Standing surface acoustic wave (SSAW)-based cell washing†
Sixing
Li
ab,
Xiaoyun
Ding
a,
Zhangming
Mao
a,
Yuchao
Chen
a,
Nitesh
Nama
a,
Feng
Guo
a,
Peng
Li
a,
Lin
Wang
d,
Craig E.
Cameron
bc and
Tony Jun
Huang
*ab
aDepartment of Engineering Science and Mechanics, The Pennsylvania State University, University Park, PA 16802, USA. E-mail: junhuang@psu.edu; Fax: 814 865 9974; Tel: 814 863 4209
bThe Molecular, Cellular and Integrative Biosciences (MCIBS) Graduate Program, The Huck Institutes of the Life Sciences, The Pennsylvania State University, University Park, PA 16802, USA
cDepartment of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA 16802, USA
dAscent Bio-Nano Technologies Inc., State College, PA 16801, USA
First published on 5th November 2014
Abstract
Cell/bead washing is an indispensable sample preparation procedure used in various cell studies and analytical processes. In this article, we report a standing surface acoustic wave (SSAW)-based microfluidic device for cell and bead washing in a continuous flow. In our approach, the acoustic radiation force generated in a SSAW field is utilized to actively extract cells or beads from their original medium. A unique configuration of tilted-angle standing surface acoustic wave (taSSAW) is employed in our device, enabling us to wash beads with >98% recovery rate and >97% washing efficiency. We also demonstrate the functionality of our device by preparing high-purity (>97%) white blood cells from lysed blood samples through cell washing. Our SSAW-based cell/bead washing device has the advantages of label-free manipulation, simplicity, high biocompatibility, high recovery rate, and high washing efficiency. It can be useful for many lab-on-a-chip applications.
Introduction
Cell and bead washing are important experimental procedures widely employed in biological studies and biomedical research. Taking fluorescent cell labeling as an example, a typical protocol requires the labeled cells to be washed after incubation with a fluorochrome.1–3 This cell-washing step is necessary to remove the unreacted fluorochrome and to optimize the staining result. In transplantation of cryopreserved and thawed hematopoietic stem cells (HSCs), washing of HSCs is required for dimethyl sulfoxide (DMSO) removal to decrease the adverse effects associated with DMSO infusion.4,5 Besides cell washing, bead washing has also been routinely used in molecular biology and immunology.6–9 For instance, multiple bead-washing steps are needed to change reagents when affinity-based DNA purification is performed using QIAEX® beads.6Conventionally, cells or beads are washed using centrifugation methods. To ensure adequate cell washing, a typical cell-washing protocol involves centrifuging the cells at 3000 × g for 10 min and for multiple rounds, which can be labor-intensive.10 In addition, cells experience high shear stress during high-speed, long-duration centrifugation processes, which may cause cell damage.11 Research data show that the percent hemolysis of red blood cells (RBCs) washed by centrifugation methods is significantly higher (~0.74%) compared with unwashed RBCs (~0.22%).12 In addition to low biocompatibility, another limitation for the centrifugation approach is its difficulty for in-line integration, which is necessary to realize automatic, micro total analysis systems (μTAS). Thus, the development of a simple, biocompatible, and continuous-flow microfluidic cell/bead washing device will address many unmet needs in cell biology and analytical chemistry.
To overcome the limitations of centrifugation-based cell-washing methods, researchers have been developing microfluidic techniques to wash cells and beads in a continuous flow.13–15 Several passive approaches have been demonstrated based on deterministic lateral displacement,16 microstructure-guided railing,17,18 hydrodynamic filtration,19–21 pinched flow fractionation,22 or inertial microfluidics.23 In these devices, cells or beads passively migrate from the original medium to the wash solution in the microchannels. However, these approaches offer limited control of cell/bead movement since the passive migration is predetermined by the geometry of the microchannel, size of the cell/bead, and/or flow conditions. In order to have better control of the cell/bead movement during the washing process, researchers have made significant efforts to use external forces to manipulate cells/beads, leading to the development of several active cell-washing techniques.24–33 Cells/beads flowing in these devices are subjected to external forces such as magnetic forces,24 dielectrophoretic (DEP) forces,25–27 or acoustic forces.28–33 As a result, beads or cells can be actively extracted from their original medium stream and placed into a wash solution. Among these cell/bead manipulation technologies, acoustic methods offer significant advantages in terms of label-free manipulation, biocompatibility, and versatility.
