching rates that was transferred from raffinose to low galactose, but to our knowledge an analogous experiment has not been performed in a wild-type strain.
In summary: persistent memory seems to be encoded by the positive feedback loops of GAL1, GAL3, and perhaps GAL2, which combine to create a bimodal switch. The GAL80 negative feedback loop apparently weakens the memory effect. Further experiments are needed to learn more about how GAL protein levels fluctuate in individual cells, either preserving or dissipating cellular memory.
Both types of memory arise from dynamic interactions between signaling proteins and the environment
Both reinduction memory and persistent memory rely on the dynamics of cytoplasmic signaling proteins. Reinduction memory has a simple primary mechanism—Gal1p is a known inducer—but it was a surprise to find it playing a critical role here, because in the conditions under which the GAL network is usually studied (initial induction in naive cells), its effect is dwarfed by that of its paralog Gal3p. Its importance to reinduction lies in its abundance in fully induced cells and its stability that allow it to persist at biologically significant levels through several cell divisions.4 The memory persists as long as Gal1p lingers in the descendants of cells that were once induced. Presumably, the initial induction level should dictate how long reinduction memory lasts, although to our knowledge this has not been tested. The SWI/SNF nucleosomal remodeling complex plays a subsidiary role in short-term reinduction memory by accelerating GAL1 transcriptional initiation. Cytoplasmic inheritance like that of Gal1p is a general mechanism of memory in other organisms,48 making the GAL network a powerful model for studying this phenomenon.Reinduction memory fades with time because it is not actively maintained. By contrast, persistent memory is perpetuated by multiple feedback loops that “lock in” the induction state, perhaps indefinitely. Positive feedback loops that produce a bimodally distributed population and preserve an earlier state of the system are a network motif that has received a great deal of attention in systems biology for its switch-like properties. Many of the biological examples of such switches are in prokaryotes and/or are synthetic,49–53 so a tractable, naturally occurring switch in a eukaryote would be a welcome addition to the menagerie. Insights into feedback regulation learned from the GAL network will extend beyond microbes. Feedback loops are an established mechanism for setting up54–56 or remembering1 stable expression patterns during animal development, and for regulating checkpoints in the cell cycle.57 They seem to form a general mechanism of cellular memory, for which the GAL network is a useful experimental model.
Conclusions
For many decades, the GAL network has been a window into mechanisms of transcriptional control and signal transduction. Recent experiments have shown that it has more to teach: not only about how cells respond to their current circumstances, but also about how they remember their pasts. Many open questions remain, not least the mystery of why, whether, and under what circumstances galactose memory is functionally important in the ecology of S. cerevisiae. One possible benefit of persistent memory of glucose and raffinose environments lies in the fact that GAL induction requires the cell to make a substantial investment in mRNA and protein synthesis. If the preferred carbon source (glucose) has recently been available, it may be a good strategy for at least some of the cell population to delay making the investment in case the preferred sugar becomes available again. If cells are coming from a poorer carbon source (raffinose, glycerol), the benefit of the prior metabolic strategy is lower so the choice is clear, and the whole population invests immediately in taking advantage of the new galactose resource. Our limited knowledge of microbial ecology currently prevents us from definitively answering the question of why yeast displays galactose memory. Nevertheless, the abundant resources available for the GAL network have made it possible to study how memory works in this model system. As new experiments test and develop the concepts that have emerged from the study of the GAL network, this classic pathway will continue to shape our thinking about cellular memory.Conflicts of interest
The authors declare that they have no conflict of interest.Glossary
Chromatin remodeling: changes to DNA-bound nucleosomes, such as adding or removing small molecules to histones or replacing one histone with another, that can affect transcription of nearby genes.Carbon source: a nutrient molecule, such as a sugar, metabolized by the cell to provide energy and carbon for the new molecules it must synthesize.
Constitutive promoter: a promoter that drives continual transcription, making the expression level of its target gene approximately constant.
Reinduction memory: faster reinduction of the GAL network upon its second exposure to galactose, as a result of the first exposure. Reinduction memory affects transient induction, not steady-state induction levels.
Persistent memory: a population of cells adopting an apparently steady-state distribution of induction states that is different (e.g., bimodal/unimodal) depending on what carbon source it was previously grown in.
Steady state: an equilibrium condition in which the concentrations of molecules are not changing with time.
Bimodal distribution: a population with two peaks, indicating two distinct subgroups with different values of the characteristic being measured (e.g., fluorescence intensity).
Unimodal distribution: a population with a single peak, indicating that all members of the population share approximately the same value for the characteristic being measured (e.g., fluorescence intensity).
Graded induction: the position of the ON peak changes gradually with the conditions (glucose/galactose) or with time.
Bistable: a system characterized by two widely separated states that are each stable; that is, a cell in one state tends to return to that state when perturbed or in some new conditions, creating memory. Positive feedback loops are a common mechanism for creating bistability.
