Open Access Article
Open Access ArticleA fully integrated microdevice for biobarcode assay based biological agent detection†
Minkyung
Cho
,
Soyi
Chung
,
Yong Tae
Kim
,
Jae Hwan
Jung
,
Do Hyun
Kim
and
Tae Seok
Seo
*
Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-ro, Yuseong-gu, Daejeon 305-701, Republic of Korea. E-mail: seots@kaist.ac.kr
First published on 18th May 2015
Abstract
An integrated microdevice, consisting of a micropump, a passive mixer, a magnetic separation chamber, and a microcapillary electrophoretic channel, was constructed for biobarcode assay based multiplex biological agent detection in a sample-to-answer-out manner within 30 min with high sensitivity.
Bioterrorism and biowarfare cause widespread and critical social damages such as diseases, economic loss, and casualties by biological agents.1,2 Since even trace amounts of biological agents can bring about serious results, their detection in an early stage is of importance to guard public safety.3,4 Despite simple operation of a lateral flow screening assay, the current commercial assay kits still suffer from low sensitivity and low multiplexity. Considering the fatality rate of biological agents at low quantities, an advanced diagnostic tool, which can surpass the performance of the existing kits, should be developed.
Biosensing with DNA/nanoparticle hybrids,5 especially, the biobarcode assay has demonstrated its excellent capability of detecting a low number of cells, nucleic acids and proteins.6–15 In this method, the target pathogens are sandwiched by two particle probes (the antibody labeled magnetic particles, and the antibody and barcode DNA labeled particles), the complexes are separated by an external magnet, and then the barcode DNA is analyzed to identify the target.16 Since even a single particle can hold abundant barcode DNA molecules and variation in the length and sequence of DNA can be controlled, sensitive and multiplex pathogen detection by the biobarcode assay is feasible. Our group has demonstrated a highly sensitive and multiplex biological agent detection by combining the off-chip based biobarcode assay with on-chip capillary electrophoresis in the previous work.6 In this study, we constructed an advanced lab-on-a-chip platform to fully integrate whole processes of the biobarcode assay to diagnose biological agents with low sample consumption, high speed, and point-of-care testing capability. Five biological agents (Bacillus anthracis, Francisella tularensis, Yersinia pestis, Vaccinia virus (VV), Botulinum toxin A (BoNT/A)) were targeted, and monoplex as well as multiplex detection was performed. Regarding the detection method for the released barcode DNAs, we employed microcapillary electrophoresis (μCE) due to its sensitive, rapid, and accurate analytical capability.17 The μCE on a chip is typically operated with four reservoirs (for sample, waste, cathode, and anode) under optimized voltage conditions, and a nanoliter sample volume is separated for generating the amplicon peaks in the electropherogram. The high performance of μCE on a chip has been demonstrated in the fields of forensic human identification, DNA sequencing, and single nucleotide polymorphism analysis.18–21 In particular, to improve the accuracy of peak assignment in the electropherogram, we added bracket ladders which enable us to identify the target peak with high fidelity by calculating the relative elution time ratio. Note that the entire operation could be completed in less than 30 min. Thus, our novel on-chip methodology with a sample-in-answer-out capability would be superior to conventional methods such as enzyme-linked immunosorbent assay or polymerase chain reaction which requires long analysis time, manual operation, bulky instrumentation, and expertise skill.
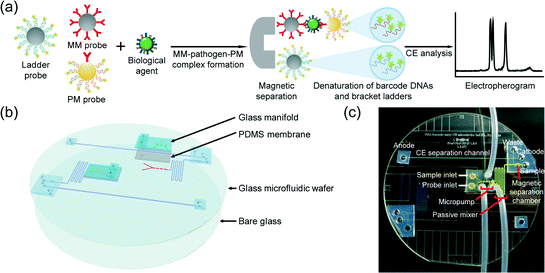
First, two bracket ladder labeled magnetic particles, magnetic microparticles (MMs) and polystyrene microparticles (PMs) for each biological agent were prepared. The lengths of the bracket ladders were 15 bp and 45 bp, and 20, 25, 30, 35, and 40 bp long barcode DNAs were designed for targeting B. anthracis, F. tularensis, Y. pestis, VV, and BoNT/A, respectively. A detailed procedure is described in the ESI.†Fig. 1a shows the overall scheme for the biobarcode assay based biological agent detection. The procedure consists of the conjugation between the biological agent and the MM/PM probes, the separation of the MM–biological agent–PM complexes and the bracket ladder probes, and the analysis of the bracket ladders and barcode DNAs released from the complexes by μCE.
