A solvent replenishment solution for managing evaporation of biochemical reactions in air-matrix digital microfluidics devices†
Mais J.
Jebrail‡
a,
Ronald F.
Renzi‡
b,
Anupama
Sinha
c,
Jim
Van De Vreugde
b,
Carmen
Gondhalekar
a,
Cesar
Ambriz§
a,
Robert J.
Meagher
a and
Steven S.
Branda
*a
aBiotechnology and Bioengineering, Sandia National Laboratories, Livermore, CA, USA. E-mail: sbranda@sandia.gov; Fax: +1 925 294 3020; Tel: +1 925 294 6751
bAdvanced Systems Engineering and Deployment, Sandia National Laboratories, Livermore, CA, USA
cSystems Biology, Sandia National Laboratories, Livermore, CA, USA
First published on 1st October 2014
Abstract
Digital microfluidics (DMF) is a powerful technique for sample preparation and analysis for a broad range of biological and chemical applications. In many cases, it is desirable to carry out DMF on an open surface, such that the matrix surrounding the droplets is ambient air. However, the utility of the air-matrix DMF format has been severely limited by problems with droplet evaporation, especially when the droplet-based biochemical reactions require high temperatures for long periods of time. We present a simple solution for managing evaporation in air-matrix DMF: just-in-time replenishment of the reaction volume using droplets of solvent. We demonstrate that this solution enables DMF-mediated execution of several different biochemical reactions (RNA fragmentation, first-strand cDNA synthesis, and PCR) over a range of temperatures (4–95 °C) and incubation times (up to 1 h or more) without use of oil, humidifying chambers, or off-chip heating modules. Reaction volumes and temperatures were maintained roughly constant over the course of each experiment, such that the reaction kinetics and products generated by the air-matrix DMF device were comparable to those of conventional benchscale reactions. This simple yet effective solution for evaporation management is an important advance in developing air-matrix DMF for a wide variety of new, high-impact applications, particularly in the biomedical sciences.
Introduction
Digital microfluidics (DMF) has emerged as a powerful preparative technique for a broad range of biological and chemical applications.1 DMF enables real-time, precise, and highly flexible control over multiple samples and reagents, including solids, liquids, and harsh chemicals, without need for pumps, valves, or complex arrays of tubing. In DMF, discrete droplets of nanoliter to microliter volumes are dispensed from reservoirs onto a planar surface coated with a hydrophobic insulator, where they are manipulated (transported, split, merged, mixed) by applying a series of electrical potentials to an embedded array of electrodes.2–4 Complex reaction series can be carried out using DMF alone, or using hybrid systems in which DMF is integrated with channel-based microfluidics.5–8 Hybrid systems offer tremendous versatility; in concept, each reaction step can be executed in the microfluidics format that best accommodates it. We have developed hybrid systems in which DMF is primarily used for multiplexed routing of samples and reagents to and from channel-based microfluidic modules that are specialized to carry out all other needed functions; in this approach, DMF serves to integrate channel-based microfluidic modules with mismatched input/output requirements, obviating the need for complex networks of tubing and microvalves. Thus, advances in DMF technology can have far-reaching impact through improvement of both digital and hybrid microfluidic systems.For many applications it is most convenient to carry out DMF on an open surface, such that the matrix surrounding the droplets is ambient air. However, use of the air-matrix format necessitates accounting for droplet evaporation, especially when the droplets are subjected to high temperatures for long periods of time. In some instances evaporation is considered a desirable feature, as it can facilitate concentration and isolation of solutes of interest.9–14 In biochemical contexts, however, evaporation frequently limits the utility of air-matrix DMF, because enzymatic reactions are often highly sensitive to changes in reactant concentration. Largely for this reason, investigators have demonstrated oil-matrix DMF for biochemical applications,15–17 despite numerous drawbacks including: 1) the added complexity of incorporating gaskets or fabricated structures to contain the oil; 2) unwanted liquid–liquid extraction of reactants into the surrounding oil;18 3) incompatibility with oil-miscible liquids (e.g., organic solvents such as alcohols); and 4) unwanted dissipation of heat, which undermines localized heating and often confounds temperature-sensitive reactions. Another strategy is to place the air-matrix DMF device in a closed humidified chamber,19,20 but this often results in unwanted condensation on the DMF surface, difficult and/or limited access to the device, and need for additional laboratory space and infrastructure. In previous work we avoided these issues by transferring reaction droplets from the air-matrix DMF device to microcapillaries, where they can be heated in dedicated off-chip modules without evaporation problems,7,8 but this complicates design and manufacture of the air-matrix DMF device, introducing the added complications of microcapillary interfaces and coordination with peripheral modules.