Acknowledgements
We thank Dan Pollard and Harris Lazaris for their valuable comments. SRS is a San Diego IRACDA Fellow supported by an NIH K12 GM068524 grant award. SAR and CRL were funded by HFSP RGY0073/2010. SAR is funded by NIH: P50 GM085764 to the San Diego Center for Systems Biology. CRL is a CIHR New Investigator and is funded by CIHR GMX-191597.References
- E. H. Davidson, J. P. Rast, P. Oliveri, A. Ransick, C. Calestani, C. H. Yuh, T. Minokawa, G. Amore, V. Hinman, C. Arenas-Mena, O. Otim, C. T. Brown, C. B. Livi, P. Y. Lee, R. Revilla, A. G. Rust, Z. Pan, M. J. Schilstra, P. J. Clarke, M. I. Arnone, L. Rowen, R. A. Cameron, D. R. McClay, L. Hood and H. Bolouri, Science, 2002, 295(5560), 1669–1678 CrossRef CAS PubMed.
- N. Vladimirov and V. Sourjik, Biol. Chem., 2009, 390(11), 1097–1104 CrossRef CAS PubMed.
- D. Lohr, P. Venkov and J. Zlatanova, FASEB J., 1995, 9(9), 777–787 CAS.
- I. Zacharioudakis, T. Gligoris and D. Tzamarias, Curr. Biol., 2007, 17(23), 2041–2046 CrossRef CAS PubMed.
- S. R. Biggar and G. R. Crabtree, EMBO J., 2001, 20(12), 3167–3176 CrossRef CAS PubMed.
- M. Acar, A. Becskei and A. van Oudenaarden, Nature, 2005, 435(7039), 228–232 CrossRef CAS PubMed.
- C. Song, H. Phenix, V. Abedi, M. Scott, B. P. Ingalls, M. Kaern and T. J. Perkins, PLoS Comput. Biol., 2010, 6(3), e1000699 Search PubMed.
- J. A. Barnett, Yeast, 2004, 21(9), 703–746 CrossRef CAS PubMed.
- C. A. Sellick, R. N. Campbell and R. J. Reece, Int. Rev. Cell Mol. Biol., 2008, 269, 111–150 CAS.
- A. Traven, B. Jelicic and M. Sopta, EMBO Rep., 2006, 7(5), 496–499 CrossRef CAS PubMed.
- F. Dienert, Ann. Inst. Pasteur, 1900, 14, 139–189 Search PubMed.
- M. Johnston and M. Carlson, in The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression, ed. E. Jones, J. Pringle and J. Broach, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY 193-281, 1992 Search PubMed.
- P. J. Bhat and R. S. Iyer, J. Biosci., 2009, 34(4), 513–522 CrossRef CAS.
- M. Johnston, J. S. Flick and T. Pexton, Mol. Cell. Biol., 1994, 14(6), 3834–3841 CAS.
- J. Ramos, K. Szkutnicka and V. P. Cirillo, J. Bacteriol., 1989, 171(6), 3539–3544 CAS.
- O. Egriboz, F. Jiang and J. E. Hopper, Genetics, 2011, 189(3), 825–836 CrossRef CAS PubMed.
- O. Egriboz, S. Goswami, X. Tao, K. Dotts, C. Schaeffer, V. Pilauri and J. E. Hopper, Mol. Cell. Biol., 2013, 33(18), 3667–3674 CrossRef CAS PubMed.
- M. Rubio-Texeira, FEMS Yeast Res., 2005, 5(12), 1115–1128 CrossRef CAS PubMed.
- O. Winge and C. Roberts, C. R. Trav. Lab. Carlsberg, Ser. Physiol., 1948, 24, 263–315 Search PubMed.
- S. Kundu and C. L. Peterson, Mol. Cell. Biol., 2010, 30(10), 2330–2340 CrossRef CAS PubMed.
- D. Abramczyk, S. Holden, C. J. Page and R. J. Reece, Eukaryotic Cell, 2012, 11(3), 334–342 CrossRef CAS PubMed.
- M. Acar, J. T. Mettetal and A. van Oudenaarden, Nat. Genet., 2008, 40(4), 471–475 CrossRef CAS PubMed.
- O. S. Venturelli, H. El-Samad and R. M. Murray, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(48), E3324–E3333 CrossRef CAS PubMed.
- S. A. Ramsey, J. J. Smith, D. Orrell, M. Marelli, T. W. Petersen, P. de Atauri, H. Bolouri and J. D. Aitchison, Nat. Genet., 2006, 38(9), 1082–1087 CrossRef CAS PubMed.
- J. O. Nehlin, M. Carlberg and H. Ronne, EMBO J., 1991, 10(11), 3373–3377 CAS.
- M. Carlson, Curr. Opin. Microbiol., 1999, 2(2), 202–207 CrossRef CAS.
- G. O. Bryant, V. Prabhu, M. Floer, X. Wang, D. Spagna, D. Schreiber and M. Ptashne, PLoS Biol., 2008, 6(12), 2928–2939 CAS.
- J. Horak and D. H. Wolf, J. Bacteriol., 1997, 179(5), 1541–1549 CAS.
- M. R. Bennett, W. L. Pang, N. A. Ostroff, B. L. Baumgartner, S. Nayak, L. S. Tsimring and J. Hasty, Nature, 2008, 454(7208), 1119–1122 CrossRef CAS PubMed.