The integrated microdevice was fabricated using a glass–PDMS hybrid, and was composed of four layers: (from top to bottom) a glass manifold, a monolithic PDMS membrane, a micropatterned glass wafer, and a blank glass wafer (Fig. 1b). The glass manifold and the monolithic PDMS membrane were used to actuate micropumping, and the micropatterned glass wafer contained a microchannel and three microvalves for loading the biological sample and the particle probes (red on the top side), a micropillar incorporated passive micromixer to induce the MM–biological agent–PM complexes, a magnetic separation chamber to isolate the complexes and the bracket ladder probes, and a microchannel for μCE separation to analyze the barcode DNAs with the bracket ladders (blue on the bottom side), thereby enabling us to identify the target biological agents with a sample-in-answer-out capability. A digital image of the assembled microdevice is shown in Fig. 1c, and the microfabrication process was described in Fig. S1.†
Before loading the sample and probes in the inlets, the μCE channel was coated with a 50% dynamic coating solution in methanol for 3 min. After flushing out the coating solution, 5% (w/v) linear polyacrylamide (LPA) with 6 M urea in a 1× Tris TAPS EDTA (TTE) buffer was injected from the anode reservoir. The reservoirs of the waste, cathode, and anode were filled with a 1× TTE buffer.
30 μL of the particle probe solution (10 μL of MM, 10 μL of PM, and 5 μL of each bracket ladder probe) and 30 μL of a biological agent sample were loaded in the probe and sample inlet reservoir, respectively. All the solutions were injected into the microfluidic channel through automatic micropumping actuation. They passed through the passive mixer (total length of 11.34 cm) for 20 min to form the MM–pathogen–PM complexes. The produced complexes were captured in the magnetic separation chamber (0.3784 mm3 volume) by an external magnet, and then washed with 60 μL of a 1× TTE buffer to remove excess PM probes. The washing step was executed by loading 60 μL of 1× TTE buffer in the sample inlet and the probe inlet, and passing the buffer from the inlets to the sample reservoir via peristaltic micropump operation. A silicon rubber heater beneath the chip was heated up to 70 °C for 2 min to make denaturing conditions of the LPA as well as dehybridization of the FAM-labeled barcode ssDNAs from the MM–pathogen–PM complexes and the two bracket ladders from the bracket ladder probes. After 30 μL of 1× TTE buffer was loaded in the sample reservoir, high voltage power supply was adjusted for the CE analysis of the denatured barcode and bracket ladder DNAs. The released DNAs were moved from the sample reservoir to the injection channel toward the waste reservoir by applying 1000 V in the waste reservoir and 0 V in the sample reservoir for 15 s. Back-biasing was performed by applying an electric field of 900 V in the waste and sample reservoirs for 10 s, while 0 V for the cathode and 2100 V for the anode reservoir were applied. Separation was conducted by applying 1800 V to the anode and 0 V to the cathode reservoir, while floating was conducted for the sample and waste reservoirs. During the μCE operation, the CE channel was heated at 70 °C with a heater. The fluorescence signal of the FAM-labeled barcode DNAs and bracket ladders was monitored near the anode reservoir using a laser-induced confocal fluorescence microscope (C1si, Nikon, Japan), and the entire process was controlled by an in-house LabVIEW program. For the automatic sample loading, we employed the active peristaltic micropumping system which was composed of three serial microvalves (Fig. 2a). The microvalves from the left to the right were consecutively open and closed, so that a certain amount of the sample (0.0655 μL) could be delivered to the passive mixer channel continuously. We chose a dwell time of 150 ms for valve actuation which could load the sample and probe solution with a flow rate of 3 μL min−1.