In this report, we present a novel solution for counteracting evaporation in an air-matrix DMF device without use of a humidified chamber, which enables facile and reliable execution of biochemical reactions over a range of temperatures (4–95 °C) and incubation times (up to 1 h or more). Our solution is just-in-time replenishment of the reaction volume using pre-heated droplets of solvent. Through this approach, the reaction volume and temperature are maintained roughly constant (≤20% and ≤1 °C change, respectively) over the course of the biochemical reaction. We demonstrate that this approach enables use of an air-matrix DMF device in executing several representative biochemical reactions (RNA fragmentation, first-strand cDNA synthesis, and PCR) drawn from a gene expression analysis workflow, and show that the reaction products are essentially indistinguishable from those generated by conventional bench-scale methods. Our solution for evaporation management is an important advance in developing air-matrix DMF for a wide variety of new, high-impact applications.
Results and discussion
System design and operation
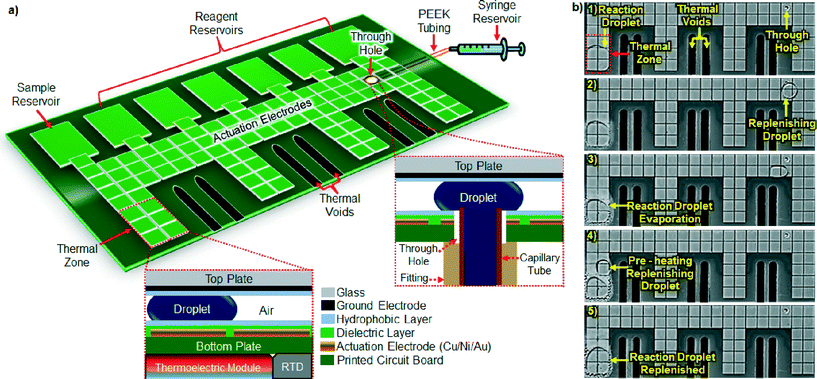
Our air-matrix DMF device was constructed from a six-layer printed circuit board (PCB).21 Multilayer PCBs have an inherent advantage over conventional single-layer devices (e.g., chrome or ITO on glass) in that electrical connections can occupy a separate layer from the actuation electrodes, affording more real estate for droplet actuation and simplifying on-chip integration of electronic components.22 Furthermore, PCB fabrication is a mature, widely available, scalable manufacturing technology that can be provided by a large number of commercial vendors, resulting in substantially lower cost per device than achieved through photolithographic patterning of chrome or ITO on glass. Fig. 1a depicts our air-matrix DMF device, which features an array of 100 actuation electrodes interfaced with seven fluid reservoirs (one for starting material, six for reaction reagents) and integrated with four thermoelectric and four resistive temperature detector (RTD) modules that provide for precise control of on-chip temperature (Fig. S1†). As shown in Fig. S2,† each module is affixed to the back side of the device, a thermal conduit channeling heat through the bottom DMF plate to a set of six embedded electrodes that serve as a thermal zone on the DMF surface. Each of the device's four thermal zones can be controlled independently of the others, such that four different on-chip temperatures can be maintained simultaneously. Each of these zones is thermally isolated from the rest of the device by voids machined into the PCB substrate. Rapid change in droplet temperature is achieved through transport across the DMF surface from one thermal zone to another. The temperature difference between the RTD on the back side of the device and a droplet within its corresponding thermal zone was measured using a fine-gauge thermocouple inserted into the droplet, and found to be 3 °C (±0.5 °C). The difference is mainly a function of the temperature drop across the PCB substrate, rather than of ambient temperature.23 To account for this temperature difference, a compensation factor was incorporated into programming of thermal zone temperature settings, to ensure that zone-localized droplets reached the desired temperature. The device also includes a through hole24,25 (0.8 mm ID) drilled into the bottom DMF plate, which enables introduction of replenishing droplets of solvent onto the device's surface from an external syringe pump reservoir.Evaporation management solution
A key drawback in operating air-matrix DMF devices is the potential for evaporation of the solvent, especially during operations that require high temperature and/or long incubation times (e.g., ≥65 °C for ≥1 min for aqueous droplets). To counteract evaporation, we have developed a solution by which the temperature-controlled biochemical reaction is periodically replenished with pre-heated droplets of solvent. As the reaction volume begins to decrease due to evaporation, a replenishing droplet is dispensed onto the device and transported to the edge of the thermal zone, where it is pre-heated for 5 s before merging with the reaction droplet (Fig. 1b). Pre-heating of replenishing droplets minimizes their impact on the reaction droplet's temperature upon merging (≤1 °C decrease; Table S1†), thereby promoting consistency in reaction kinetics. Similarly, reaction droplets were replenished with solvent upon loss of 15–20% of their volume, in order to minimize changes to solute concentration that could adversely affect reaction kinetics. Using this approach, reaction droplets of 2 μL were maintained at roughly constant volume (≤20% variation) over a wide range of temperatures (35–95 °C) (Fig. 2). Note that lower volumes (nL to pL) could be accommodated by the device by decreasing the gap spacing between the DMF plates and/or the size of actuation electrodes; smaller droplets are more vulnerable to evaporation, however, so it would be necessary to replenish them at greater frequency in order to maintain volume. In the current version of our air-matrix DMF device, evaporation is monitored visually and the reaction volume replenished “just-in-time” through manual intervention by the user. Future devices could be fitted with a sensing and feedback control system22,26 in which the reaction droplet's volume or concentration is monitored and, upon reaching a pre-determined threshold, the volume automatically reconstituted through addition of a replenishing droplet.Test applications