- B. L. Baumgartner, M. R. Bennett, M. Ferry, T. L. Johnson, L. S. Tsimring and J. Hasty, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(52), 21087–21092 CrossRef CAS PubMed.
- S. Kundu, P. J. Horn and C. L. Peterson, Genes Dev., 2007, 21(8), 997–1004 CrossRef CAS PubMed.
- J. E. Halley, T. Kaplan, A. Y. Wang, M. S. Kobor and J. Rine, PLoS Biol., 2010, 8(6), e1000401 Search PubMed.
- D. G. Brickner, I. Cajigas, Y. Fondufe-Mittendorf, S. Ahmed, P. C. Lee, J. Widom and J. H. Brickner, PLoS Biol., 2007, 5(4), e81 Search PubMed.
- M. Floer, X. Wang, V. Prabhu, G. Berrozpe, S. Narayan, D. Spagna, D. Alvarez, J. Kendall, A. Krasnitz, A. Stepansky, J. Hicks, G. O. Bryant and M. Ptashne, Cell, 2010, 141(3), 407–418 CrossRef CAS PubMed.
- J. M. Gancedo, Microbiol. Mol. Biol. Rev., 1998, 62(2), 334–361 CAS.
- J. E. J. Ferrell, Curr. Opin. Cell Biol., 2002, 14(2), 140–148 CrossRef CAS.
- P. de Atauri, D. Orrell, S. Ramsey and H. Bolouri, Syst. Biol., 2004, 1(1), 28–40 CrossRef CAS.
- M. Verma, P. J. Bhat, S. Bhartiya and K. V. Venkatesh, Eur. J. Biochem., 2004, 271(20), 4064–4074 CrossRef CAS PubMed.
- V. V. Kulkarni, V. Kareenhalli, G. A. Viswananthan and M. Riedel, Syst. Synth. Biol., 2011, 5(3–4), 97–104 CrossRef PubMed.
- R. Apostu and M. C. Mackey, J. Theor. Biol., 2012, 293, 219–235 CrossRef CAS PubMed.
- M. Acar, B. F. Pando, F. H. Arnold, M. B. Elowitz and A. van Oudenaarden, Science, 2010, 329(5999), 1656–1660 CrossRef CAS PubMed.
- Z. Cheng, F. Liu, X.-P. Zhang and W. Wang, FEBS Lett., 2008, 582, 3776–3782 CrossRef CAS PubMed.
- B. B. Kaufmann, Q. Yang, J. T. Mettetal and A. van Oudenaarden, PLoS Biol., 2007, 5(9), e239 Search PubMed.
- C. Hsu, S. Scherrer, A. Buetti-Dinh, P. Ratna, J. Pizzolato, V. Jaquet and A. Becskei, Nat. Commun., 2012, 3, 682 CrossRef PubMed.
- A. Novick and M. Weiner, Proc. Natl. Acad. Sci. U. S. A., 1957, 43(7), 553–566 CrossRef CAS.
- T. J. Perkins, A. Y. Weisse and P. S. Swain, in Quantitative biology: from molecular to cellular systems, ed. M. E. Wall, CRC Press, Boca Raton, Florida, 2012 Search PubMed.
- K. M. Hawkins and C. D. Smolke, J. Biol. Chem., 2006, 281(19), 13485–13492 CrossRef CAS PubMed.
- M. Ptashne, Curr. Biol., 2008, 18(1), R25–R27 CrossRef CAS PubMed.
- T. S. Gardner, C. R. Cantor and J. J. Collins, Nature, 2000, 403(6767), 339–342 CrossRef CAS PubMed.
- F. J. Isaacs, J. Hasty, C. R. Cantor and J. J. Collins, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(13), 7714–7719 CrossRef CAS PubMed.
- H. Kobayashi, M. Kaern, M. Araki, K. Chung, T. S. Gardner, C. R. Cantor and J. J. Collins, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(22), 8414–8419 CrossRef CAS PubMed.
- D. Chen and A. P. Arkin, Mol. Syst. Biol., 2012, 8, 620 CrossRef PubMed.
- J. Kim, K. S. White and E. Winfree, Mol. Syst. Biol., 2006, 2, 68 CrossRef PubMed.
- O. Brandman, J. E. J. Ferrell, R. Li and T. Meyer, Science, 2005, 310(5747), 496–498 CrossRef CAS PubMed.
- J. Jaeger, M. Blagov, D. Kosman, K. N. Kozlov, Manu, E. Myasnikova, S. Surkova, C. E. Vanario-Alonso, M. Samsonova, D. H. Sharp and J. Reinitz, Genetics, 2004, 167(4), 1721–1737 CrossRef CAS PubMed.
- W. Xiong and J. E. J. Ferrell, Nature, 2003, 426(6965), 460–465 CrossRef CAS PubMed.
- J. E. J. Ferrell, Curr. Biol., 2008, 18(6), R244–R245 CrossRef CAS PubMed.
- P. J. Bhat, D. Oh and J. E. Hopper, Genetics, 1990, 125(2), 281–291 CAS.
| This journal is © The Royal Society of Chemistry 2015 |