For efficient immuno-conjugation between the biological agents and particle probes, we designed a serpentine microchannel in which micropillar structures were incorporated. We controlled the microdot diameter embedded in the passive mixer in the mask design and the wet etching time to produce 200 μm dia. micropillars, which led to turbulent mixing of the sample and probe solutions, effectively forming the complexes (Fig. 2b). As shown in Fig. 2a, the input yellow and blue solutions were blended to reveal the green solution in the serpentine channel, demonstrating high mixing efficiency.
The MM–pathogen–PM complexes and the bracket ladder probes were captured by an external magnet which was located beneath the capture chamber (Fig. 2c). The image of the captured particle probes was enlarged in Fig. 2d. Since a number of particle probes could be isolated, the barcode DNAs and the bracket ladders could generate discernible CE peaks in the electropherogram. Fig. 2e shows a typical μCE channel with a cross design. Through optimized power supplies in the four reservoirs, we could isolate the sample in the intersection part. Fig. 2e shows the scheme of the injection, back-biasing, and separation with supplied voltages, and the corresponding fluorescence images in the intersection of the μCE channel. In particular, the back-biasing step was necessary to isolate the sample plug at the intersection part without the excess sample leaking into the separation channel, which could cause peak broadening. Since the CE channel dimensions were 280 μm × 100 μm × 7 cm (W × H × L), the separated DNA sample volume was roughly 7 nL. The FAM-labeled barcode DNAs and bracket ladders were denatured by heating at 70 °C, and the CE analysis was completed in 3 min. The total analysis time from the sample and probe injection to the laser-induced fluorescence detection of the barcode and bracket ladder DNAs was 30 min.
We designed 15 and 45 bp DNAs for the short and long bracket ladder, respectively, while the 20, 25, 30, 35, 40 bp-long barcode DNAs indicate B. anthracis, F. tularensis, Y. pestis, VV, and BoNT/A (Table S1†). Although the elution time of DNA in the electropherogram mainly depends on its length, the absolute elution time could be varied according to gel matrix conditions, temperature, humidity, and CE operator, causing the possibility of false peak assignments. To improve the accuracy of the peak assignments, we employed the relative elution time ratio (Relu) by using the two bracket ladders, instead of using the absolute elution time. Since the 15 bp and 45 bp bracket ladders are the shortest and the longest among the used DNAs and are labeled to the magnetic beads, the peak of the biobarcode DNAs should always appear in the middle of the two bracket ladders. The Relu was calculated by dividing the elution time difference between the barcode DNA and the 15 bp bracket ladder by the elution time difference between the two bracket ladders. As shown in Table S2,† the elution time ratio of the 20, 25, 30, 35, 40 bp barcode DNAs was 0.1651, 0.3331, 0.5499, 0.6789, and 0.8547, which corresponded to B. anthracis, F. tularensis, Y. pestis, VV, and BoNT/A, respectively.
We performed monoplex B. anthracis, F. tularensis and VV detection on the proposed integrated microdevice (Fig. 3a). The corresponding 20, 25, and 35 bp FAM-labeled barcode DNAs of B. anthracis, F. tularensis and VV were found between the two bracket ladders with an Relu of 0.15, 0.35 and 0.66, respectively, which was consistent with the reference values in Table S2.† As the target concentration increased, the target peak intensity compared to that of the bracket ladders gradually augmented. Even 102 CFU mL−1 (or PFU mL−1) which is equivalent to 3 CFU (or PFU) of the target biological agents could generate a distinguishable target peak in the electropherogram. Thus, the biobarcode assay incorporated microdevice can successfully execute monoplex biological agent detection with high sensitivity and accuracy in a short time. The relative CE peak intensity linearly increased in proportion to the logarithmic pathogen concentration (Fig. S3†).