Recently, our group has utilized standing surface acoustic waves (SSAWs) to accomplish label-free manipulation of various micro/nano-objects (e.g., beads, cells, droplets, microorganisms, and nanowires).34–39 Particularly, a unique configuration of tilted-angle standing surface acoustic wave (taSSAW) was introduced recently, in which the interdigital transducers (IDTs) were inclined at a specific angle to the flow direction.40 Compared with previous SSAW approaches where the IDTs are aligned in parallel with the flow direction,41–43 this taSSAW approach offers a significantly larger lateral displacement for particles. As a result, cells/beads can be effectively separated based on differences in size, density, and/or compressibility.
In this article, we explore the potential application of this taSSAW approach in the development of a SSAW-based cell/bead washing device. We first tested washing 10 μm beads and characterized the performance of our device for bead washing. By optimizing the input voltage, we were able to wash beads with high recovery rate (>98%) and high washing efficiency (>97%). For the demonstration of cell washing, we investigated the purification of white blood cells (WBCs) from lysed blood samples. Preparation of WBCs from whole blood through RBC lysis is widely employed in immunology and clinical diagnosis.44,45 After RBC lysis, WBC washing is necessary to remove the lysis buffer which can be detrimental to WBCs during long-term exposure. In addition, cell debris produced during RBC lysis also needs to be removed to reduce interference and background noise during further WBC analysis. For the optimization of WBC washing, we considered the differences between WBCs and debris in terms of size, density and compressibility and conducted numerical simulations to evaluate washing performances under different input voltages. After numerical simulations, we demonstrated that our SSAW device was able to isolate WBCs from lysed blood samples with high-purity (>97%) while preserving cell integrity. The technique described here is expected to be an ideal continuous-flow cell/bead washing module and has great potential in the development of μTAS.46,47
Working mechanism
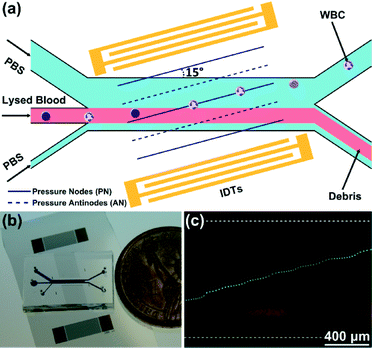
Our SSAW-based cell/bead washing device, shown in Fig. 1, is fabricated by bonding a polydimethylsiloxane (PDMS) microchannel to a piezoelectric substrate; the microchannel is placed between a pair of IDTs and inclined at a specific angle (15°) relative to the IDTs. As shown in Fig. 1(a), when a radio frequency (RF) signal is applied to the pair of IDTs, surface acoustic waves (SAWs) are generated from both IDTs. The SAWs propagate in opposite directions on the substrate surface and leak into the liquid inside the microchannel. The interference between them forms a SSAW field and causes pressure fluctuations in the liquid. As a result, a periodic distribution of pressure nodes (regions of minimum pressure amplitude) and pressure antinodes (regions of maximum pressure amplitude) is formed inside the microchannel. Particles flowing into the SSAW field experience the acoustic radiation force (Fr) and Stokes drag force (Fd), which can be expressed as48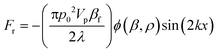 | (1) |
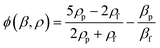 | (2) |
| Fd = −6πηRp(up − uf) | (3) |
Thus, we can study particle movement in a SSAW field based on Newton's second law:
| Fr + Fd = ma | (4) |
Since the microchannel is inclined at a specific angle to the IDTs, cells or beads flowing into the SSAW field will deviate from their original medium stream due to the competition of the acoustic radiation force and Stokes drag force, as shown in Fig. 1(c). As a result, cells or beads can be washed out from the original medium and collected through the upper outlet.