We evaluated the utility of our evaporation management solution in supporting execution of several high-value biochemical reactions (RNA fragmentation, first-strand cDNA synthesis, and PCR) in our air-matrix DMF device. Each of these reactions requires high temperature and/or extended incubation, which has confounded their implementation in DMF format without use of an oil matrix or a humidified chamber to counteract evaporation.For RNA fragmentation, three sample droplets [0.5 μL each, containing 180 ng μL−1 of total RNA purified from human peripheral blood mononuclear cells (PBMC)] and one droplet of 4× fragmentation buffer (0.5 μL) were dispensed from their respective reservoirs, mixed together on the DMF device surface, and the resulting reaction droplet (2 μL containing 270 ng of RNA and 1× fragmentation buffer) was transported to a thermal zone for incubation at 94 °C for 3 min. Without solvent replenishment, the reaction droplet dried completely in <2 min (data not shown). To counteract this, a total of six droplets of solvent (nuclease-free distilled water; 0.5 μL each) were used to replenish the reaction volume over the course of the incubation period. The reaction was then cooled to 4 °C and terminated upon addition of a droplet of stop solution (0.5 μL). We compared the performance of the on-chip reaction with that of the conventional benchscale reaction, which used larger volumes but identical final concentrations of RNA (18 μL at 15 ng μL−1; 270 ng RNA final) and fragmentation buffer (2 μL of 10×; 1× fragmentation buffer final), and was terminated upon addition of an appropriately-scaled droplet of stop solution (2 μL). The benchscale reaction was carried out in a microcentrifuge tube that was temperature controlled using a thermal cycler with a heated lid. Size distribution analysis of control RNA samples, which were not fragmented, indicated a broad range of product sizes, with prominent peaks at ~1900 nt and ≥4000 nt, corresponding to the 18S and 28S ribosomal RNA (rRNA) (1868 nt and 5025 nt, respectively) (Fig. 3a). In contrast, DMF-mediated RNA fragmentation generated products of notably smaller size (predominantly 40–1000 nt, peaking at ~150 nt), leaving no trace of the rRNA peaks (Fig. 3b). This size distribution profile was indistinguishable from that generated via RNA fragmentation by conventional means (Fig. 3c). These results indicate that RNA fragmentation through use of the air-matrix DMF device and evaporation management was successful, and generated reaction products of essentially identical size to those generated by the conventional benchscale method.
For first-strand cDNA synthesis using the Peregrine approach,27 a droplet of pre-fragmented human PBMC total RNA (0.5 μL) and a droplet containing primer PP_RT (0.5 μL) were dispensed from their respective reservoirs, mixed together on the DMF surface, and the resulting reaction droplet (1 μL) transported to the thermal zone. The reaction droplet was incubated at 65 °C for 2 min to denature the RNA, then immediately cooled to 4 °C. At this point, three droplets of master mix (0.5 μL each, containing buffer, nucleotides, and reverse transcriptase) were dispensed onto the DMF surface, merged with the 1 μL droplet, and the reaction incubated at 25 °C for 3 min followed by 42 °C for 1 min. Next, a droplet containing primer PP_TS (0.5 μL) was merged with the reaction droplet, and incubation continued at 42 °C for 1 h. Finally, the reaction was terminated by incubating at 70 °C for 5 min. The reaction volume (2 μL) was maintained through addition of 13 replenishing droplets of nuclease-free distilled water (0.5 μL each) over the course of the experiment; nine replenishing droplets were added during the incubation at 42 °C for 1 h, and four during the incubation at 70 °C for 5 min. We found that this DMF-mediated approach to first-strand cDNA synthesis generated yields that were roughly equivalent those generated by the conventional benchscale approach, as measured using a product-specific qPCR-based assay27 (Fig. 4a). Moreover, the DMF-generated first-strand cDNA products successfully served as template for second-strand cDNA synthesis, such that the yield and size distribution profile of double-stranded cDNA products were comparable to those generated from conventionally-prepared first-strand cDNA (Fig. 4b).