Encouraged by the success of the monoplex biological agent detection, we furthermore conducted multiplex analysis on the same platform. For multiplex biological agent detection, we first prepared a probe mixture solution and a pathogen mixture solution. The volume of the probe mixture solution is 120 μL, including 10 μL of each bracket ladder probe solution, 10 μL of each MM probe solution, and 10 μL of each PM probe solution. The volume of the sample mixture solution is 50 μL, including 10 μL of B. anthracis, F. tularensis, and Y. pestis biological agent solution (103 CFU mL−1), 10 μL of VV (103 PFU mL−1), and 10 μL of BoNT/A (2 ng mL−1). Thirty μL of the probe mixture solution was loaded in the probe reservoir and 30 μL of the biological agent mixture solution (which is equivalent to 6 CFU of B. anthracis, F. tularensis, and Y. pestis, 6 PFU of VV and 12 pg of BoNT/A) was added to the sample reservoir. Then, the same experimental procedure as described above was followed.
Fig. 3b(i) shows the CE results for duplex B. anthracis and F. tularensis detection by displaying the two target peaks in the electropherogram. Triplex detection for B. anthracis, F. tularensis, and Y. pestis was also successfully conducted as shown in Fig. 3b(ii). Even when the five biological agents were targeted, all the matched peaks were produced (Fig. 3b(iii)). The 5 bp difference of the barcode DNAs was enough to separate the adjacent CE peaks with a good baseline, and the peak assignment was carried out with ease and accuracy due to the presence of the bracket ladders. Thus, the control of the barcode DNA length enables us to perform multiplex detection in the CE analysis, and the multiplexing capability can be expanded by using even longer DNA molecules.
To demonstrate the potential of the real sample analysis on our platform, we prepared the biological agent solution in serum or with the presence of the other pathogens. Fig. 3c(i) is the resultant electropherogram of F. tularensis which was suspended in a serum medium. 100 μL of 104 CFU mL−1F. tularensis was mixed with 100 μL of serum medium, and 30 μL of the mixture solution was used as a sample solution that contains 150 CFU of F. tularensis. The Relu of the present peak confirmed that the biological agent in the sample was F. tularensis. Fig. 3c(ii) is the detection result of F. tularensis (6 CFU) in the presence of other pathogens as interferents. Ten μL of 103 CFU mL−1B. anthracis, F. tularensis, and Y. pestis, 10 μL of 103 PFU mL−1 VV, and 10 μL of 2 ng mL−1 BoNT/A were mixed and 30 μL of the solution was used as a sample. Only the barcode probe set for targeting F. tularensis was added and the same procedure as described above was followed. The desired peak was the only one found in the graph, whose Relu matched with 0.3331, thereby demonstrating the applicability for real sample analysis with high specificity on our proposed diagnostic tool.
Conclusions
We successfully demonstrated the highly sensitive, multiplex, and rapid biological agent detection on a biobarcode assay incorporated microdevice. The integrated microdevice can perform micropumping actuation for sample loading, passive mixing to efficiently induce antigen–antibody interactions, magnetic separation to purify the target–probe complexes, and μCE to analyze the barcode DNAs. Addition of the bracket ladders helps us to interpret the target peak with high fidelity. Not only the monoplex biological agents were detected with a limit-of-detection of 102 CFU mL−1, but also multiplex biological agents could be simultaneously identified. Moreover, the pathogen in serum medium or with the presence of other pathogen interferents could be verified. All the processes from the sample injection to detection were completed in 30 min. In combination with miniaturized peripheral hardware, our integrated microsystem can provide an advanced bioassay platform for point-of-care tests and early diagnosis of biological agents in the future.22–24Acknowledgements
This research was supported by Center for BioNano Health-Guard funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea as a Global Frontier Project (H-GUARD_2013M3A6B2078964), and the Engineering Research Center of Excellence Program of Korea Ministry of Science, ICT & Future Planning (MSIP)/National Research Foundation of Korea (NRF) (2014R1A5A1009799).Notes and references
- S. Syal, Curr. Sci., 2008, 95, 1665–1666 Search PubMed.
- J. Taylor, D. Margaritis, Z. Nasir, H. Borrion and K. Lai, J. Bioterrorism Biodef., 2013, 4, 123 Search PubMed.