Methods
Device fabrication
Fig. 1(b) shows an optical image of our SSAW-based cell/bead washing device. To fabricate the device, we first deposited a double layer of chrome and gold (Cr/Au, 50 Å/500 Å) on a photoresist-patterned lithium niobate (LiNbO3) wafer (128° Y-cut, 500 μm thick, and double-side polished) using an e-beam evaporator (RC0021, Semicore, USA). Then we used a lift-off technique to expose the pair of IDTs with a period of 200 μm and width of 8 mm. The PDMS microchannel (1 mm wide in the main channel) with three inlets (650, 300, and 150 μm wide, respectively) and two outlets (300 μm wide) was fabricated by standard soft-lithography using SU-8 photoresist (channel height is 75 μm). A Harris Uni-Core 0.75 mm punch was used to drill holes for inlets and outlets. For device bonding, we first placed the PDMS microchannel and the LiNbO3 substrate in a plasma cleaner (PDC001, Harrick Plasma, USA) for 3 min. Immediately after plasma treatment, we aligned the PDMS microchannel with markers on the LiNbO3 substrate in between the IDTs and bonded with a 15° tilt angle between the microchannel and the IDTs. After bonding, we cured the whole device at 65 °C overnight before use.Sample preparation
Both bead and cell samples were used in our experiments. For illustrating bead deviation in the SSAW field, 1 μl of 9.77 μm polystyrene microparticles (Dragon Green, Bangs Laboratory, USA) was diluted in 500 μl of 100 μM Rhodamine B (Sigma, USA) solution with 0.1% sodium dodecyl sulfate (SDS, Sigma, USA) (final bead concentration was around 1.5 × 104 ml−1). For the demonstration of SSAW-based bead washing, 2 μl of 9.77 μm (Dragon Green) and 0.87 μm (Rhodamine WT) polystyrene microparticles (Bangs Laboratory, USA) were mixed in 500 μl of 0.1% SDS solution (final bead concentrations were around 3 × 104 ml−1 and 4 × 107 ml−1, respectively). To characterize the recovery rate and washing efficiency, we mixed 10 μl of 10 μm polystyrene microparticles (Sigma, USA) with 2 μl of 0.87 μm polystyrene microparticles (Rhodamine WT, Bangs Laboratory, USA) in 500 μl of 0.1% SDS solution (final bead concentrations were around 4 × 106 ml−1 and 4 × 107 ml−1, respectively).Human whole blood was purchased from Zen-Bio, Inc. To prepare the cell sample, 1 ml of blood was mixed with 10 ml of 1× RBC Lysis Buffer (eBioscience, USA) for 3 min to lyse most of the red blood cells (RBCs). Then the prepared cell sample was centrifuged at 400 × g for 5 min, resuspended in 1 ml of 4% paraformaldehyde in phosphate-buffered saline (PBS) solution (Santa Cruz Biotechnology, USA), and fixed at room temperature for 30 min. After fixation, the cell sample was centrifuged again and resuspended in 1 ml of PBS (Gibco, Life Technologies, USA) for use in experiments (final concentration of WBCs was around 5 × 106 ml−1).
Experimental setup
All the experiments were conducted on the stage of an inverted microscope (Eclipse Ti-U, Nikon, Japan). To eliminate the double images caused by using a double-side polished LiNbO3 substrate, we placed a polarizer in the light path and adjusted it to a certain angle during the experiments. To generate the SSAW, we applied amplified RF signals at desired frequencies and input voltages to the IDTs using an RF signal function generator (E4422B, Agilent, USA) and a power amplifier (100A250A, Amplifier Research, USA). A digital phosphor oscilloscope (load set at 1 MΩ) (DPO4104, Tektronix, USA) was used to measure the input voltages. In all of our experiments, the frequencies of the applied RF signals were in the range of 19.40 to 19.60 MHz and the input voltages were in the range of 20 to 40 Vpp, unless specified otherwise. Bead or cell samples prepared in 1 ml plastic syringes (Becton Dickinson, USA) were injected into the center inlet through polyethylene tubing (Becton Dickinson, USA) using syringe pumps (neMESYS, cetoni GmbH, Germany). Sheath flows (0.1% SDS solution for bead experiments and PBS for cell experiments) were introduced through two side inlets. Sheath flow introduced through the upper inlet also acted as a wash solution for collecting washed cells/beads.Data acquisition and analysis
A fast camera (SA4, Photron, Japan) and a CCD camera (CoolSNAP HQ2, Photometrics, USA) were connected to the microscope for data acquisition. To calculate recovery rate and washing efficiency, a hemocytometer was used to measure the concentrations of 10 μm and 0.87 μm beads in collected samples. For the characterization of WBC washing, flow cytometry results were collected using a Beckman Coulter FC500 Flow Cytometer and analyzed using FlowJo software (Tree Star, USA). Image J (NIH, Bethesda, MD, USA) was used for video processing.Results and discussion
Demonstration and optimization of SSAW-based bead washing
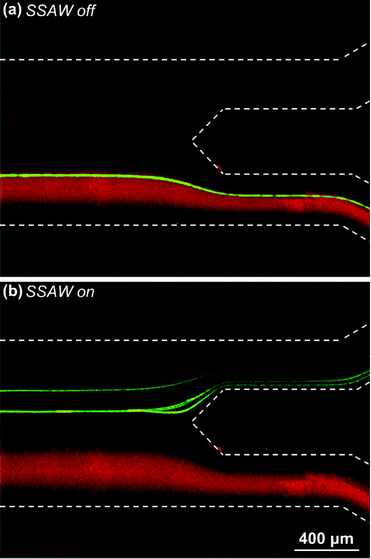
In order to evaluate the taSSAW approach for continuous-flow cell/bead washing, we first demonstrated SSAW-based bead washing. In this experiment, we mixed 9.77 μm green fluorescent microparticles with 0.87 μm red fluorescent microparticles and introduced them into the center inlet. Here, the 9.77 μm beads represented the target beads to be washed out, and the 0.87 μm beads represented the original medium. Fluorescence images of the outlet region were taken when the SSAW was off and on (Fig. 2). When the SSAW was off, the 9.77 μm beads flowed within the original medium stream and exited through the lower outlet. Conversely, when the SSAW was on, the 9.77 μm beads were washed out from the original medium, exiting through the upper outlet in the wash solution.During our experiment, we found that the input voltage clearly affected bead-washing results. If the input voltage was too low, the generated acoustic radiation force was too weak to deflect all the target beads from the original medium stream. As a result, only a portion of target beads were washed into the upper outlet. In contrast, when the input voltage was too high, the acoustic streaming effect could become obvious, and mixing between the wash solution and original medium occurred.49 Under this circumstance, bead-washing performance was reduced as some of the small beads were extracted, exiting through the upper outlet with the target beads.