For PCR, three droplets (0.5 μL each) respectively containing master mix (buffer, nucleotides, and DNA polymerase), primers, and template (single-stranded genomic DNA from bacteriophage M13mp18) were dispensed from separate reservoirs and mixed together on the DMF surface. The resulting reaction droplet (1.5 μL) was shuttled between three thermal zones for temperature cycling: 95 °C for 45 s, then 33 cycles of 95 °C for 20 s, 50 °C for 30 s, and 68 °C for 45 s; and finally 68 °C for 5 min. The reaction volume was maintained through addition of 33 replenishing droplets of nuclease-free distilled water (0.5 μL each) over the course of the experiment (~45 min), adding one droplet upon completion of each temperature cycle. We found that the DMF-mediated approach generated a PCR product that was of the expected size (200 bp) and comparable to that generated by conventional PCR, though with slightly reduced yield, as indicated by gel electrophoresis analysis (Fig. 5).
Conclusions
We present a simple and effective solution for managing evaporation in an air-matrix DMF device without use of oil, a humidified chamber, or off-chip heating. Our results indicate that just-in-time replenishment of the reaction volume with pre-heated droplets of solvent enabled DMF-mediated execution of three different biochemical reactions requiring temperatures of ≥68 °C and/or incubation times of ≥3 min. In each case, the reaction kinetics and products generated by the air-matrix DMF device were comparable to those generated by conventional benchscale methods. We anticipate that this new approach will greatly improve the utility of air-matrix DMF in executing a broad variety of biochemical reactions, and enable facile, on-chip integration of multi-step workflows for new applications in the biomedical sciences.Experimental section
Reagents and materials
Fluorinert FC-40, Pluronic F127, and ethanol were purchased from Sigma Chemical (St. Louis, MO); Teflon-AF was from DuPont (Wilmington, DE); thermoelectric modules were from Laird Technologies (OptoTEC Series OT08-08-F0-0303; Earth City, MO); resistive temperature detectors (RTD) were from US Sensor (PPG102A1; Orange, CA); human peripheral blood mononuclear cells (PBMC) were from Astarte Biologics (Bellevue, WA); 10× NEBNext RNA fragmentation buffer and stop solution were from New England Biolabs (Ipswich, MA); and RNaseZAP solution was from Life Technologies (Grand Island, NY). For PCR, single-stranded genomic DNA from bacteriophage M13mp18 was purchased from New England Biolabs (Ipswich, MA); primers designed for amplification of a 200 bp region (positions 4905-5104) of the M13mp18 genome (Genbank X02513.1), of sequences TGTTAATACTGACCGCCTCAC (forward) and CAGTAATAAAAGGGACATTCTGGC (reverse), were purchased from Integrated DNA Technologies (Coralville, IA); and Hot Start Taq-driven PCR reagents were from New England Biolabs (Ipswich, MA). Molecular biology-grade organic solvents and nuclease-free water were used throughout.Device fabrication and assembly
The bottom plate of the DMF device was designed in CAD systems, and Gerber files were outsourced to World International Printed Circuit Board (PCB) (Orange, CA) for fabrication. Briefly, a six-layer PCB substrate bearing copper electrodes (43 μm thick) plated with nickel (185 μm) and gold (3.6 μm) were formed by conventional photolithography and etching,21 and were coated with soldermask (~15 μm) as the dielectric. The substrate featured an array of one-hundred actuation electrodes (each 1.6 × 1.6 mm) connected to six reservoir electrodes (each 3.5 × 5.5 mm) and a waste reservoir (5.2 × 5.5 mm), with inter-electrode gaps of 80 μm, thermal voids (each 1.0 × 7.0 mm) for temperature isolation, and a through hole (0.8 mm ID) that enabled delivery of replenishing droplets to the interior surface of the substrate. The electrical contact pads were masked with polyimide tape (DuPont; Hayward, CA), and the substrate was spin-coated with a 50 nm layer of Teflon-AF (1% wt/wt in Fluorinert FC-40, 1500 rpm for 30 s) and then baked at 100 °C for 5 h. Access for electrical contact was re-established by removing the polyimide tape. For temperature cycling and detection, a thermoelectric module (Laird; Schaumburg, IL) and a resistive temperature detector (RTD) (US Sensor; Orange, CA) were adhered to gold plated copper pads on the back side of the substrate (Fig. S1†), using thermally conductive epoxy. These pads, in turn, were thermally coupled to the thermal zone electrodes on the front side of the substrate by means of drilled through holes filled with conductive epoxy. The top plate of the DMF device, consisting of a glass substrate coated