- A. Goldenberg, G. Shmueli, R. A. Caruana and S. E. Fienberg, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5237–5240 CrossRef CAS PubMed.
- L. D. Rotz, A. S. Khan, S. R. Lillibridge, S. M. Ostroff and J. M. Hughes, Emerging Infect. Dis., 2002, 8, 225–230 CrossRef PubMed.
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607–609 CrossRef CAS PubMed.
- M. Cho, S. Chung, J. H. Jung, G. Rhie, J. H. Jeon and T. S. Seo, Biosens. Bioelectron., 2014, 61, 172–176 CrossRef CAS PubMed.
- Y. Gao, A. W. Lam and W. C. Chan, ACS Appl. Mater. Interfaces, 2013, 5, 2853–2860 CAS.
- H. D. Hill and C. A. Mirkin, Nat. Protoc., 2006, 1, 324–336 CrossRef CAS PubMed.
- J. H. Jung, G. Y. Kim and T. S. Seo, Lab Chip, 2011, 11, 3465–3470 RSC.
- H. Lee, J. E. Park and J. M. Nam, Nat. Commun., 2014, 5, 3367 Search PubMed.
- B. K. Oh, J. M. Nam, S. W. Lee and C. A. Mirkin, Small, 2006, 2, 103–108 CrossRef CAS PubMed.
- H. Yin, M. Jia, S. Yang, S. Wang and J. Zhang, Toxicon, 2012, 59, 12–16 CrossRef CAS PubMed.
- D. Zhang, M. C. Huarng and E. C. Alocilja, Biosens. Bioelectron., 2010, 26, 1736–1742 CrossRef CAS PubMed.
- J. M. Nam, S. J. Park and C. A. Mirkin, J. Am. Chem. Soc., 2002, 124, 3820–3821 CrossRef CAS PubMed.
- C. S. Thaxton, H. D. Hill, D. G. Georganopoulou, S. I. Stoeva and C. A. Mirkin, Anal. Chem., 2005, 77, 8174–8178 CrossRef CAS PubMed.
- J. M. Nam, C. S. Thaxton and C. A. Mirkin, Science, 2003, 301, 1884–1886 CrossRef CAS PubMed.
- S. J. Kim, S. J. Choi, R. Neelamegam and T. S. Seo, BioChip J., 2010, 4, 42–48 CrossRef.
- R. G. Blazej, P. Kumaresan, S. A. Cronier and R. A. Mathies, Anal. Chem., 2007, 79, 4499–4506 CrossRef CAS PubMed.
- J. Y. Choi, Y. T. Kim, J. Ahn, K. S. Kim, D. G. Gweon and T. S. Seo, Biosens. Bioelectron., 2012, 35, 327–334 CrossRef CAS PubMed.
- Y. T. Kim, J. Y. Choi, Y. Chen and T. S. Seo, RSC Adv., 2013, 3, 8461–8467 RSC.
- P. Liu, J. R. Scherer, S. A. Greenspoon, T. N. Chiesl and R. A. Mathies, Forensic Sci. Int.: Genet., 2011, 5, 484–492 CrossRef CAS PubMed.
- J. Y. Choi, Y. T. Kim, J. Y. Byun, J. Ahn, S. Chung, D. G. Gweon, M. G. Kim and T. S. Seo, Lab Chip, 2012, 12, 5146–5154 RSC.
- Y. T. Kim, Y. Chen, J. Y. Choi, W. J. Kim, H. M. Dae, J. Jung and T. S. Seo, Biosens. Bioelectron., 2012, 33, 88–94 CrossRef CAS PubMed.
- A. Fernández-la-Villa, D. Sánchez-Barragán, D. F. Pozo-Ayuso and M. Castaño-Álvarez, Electrophoresis, 2012, 33, 2733–2742 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The sequence information of the barcode DNAs and bracket ladders, the fabrication process of the integrated microdevice, and the calculation of the relative elution time ratio of barcode DNAs. See DOI: 10.1039/c5lc00355e |
| This journal is © The Royal Society of Chemistry 2015 |