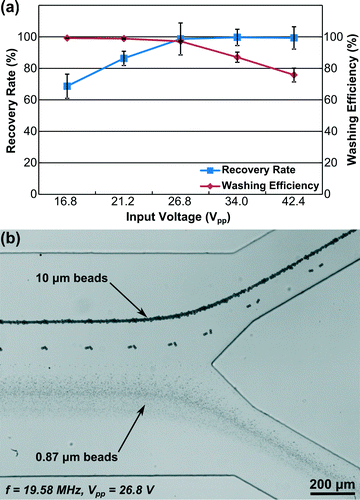
In order to optimize the input voltage for bead washing, we conducted an experiment to study the impact of input voltage on the bead-washing performance. In this experiment, we mixed 10 μm beads and 0.87 μm beads in 0.1% SDS solution as the sample. The flow rates of the upper inlet, center inlet, and lower inlet were 5, 1, and 2 μl min−1, respectively. The flow rates of the two outlets were controlled equally with the syringe pump in withdrawal mode. To evaluate the device performance, we examined both recovery rate and washing efficiency. Recovery rate is defined as the percentage of 10 μm beads that are washed out and collected through the upper outlet and washing efficiency is defined as the percentage of 0.87 μm beads exiting through the lower outlet. Before the SSAW was applied, all of the beads flowed out through the lower outlet, and the concentrations of 10 μm and 0.87 μm beads from the lower outlet were measured as references. Then, we applied 19.58 MHz RF signals with five different input voltages (16.8, 21.2, 26.8, 34.0, and 42.4 Vpp) to the IDTs, collected samples from the upper outlet, and measured the concentrations of 10 μm and 0.87 μm beads. All sample collections and corresponding measurements were repeated three times for each input voltage.
Fig. 3(a) shows the calculated recovery rate and washing efficiency at each input voltage. When we increased the input voltage from 16.8 to 26.8 Vpp, the recovery rate increased, indicating that more 10 μm beads were washed out. At 26.8 Vpp, the acoustic radiation force was already strong enough to wash almost all of the 10 μm beads from the medium, rendering subsequent changes in the recovery rate due to increases in the input voltage negligible. The influence of input voltage on washing efficiency followed an opposite trend. At low voltages (16.8 and 21.2 Vpp), the washing efficiency was almost 100%. From 26.8 to 42.4 Vpp, the washing efficiency decreased dramatically as significantly more 0.87 μm beads were extracted with the 10 μm beads through the upper outlet. Considering the balance between recovery rate and washing efficiency, the optimized input voltage at this frequency and flow condition should be 26.8 Vpp for bead washing, where we can achieve around 98.7% recovery rate and 97.2% washing efficiency.
A video was recorded at this optimized input voltage and is available online as Video S1 (ESI†). A stacked image of this video is shown as Fig. 3(b). From Fig. 3(b) and Video S1,† we can see that all of the 10 μm beads exit through the upper outlet and almost all of the 0.87 μm beads exit through the lower outlet. This result agrees with our characterization of recovery rate and washing efficiency, and illustrates the excellent performance of our SSAW-based bead washing device.