uniformly with unpatterned indium tin oxide (ITO) (Delta Technologies Ltd; Stillwater, MN), was spin-coated with 50 nm of Teflon-AF, as described above.DMF devices were assembled in frames identical to those described previously.7 DMF frames that maintain a 185 or 400 μm gap spacing between the top and bottom substrates were used to accommodate different droplet volumes. Reagents were loaded into their designated reservoirs using pipetmen, prior to assembly of the device; for RNA fragmentation and first-strand cDNA synthesis, the reservoir volume was 3.6 μL, whereas for PCR the reservoir volumes were 4 μL (master mix), 2 μL (primers) and 2 μL (template), respectively. For introduction of replenishing droplets onto the DMF device, a 360/150 μm OD/ID PEEK tube (IDEX; Oak Harbor, WA) connected to a syringe pump was inserted in the through hole from the back side of the device and immobilized using custom fittings (Fig. S1†). Replenishing droplets were driven onto the device using a stepper motor equipped with a computer-controlled electronic interface responding to ASCII commands (Allmotion; Union City, CA).
Temperature control of the thermoelectric module was achieved using a custom control board with a microcontroller (PICAXE 14M2; Digi-Key Electronics; Thief River Fall, MN) that measured the temperature of the substrate and provided both a pulse width modulated and polarity control output to enable the thermoelectric module to heat and cool. Droplet actuation in the DMF device was achieved by applying driving potentials of 80–100 Vrms generated by amplifying (Digi-Key Electronics; Thief River Falls, MN) the output of a function generator (Trek; Medina, NY) operating at 18 kHz. Droplet movements in the device were visualized using an MVX10 microscope (Olympus; Center Valley, PA) equipped with a high-speed QIClick digital camera (Qimaging; Surrey, Canada) with a field of view of ~35 × 26 mm2. Video stills were captured to PC using a commercial software package (Streampix5; Norpix; Montreal, Canada). Thermal images and their thermal traces were captured using a Thermographic IR camera (ThermoProTMTP8; Guide Infrared; Wuhan, China) after allowing the temperature of thermal zone electrodes to equilibrate for 10 s.
To combat evaporation in the air-matrix DMF device, reaction volumes were replenished using a neat solvent droplet, which was dispensed onto the DMF device, actuated to the thermal zone, pre-heated for 5 s, and then merged with the reaction droplet to reconstitute its volume. Measurement of reaction droplet volume after replenishment was accomplished by measuring the two-dimensional area of the droplet in video stills using ImageJ software (National Institutes of Health; Bethesda, MD) and multiplying by the gap spacing between the DMF plates. This common approach assumes that the droplet is cylindrical, though the hydrophobic surface and spacing of DMF plates can cause the sidewalls of the droplet to bulge outward; given the measured dimensions of the droplets and device, however, the error potentially introduced in assuming a cylindrical shape would not exceed 10% of the droplet volume measurements. In future iterations of the device, feedback systems based on capacitance and resistance measurements would further improve the accuracy of droplet volume measurements. Temperature change in the reaction droplet before and after replenishment was measured using a fine-gauge thermocouple inserted into the reaction droplet. For all other analyses, the top DMF plate was removed and the samples collected by pipet for off-chip evaluation.
RNA extraction
For extraction of total RNA from human PBMC, 5–10 × 106 cells were centrifuged at 1000 rpm at 4 °C for 5 min, and re-suspended in 1 ml of RNAzol (Molecular Research Center; Cincinnati, OH), followed by dilution with 400 μl of water. After incubation at room temperature (RT) for 15 min, the samples were centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 15 min, and ~800 μl of the aqueous phase from each tube were transferred to a new 2 ml tube and mixed 1
000 rpm at 4 °C for 15 min, and ~800 μl of the aqueous phase from each tube were transferred to a new 2 ml tube and mixed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with ethanol. Purified total RNA was recovered using the Direct-zol kit (Zymo Research; Irvine, CA), following the manufacturer's instructions and eluting in 10 μL of water. RNA yield was quantified using a Qubit 2.0 fluorimeter (Life Technologies; Carlsbad, CA), and fragment size distribution was assessed using a 2100 Bioanalyzer equipped with an RNA Nano 6000 Chip (Agilent; Santa Clara, CA). RNA samples were stored at −80 °C.