Numerical simulations of particle trajectories
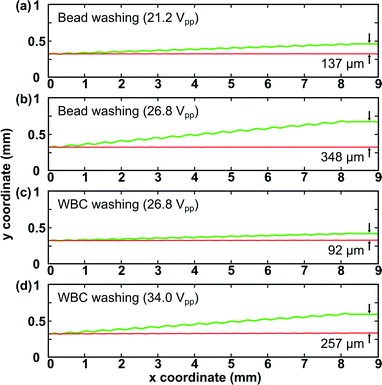
Before applying our SSAW device for cell washing, we conducted numerical simulations of particle trajectories using a 2D model where the particle motion was governed by the acoustic radiation force and Stokes drag force. The particle trajectory was obtained via MATLAB by considering these two forces and particle acceleration in the SSAW field. A calibration was performed to connect the simulation model with experimental conditions. The acoustic pressure p0 in eqn (1) can be expressed as ρfcfωA, where cf (taken to be 1495 m s−1), ω (2πf, where f is the excitation frequency of SAW), and A are sound speed in the fluid, angular velocity, and acoustic vibration amplitude, respectively. The vibration amplitude of SAW is proportional to the input voltage on the IDTs. Thus, the acoustic pressure is linearly dependent on the input voltage. In the calibration, we first measured the separation distance between 9.77 μm and 0.87 μm beads obtained in an experiment under a certain input voltage. Then the vibration amplitude A that could result in similar separation distance between 9.77 μm and 0.87 μm beads in the simulation was considered to be the actual vibration amplitude excited by this specific input voltage. According to the linear relation between the amplitude A and input voltage, the vibration amplitudes excited by other input voltages can be found as well. After calibration, the vibration amplitudes actuated by input voltages of 21.2, 26.8, and 34.0 Vpp were found to be 0.87, 1.10, and 1.40 nm, respectively. The simulations for cases under these input voltages were performed by inputting vibration amplitudes into the model accordingly.Fig. 4 shows the predicted particle trajectories in bead/cell washing under different input voltages. The x-axis is along the lower channel wall while the y-axis indicates the channel width. For bead washing, the 9.77 μm and 0.87 μm polystyrene beads have the same density (1.05 g cm−3) and compressibility (2.16 ×10−10 Pa−1) but different sizes.40,50 The simulation results in Fig. 4(a–b) indicate that at 21.2 Vpp, the separation distance between 9.77 μm and 0.87 μm beads is 137 μm while it increases to 348 μm at 26.8 Vpp. The large separation distance at 26.8 Vpp is expected to wash 9.77 μm beads out with high recovery rate and high washing efficiency, validated by our experimental results (Fig. 3).
In lysed blood samples, the average diameter, density, and compressibility of WBCs are 12 μm, 1.082 g cm−3, and 3.99 ×10−10 Pa−1, respectively.40,50 Through microscopic examination of the lysed blood sample, we measured the average diameter of debris present in the sample to be around 4 μm. The density and compressibility of debris were approximated using the values of RBCs (1.093 g cm−3 and 3.38 × 10−10 Pa−1).51 We first examined the predicted cell trajectories at the optimized input voltage for bead washing (26.8 Vpp) in our simulation. The simulation result in Fig. 4(c) shows that the separation distance between WBCs and debris is only 92 μm at 26.8 Vpp. One major reason is that WBCs have a much lower acoustic contrast factor (ϕ = 0.20) than 9.77 μm beads (ϕ = 0.57), consequently experiencing smaller acoustic radiation forces at the same input voltage. Therefore, to optimize WBC washing performance, higher input voltage is needed. Fig. 4(d) shows that at 34.0 Vpp, the separation distance between WBCs and debris is expected to be 257 μm, which should be sufficient to wash WBCs out with good performance.