1 with ethanol. Purified total RNA was recovered using the Direct-zol kit (Zymo Research; Irvine, CA), following the manufacturer's instructions and eluting in 10 μL of water. RNA yield was quantified using a Qubit 2.0 fluorimeter (Life Technologies; Carlsbad, CA), and fragment size distribution was assessed using a 2100 Bioanalyzer equipped with an RNA Nano 6000 Chip (Agilent; Santa Clara, CA). RNA samples were stored at −80 °C.
RNA fragmentation
DMF-mediated RNA fragmentation was implemented in three steps. First, three droplets (0.5 μL each) containing 180 ng μL−1 of human PBMC total RNA (270 ng RNA final) and a droplet (0.5 μL) of diluted 10× NEBNext fragmentation buffer (New England Biolabs; Ipswitch, MA) (4× final) were dispensed from their respective reservoirs, mixed on the DMF surface for 10 s, and transported to a thermal zone. Second, the reaction droplet (2 μL; 270 ng RNA and 1× fragmentation buffer final) was incubated at 94 °C for 3 min. Finally, the reaction was cooled to 4 °C, and RNA fragmentation was terminated by supplementing the reaction with a droplet (0.5 μL) of NEBNext stop solution (New England Biolabs; Ipswitch, MA). The reaction volume was maintained through addition of six replenishing droplets of nuclease-free distilled water (0.5 μL each) over the course of the experiment. For RNA fragmentation using the conventional benchscale method, processing was identical except for the volumes [18 μL of 15 ng μL−1 RNA (270 ng RNA final), 2 μL of 10× fragmentation buffer (1× final), and 2 μL of stop solution] and that incubations were carried out in microcentrifuge tubes heated by a conventional thermocycler. In both cases, RNA fragmentation reaction products were purified using the Zymo RNA Clean and Concentrator-5 system (Zymo Research; Irvine, CA), following the manufacturer's general procedure and eluting in 5 μl of nuclease-free distilled water. RNA fragment size distributions were analyzed using an RNA Nano 6000 Chip on a 2100 Bioanalyzer (Agilent; Santa Clara, CA).cDNA synthesis
First-strand cDNA synthesis was accomplished through DMF or benchscale implementation of the Peregrine method.27 For DMF-mediated cDNA synthesis, a five-step protocol was developed. First, a 0.5 μL droplet of fragmented human PBMC total RNA (100 ng) and a 0.5 μL droplet of primer PP_RT (25 μM) were dispensed from their respective reservoirs, merged and mixed on the DMF surface, and the 1 μL droplet transported to a thermal zone. Second, the droplet was incubated at 65 °C for 2 min, and then immediately cooled to 4 °C. Third, three droplets of master mix (0.5 μL each, containing 45% of SMARTScribe 5× First-Strand Buffer (Clontech; Mountain View, CA), 5.5% of 20 mM DTT, 22% of 10 mM dNTP mix, 5.5% of RiboGuard RNase inhibitor (Epicentre; Madison, WI) and 22% of SMARTScribe Reverse Transcriptase (Clontech; Mountain View, CA), as well as Pluronic F127 at 0.1% w/v) were dispensed onto the DMF surface, merged with the 1 μL droplet, and the reaction incubated at RT for 3 min followed by 42 °C for 1 min. Fourth, a 0.5 μL droplet of primer PP_TS (12 μM) was merged with the reaction droplet, and incubation continued at 42 °C for 1 h. Finally, the reaction was terminated by incubating at 70 °C for 5 min. In all cases, temperature changes were carried out by shuttling the reaction droplet between thermal zones set at the desired temperatures, as described above. The reaction volume was maintained through addition of 13 replenishing droplets of nuclease-free distilled water (0.5 μL each) over the course of the experiment. For first-strand cDNA synthesis using the conventional benchscale method, processing was identical except for the volumes (3.5 μL of fragmented RNA, 1 μL of primer PP_RT, 4.5 μL of master mix, and 1 μL of primer PP_TS) and that incubations were carried out in microcentrifuge tubes heated by a conventional thermocycler. In both cases, first-strand cDNA synthesis reaction products were purified using AMPure XP beads (Beckman Coulter Genomics; Danvers, MA), using 1.8× volumes and eluting in 10–20 μl of nuclease-free distilled water, following the manufacturer's protocol. A qPCR-based assay was used to determine the number of PCR cycles needed for optimal production of high-quality double-stranded cDNA libraries from first-strand cDNA synthesis reaction products.27 After diluting the first-strand cDNA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in nuclease-free water, 1 μl of the dilution was combined with 5 μl of SsoFast EvaGreen SuperMix (Bio-Rad; Hercules, CA), 3 μl of nuclease-free water, 0.5 μl of 10 μM primer PP_P1 (5′-CAGGACGCTGTTCCGTTCTATGGG-3′), and 0.5 μl of 10 μM primer PP_P2 (5′-CAGACGTGTGCTCTTCCGATC T-3′). The assays were run in quadruplicate on a CFX96 qPCR machine (Bio-Rad; Hercules, CA), using the following cycle parameters: 95 °C for 45 s, followed by 25 cycles of 95 °C for 5 s and 60 °C for 30 s. The cycle number at which fluorescence intensity exceeded the detection threshold [i.e., the cycle threshold (Ct)] was identified as optimal for production of double-stranded cDNA libraries from the undiluted first-strand cDNA synthesis reaction products. The yields and size distribution profiles of cDNA libraries were analyzed using a High Sensitivity DNA Assay Chip on a 2100 Bioanalyzer (Agilent; Santa Clara, CA).
10 in nuclease-free water, 1 μl of the dilution was combined with 5 μl of SsoFast EvaGreen SuperMix (Bio-Rad; Hercules, CA), 3 μl of nuclease-free water, 0.5 μl of 10 μM primer PP_P1 (5′-CAGGACGCTGTTCCGTTCTATGGG-3′), and 0.5 μl of 10 μM primer PP_P2 (5′-CAGACGTGTGCTCTTCCGATC T-3′). The assays were run in quadruplicate on a CFX96 qPCR machine (Bio-Rad; Hercules, CA), using the following cycle parameters: 95 °C for 45 s, followed by 25 cycles of 95 °C for 5 s and 60 °C for 30 s. The cycle number at which fluorescence intensity exceeded the detection threshold [i.e., the cycle threshold (Ct)] was identified as optimal for production of double-stranded cDNA libraries from the undiluted first-strand cDNA synthesis reaction products. The yields and size distribution profiles of cDNA libraries were analyzed using a High Sensitivity DNA Assay Chip on a 2100 Bioanalyzer (Agilent; Santa Clara, CA).
PCR
Single-stranded genomic DNA from bacteriophage M13mp18 was diluted in nuclease-free water to a concentration of 250 pg μL−1. The forward and reverse primers (each 500 μM in 10 mM Tris–HCl), designed for amplification of a 200 bp region (positions 4905-5104) of the M13mp18 genome, were mixed in equimolar ratio and diluted in nuclease-free water to generate a 4× stock solution (4 μM per primer). PCR reactions were assembled using Hot Start Taq 2× Master Mix (New England Biolabs; Ipswitch, MA) supplemented with 0.025 units μL−1 of Hot Start Taq polymerase (New England Biolabs; Ipswitch, MA), effectively doubling the Taq concentration in the 2× Master Mix. For PCR on the DMF device, droplets of master mix, primers, and template (0.5 μL each) were dispensed from their respective reservoirs, merged and mixed on the DMF surface, and transported to thermal zones for temperature cycling (Table S1†): 95 °C for 45 s; then 33 cycles of 95 °C for 20 s, 50 °C for 30 s, and 68 °C for 45 s; and finally 68 °C for 5 min. Replenishing droplets (0.5 μL each) were added to the reaction droplet at the end of each 95 °C step. For conventional PCR, the reaction mixture composition was identical but scaled up to 20 μL total, and temperature cycling was identical but accomplished using a conventional bench-top thermocycler (CFX96; Bio-Rad; Hercules, CA). PCR products were analyzed by gel electrophoresis, using 2% agarose gels in the E-Gel electrophoresis system (Life Technologies; Carlsbad, CA).Acknowledgements
This work was funded by the Sandia National Laboratories (SNL) Laboratory-Directed Research and Development (LDRD) program (grant 158814). SNL is a multi-program laboratory managed and operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the U.S. Department of Energy's National Nuclear Security Administration under contract DE-AC04-94AL85000.References
- M. J. Jebrail, M. S. Bartsch and K. D. Patel, Lab Chip, 2012, 12, 2452–2463 RSC.
- M. G. Pollack, R. B. Fair and A. D. Shenderov, Appl. Phys. Lett., 2000, 77, 1725–1726 CrossRef CAS PubMed.
- J. Lee, H. Moon, J. Fowler, T. Schoellhammer and C. J. Kim, Sens. Actuators, A, 2002, 95, 259–268 CrossRef CAS.
- A. R. Wheeler, Science, 2008, 322, 539–540 CrossRef CAS PubMed.