SSAW-based WBC washing
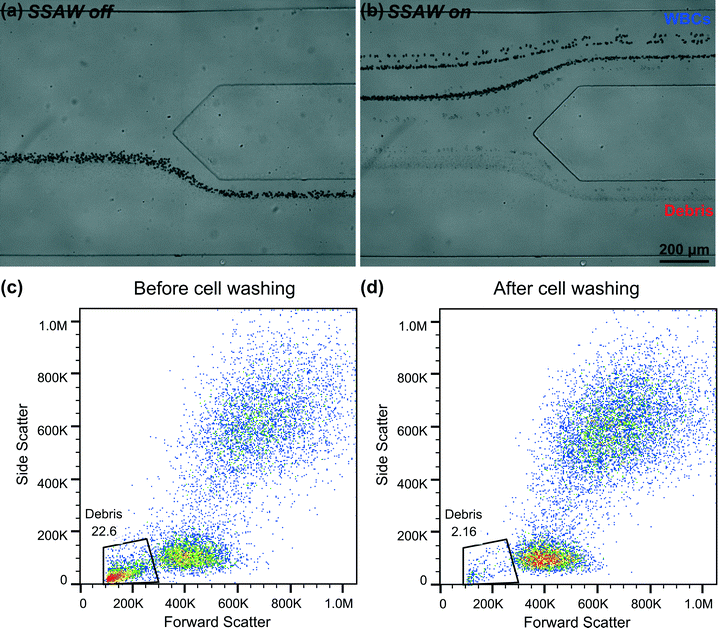
After optimizing the input voltage for WBC washing through numerical simulations, we conducted experiments to purify WBCs from lysed blood samples using our SSAW-based cell washing device. In our experiment, the flow conditions and excitation frequency were kept the same as in our optimized bead-washing experiment while an input voltage of 34.0 Vpp (instead of 26.8 Vpp) was used based on our simulation results. Under the microscope, WBCs appeared as large, dark spheres and were easy to focus. Apart from WBCs, we also found a considerable amount of small objects with various sizes and lighter color, which were debris produced during sample preparation. As shown in Fig. 5(a) and Video S2 (ESI†), when there was no SSAW applied, all of the WBCs exited the microchannel through the lower outlet, mixed with debris. Fig. 5(b) and Video S3 (ESI†) show that when the SSAW was applied at the input voltage of 34.0 Vpp, almost all of the WBCs were washed out and collected through the upper outlet, while the debris remained in the original flow stream, exiting through the lower outlet.For the characterization of WBC washing, we collected WBC samples before and after applying SSAW, and analyzed the samples using flow cytometry [Fig. 5(c–d)]. After SSAW-based cell washing, the percentage of debris present in the WBC sample decreased from 22.6% to 2.16%. Therefore, we were able to collect WBCs in clean PBS solution with more than 97% purity. To evaluate the biocompatibility of our SSAW-based cell washing device, we also measured the viability of WBCs collected after cell washing. The result indicates that the viability of WBCs was not compromised after cell washing (Fig. S1, ESI†). Collectively, these results demonstrate that our SSAW-based cell washing device can indeed take advantage of the difference in size and physical properties between WBCs and debris to successfully purify WBCs from lysed blood samples with good performance and high biocompatibility.
Conclusions
In conclusion, we have applied our recently introduced taSSAW approach to develop and test a SSAW-based cell/bead washing platform. Under the optimized condition, our SSAW device shows excellent performance with >98% recovery rate and >97% washing efficiency for bead washing. Numerical simulations were performed taking into account the properties of WBCs and debris (size, density, and compressibility) in order to apply our SSAW device for cell washing. The successful application of our device for cell washing is demonstrated by high-purity (>97%) WBC preparation. With the advantages of label-free manipulation, simplicity, high biocompatibility, high recovery rate, high washing efficiency, and compatibility with other on-chip components, our SSAW-based cell-washing technique offers an excellent option for on-chip, continuous-flow cell/bead washing and can be used for many lab-on-a-chip devices.52–55Acknowledgements
We thank Joey Rufo and Robert Shreiner for manuscript revision. This research was supported by National Institutes of Health (1 R01 GM112048-01A1 and 1R33EB019785-01), NIH Clinical and Translational Science Award, Huck Innovative & Transformational Seed (HITS) Fund, CTSI grant (UL1 TR000127), US Department of Agriculture (USDA/NRI), and the Penn State Center for Nanoscale Science (MRSEC) under Grant DMR-0820404. S. L. and C. E. C. were funded in part by grant AI45818 from NIAID, NIH. Components of this work were conducted at the Penn State node of the NSF-funded National Nanotechnology Infrastructure Network (NNIN).References
- E. C. Butcher and I. L. Weissman, J. Immunol. Methods, 1980, 37, 97–108 CrossRef CAS.
- X. Zheng, L. S.-L. Cheung, J. A. Schroeder, L. Jiang and Y. Zohar, Lab Chip, 2011, 11, 3269–3276 RSC.
- X. Zheng, L. S.-L. Cheung, J. A. Schroeder, L. Jiang and Y. Zohar, Lab Chip, 2011, 11, 3431–3439 RSC.
- A. Sánchez-Salinas, V. Cabañas-Perianes, M. Blanquer, M. J. Majado, C. L. Insausti, J. Monserrat, M. V. Sánchez-Ibáñez, P. Menchón, A. García-Hernández, J. Gómez-Espuch, A. Morales and J. M. Moraleda, Transfusion, 2012, 52, 2382–2386 CrossRef PubMed.
- B. Calmels, P. Houzé, J.-C. Hengesse, T. Ducrot, C. Malenfant and C. Chabannon, Bone Marrow Transplant., 2003, 31, 823–828 CrossRef CAS PubMed.