- M. W. L. Watson, M. J. Jebrail and A. R. Wheeler, Anal. Chem., 2010, 82, 6680–6686 CrossRef CAS PubMed.
- M. J. Jebrail, A. Sinha, S. Vellucci, R. F. Renzi, C. Ambriz, C. Gondhalekar, J. S. Schoeniger, K. D. Patel and S. S. Branda, Anal. Chem., 2014, 86, 3856–3862 CrossRef CAS PubMed.
- H. Kim, M. J. Jebrail, A. Sinha, Z. W. Bent, O. D. Solberg, K. P. Williams, S. A. Langevin, R. F. Renzi, J. L. Van De Vreugde, R. J. Meagher, J. S. Schoeniger, T. W. Lane, S. S. Branda, M. S. Bartsch and K. D. Patel, PLoS One, 2013, 8, e68988 CAS.
- A. Sinha, M. J. Jebrail, H. Kim, K. D. Patel and S. S. Branda, J. Visualized Exp., 2013, e50597 CAS.
- M. J. Jebrail and A. R. Wheeler, Anal. Chem., 2009, 81, 330–335 CrossRef CAS PubMed.
- N. A. Mousa, M. J. Jebrail, H. Yang, M. Abdelgawad, P. Metalnikov, J. Chen, A. R. Wheeler and R. F. Casper, Sci. Transl. Med., 2009, 1, 1ra2 Search PubMed.
- M. J. Jebrail, A. H. C. Ng, V. Rai, R. Hili, A. K. Yudin and A. R. Wheeler, Angew. Chem., Int. Ed., 2010, 49, 8625–8629 CrossRef CAS PubMed.
- W. C. Nelson, I. Peng, G. A. Lee, J. A. Loo, R. L. Garrell and C. J. Kim, Anal. Chem., 2010, 82, 9932–9937 CrossRef CAS PubMed.
- P. Y. Keng, S. Chen, H. Ding, S. Sadeghi, G. J. Shah, A. Dooraghi, M. E. Phelps, N. Satyamurthy, A. F. Chatziioannou, C. J. Kim and R. M. Van Dam, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 690–695 CrossRef CAS PubMed.
- M. J. Jebrail, N. Assem, J. M. Mudrik, M. D. M. Dryden, K. Lin, A. K. Yudin and A. R. Wheeler, J. Flow Chem., 2012, 2, 103–107 CrossRef CAS.
- M. G. Pollack, A. D. Shenderov and R. B. Fair, Lab Chip, 2002, 2, 96–101 RSC.
- D. Brassard, L. Malic, F. Normandin, M. Tabrizian and T. Veres, Lab Chip, 2008, 8, 1342–1349 RSC.
- M. J. Jebrail and A. R. Wheeler, Curr. Opin. Chem. Biol., 2010, 14, 574–581 CrossRef CAS PubMed.
- M. Abdelgawad, S. L. S. Freire, H. Yang and A. R. Wheeler, Lab Chip, 2008, 8, 672–677 RSC.
- I. Barbulovic-Nad, S. H. Au and A. R. Wheeler, Lab Chip, 2010, 10, 1536–1542 RSC.
- S. Au, S. C. Shih and A. Wheeler, Biomed. Microdevices, 2011, 13, 41–50 CrossRef CAS PubMed.
- J. Gong and C. J. Kim, J. Microelectromech. Syst., 2008, 17, 257–264 CrossRef PubMed.
- J. Gong and C. J. Kim, Lab Chip, 2008, 8, 898–906 RSC.
- C. Mauney, Thermal Considerations for Surface Mount Layouts, Texas Instruments, 2005 Search PubMed.
- G. J. Shah, H. Ding, S. Sadeghi, S. Chen, C. J. Kim and R. M. van Dam, Lab Chip, 2013, 13, 2785–2795 RSC.
- H. Ren, R. B. Fair and M. G. Pollack, Sens. Actuators, B, 2004, 98, 319–327 CrossRef CAS PubMed.
- S. C. Shih, R. Fobel, P. Kumar and A. R. Wheeler, Lab Chip, 2011, 11, 535–540 RSC.
- S. A. Langevin, Z. W. Bent, O. D. Solberg, D. J. Curtis, P. D. Lane, K. P. Williams, J. S. Schoeniger, A. Sinha, T. W. Lane and S. S. Branda, RNA Biol., 2013, 10, 502–515 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4lc00703d |
| ‡ Authors contributed equally to this work. |
| § Author Present Address: C.A.: Stanford University, Stanford, California, USA. |
| This journal is © The Royal Society of Chemistry 2015 |