- J. Sambrook, E. F. Fritsch and T. Maniatis, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York, 1989 Search PubMed.
- B. Keeley, A. Stark, T. R. Pisanic, R. Kwak, Y. Zhang, J. Wrangle, S. Baylin, J. Herman, N. Ahuja, M. V. Brock and T.-H. Wang, Clin. Chim. Acta, 2013, 425, 169–175 CrossRef CAS PubMed.
- D. A. A. Vignali, J. Immunol. Methods, 2000, 243, 243–255 CrossRef CAS.
- K. Choi, A. H. C. Ng, R. Fobel, D. A. Chang-Yen, L. E. Yarnell, E. L. Pearson, C. M. Oleksak, A. T. Fischer, R. P. Luoma, J. M. Robinson, J. Audet and A. R. Wheeler, Anal. Chem., 2013, 85, 9638–9646 CrossRef CAS PubMed.
- W. B. Lockwood, R. W. Hudgens, I. O. Szymanski, R. A. Teno and A. D. Gray, Transfusion, 2003, 43, 1527 CrossRef.
- B. W. Peterson, P. K. Sharma, H. C. van der Mei and H. J. Busscher, Appl. Environ. Microbiol., 2012, 78, 120–125 CrossRef CAS PubMed.
- S. K. Harm, J. S. Raval, J. Cramer, J. H. Waters and M. H. Yazer, Transfus. Med., 2012, 22, 181–185 CrossRef CAS PubMed.
- M. D. Tarn, M. J. Lopez-Martinez and N. Pamme, Anal. Bioanal. Chem., 2014, 406, 139–161 CrossRef CAS PubMed.
- X. Li, W. Chen, G. Liu, W. Lu and J. Fu, Lab Chip, 2014, 14, 2565–2575 RSC.
- Z. T. F. Yu, K. M. Aw Yong and J. Fu, Small, 2014, 10, 1687–1703 CrossRef CAS PubMed.
- K. J. Morton, K. Loutherback, D. W. Inglis, O. K. Tsui, J. C. Sturm, S. Y. Chou and R. H. Austin, Lab Chip, 2008, 8, 1448–1453 RSC.
- R. D. Sochol, S. Li, L. P. Lee and L. Lin, Lab Chip, 2012, 12, 4168–4177 RSC.
- C. Kantak, S. Beyer, L. Yobas, T. Bansal and D. Trau, Lab Chip, 2011, 11, 1030–1035 RSC.
- K. Toyama, M. Yamada and M. Seki, Biomed. Microdevices, 2012, 14, 751–757 CrossRef CAS PubMed.
- V. VanDelinder and A. Groisman, Anal. Chem., 2007, 79, 2023–2030 CrossRef CAS PubMed.
- M. Yamada, J. Kobayashi, M. Yamato, M. Seki and T. Okano, Lab Chip, 2008, 8, 772–778 RSC.
- Y.-Y. Chiang and J. West, Lab Chip, 2013, 13, 1031–1034 RSC.
- D. R. Gossett, H. T. K. Tse, J. S. Dudani, K. Goda, T. A. Woods, S. W. Graves and D. Di Carlo, Small, 2012, 8, 2757–2764 CrossRef CAS PubMed.
- S. A. Peyman, A. Iles and N. Pamme, Chem. Commun., 2008, 1220–1222 RSC.
- S. Park, Y. Zhang, T.-H. Wang and S. Yang, Lab Chip, 2011, 11, 2893–2900 RSC.
- G.-B. Lee, H.-C. Wu, P.-F. Yang and J. D. Mai, Lab Chip, 2014, 14, 2837–2843 RSC.
- R. Tornay, T. Braschler, N. Demierre, B. Steitz, A. Finka, H. Hofmann, J. A. Hubbell and P. Renaud, Lab Chip, 2008, 8, 267–273 RSC.
- J. J. Hawkes, R. W. Barber, D. R. Emerson and W. T. Coakley, Lab Chip, 2004, 4, 446–452 RSC.
- P. Augustsson, L. B. Åberg, A.-M. K. Swärd-Nilsson and T. Laurell, Microchim. Acta, 2008, 164, 269–277 CrossRef.
- F. Petersson, A. Nilsson, H. Jönsson and T. Laurell, Anal. Chem., 2005, 77, 1216–1221 CrossRef CAS PubMed.
- M. Nordin and T. Laurell, Lab Chip, 2012, 12, 4610–4616 RSC.
- M. Antfolk, P. B. Muller, P. Augustsson, H. Bruus and T. Laurell, Lab Chip, 2014, 14, 2791–2799 RSC.
- D. Carugo, T. Octon, W. Messaoudi, A. Fisher, M. Carboni, N. R. Harris, M. Hill and P. Glynne-Jones, Lab Chip, 2014, 14, 3830–3842 RSC.
- X. Ding, S.-C. S. Lin, B. Kiraly, H. Yue, S. Li, I.-K. Chiang, J. Shi, S. J. Benkovic and T. J. Huang, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11105–11109 CrossRef CAS PubMed.
- X. Ding, S.-C. S. Lin, M. I. Lapsley, S. Li, X. Guo, C. Y. Chan, I.-K. Chiang, L. Wang, J. P. McCoy and T. J. Huang, Lab Chip, 2012, 12, 4228–4831 RSC.
- S. Li, X. Ding, F. Guo, Y. Chen, M. I. Lapsley, S.-C. S. Lin, L. Wang, J. P. McCoy, C. E. Cameron and T. J. Huang, Anal. Chem., 2013, 85, 5468–5474 CrossRef CAS PubMed.
- J. Shi, X. Mao, D. Ahmed, A. Colletti and T. J. Huang, Lab Chip, 2008, 8, 221–223 RSC.
- J. Shi, H. Huang, Z. Stratton, Y. Huang and T. J. Huang, Lab Chip, 2009, 9, 3354–3359 RSC.
- Y. Chen, X. Ding, S.-C. S. Lin, S. Yang, P.-H. Huang, N. Nama, Y. Zhao, A. A. Nawaz, F. Guo, W. Wang, Y. Gu, T. E. Mallouk and T. J. Huang, ACS Nano, 2013, 7, 3301–3314 Search PubMed.
- X. Ding, Z. Peng, S.-C. S. Lin, M. Geri, S. Li, P. Li, Y. Chen, M. Dao, S. Suresh and T. J. Huang, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12992–12997 CrossRef CAS PubMed.
- J. Nam, H. Lim, C. Kim, J. Y. Kang and S. Shin, Biomicrofluidics, 2012, 6, 24120 CrossRef PubMed.
- Y. Ai, C. K. Sanders and B. L. Marrone, Anal. Chem., 2013, 85, 9126–9134 CrossRef CAS PubMed.
- J. Nam, H. Lim, D. Kim and S. Shin, Lab Chip, 2011, 11, 3361–3364 RSC.
- X. Bossuyt, G. E. Marti and T. A. Fleisher, Cytometry, 1997, 30, 124–133 CrossRef CAS.
- S. Chow, D. Hedley, P. Grom, R. Magari, J. W. Jacobberger and T. V. Shankey, Cytometry, Part A, 2005, 67, 4–17 CrossRef PubMed.
- A. J. Hughes and A. E. Herr, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 21450–21455 CrossRef CAS PubMed.
- A. J. Hughes, R. K. C. Lin, D. M. Peehl and A. E. Herr, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 5972–5977 CrossRef CAS PubMed.
- K. Yosioka and Y. Kawasima, Acustica, 1955, 5, 167–173 Search PubMed.
- M. Wiklund, R. Green and M. Ohlin, Lab Chip, 2012, 12, 2438–2451 RSC.
- F. Petersson, L. Aberg, A.-M. Swärd-Nilsson and T. Laurell, Anal. Chem., 2007, 79, 5117–5123 CrossRef CAS PubMed.
- M. Toubal, M. Asmani, E. Radziszewski and B. Nongaillard, Phys. Med. Biol., 1999, 44, 1277–1287 CrossRef CAS.
- V. N. Goral, C. Zhou, F. Lai and P. K. Yuen, Lab Chip, 2013, 13, 1039–1043 RSC.
- V. N. Goral, Y.-C. Hsieh, O. N. Petzold, J. S. Clark, P. K. Yuen and R. A. Faris, Lab Chip, 2010, 10, 3380–3386 RSC.
- S.-C. S. Lin, X. Mao and T. J. Huang, Lab Chip, 2012, 12, 2766–2770 RSC.
- X. Ding, P. Li, S.-C. S. Lin, Z. S. Stratton, N. Nama, F. Guo, D. Slotcavage, X. Mao, J. Shi, F. Costanzo and T. J. Huang, Lab Chip, 2013, 13, 3626–3649 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1 shows the viability test on washed WBCs. Video S1 shows the bead washing result at the optimized condition (19.58 MHz and 26.8 Vpp). Videos S2 and S3 illustrate the outlet region of WBC washing experiment when the SSAW was off and on (19.58 MHz and 34.0 Vpp). See DOI: 10.1039/c4lc00903g |
| This journal is © The Royal Society of Chemistry 2015 |