Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
On the translocation of bacteria and their lipopolysaccharides between blood and peripheral locations in chronic, inflammatory diseases: the central roles of LPS and LPS-induced cell death†
Douglas B.
Kell
*a and
Etheresia
Pretorius
*b
aSchool of Chemistry and The Manchester Institute of Biotechnology, The University of Manchester, 131, Princess St, Manchester M1 7DN, Lancs, UK. E-mail: dbk@manchester.ac.uk; Web: http://dbkgroup.org/ Tel: +44 (0)161 306 4492
bDepartment of Physiology, Faculty of Health Sciences, University of Pretoria, Arcadia 0007, South Africa. E-mail: resia.pretorius@up.ac.za; Tel: +27 12 420 2864
First published on 1st September 2015
Abstract
We have recently highlighted (and added to) the considerable evidence that blood can contain dormant bacteria. By definition, such bacteria may be resuscitated (and thus proliferate). This may occur under conditions that lead to or exacerbate chronic, inflammatory diseases that are normally considered to lack a microbial component. Bacterial cell wall components, such as the endotoxin lipopolysaccharide (LPS) of Gram-negative strains, are well known as potent inflammatory agents, but should normally be cleared. Thus, their continuing production and replenishment from dormant bacterial reservoirs provides an easy explanation for the continuing, low-grade inflammation (and inflammatory cytokine production) that is characteristic of many such diseases. Although experimental conditions and determinants have varied considerably between investigators, we summarise the evidence that in a great many circumstances LPS can play a central role in all of these processes, including in particular cell death processes that permit translocation between the gut, blood and other tissues. Such localised cell death processes might also contribute strongly to the specific diseases of interest. The bacterial requirement for free iron explains the strong co-existence in these diseases of iron dysregulation, LPS production, and inflammation. Overall this analysis provides an integrative picture, with significant predictive power, that is able to link these processes via the centrality of a dormant blood microbiome that can resuscitate and shed cell wall components.
Insight, innovation, integrationThe Biological Insight of this manuscript is that while dormant bacteria, including those in blood, are normally unnoticed (as they are invisible to conventional methods of culture), they can by definition be resuscitated and then proliferate for at least a certain number of generations before possibly returning to a state of dormancy. This allows a continuing production and shedding of potent inflammatory agents such as the lipopolysaccharide (LPS) characteristic of the Gram-negative cell wall. Well-established pathways link LPS (sensu lato) to inflammatory cytokine production, and to cell death via apoptosis, programmed necrosis, and pyroptosis, with the accompanying microparticle formation known to occur with these cell death mechanisms. Cytokine-mediated cell death mechanisms that permit both (i) the translocation of bacteria between blood and other tissues, and (ii) localised proliferation leading to inflammation and cell death, are likely to be a major component of the various disease manifestations involved. One established requirement for bacterial resuscitation and proliferation comes from the need for available iron. The Technological Innovation is the use of advanced microscopy techniques to detect these dormant bacteria as well as microparticle formation. The Benefit of Integration comes (i) from bringing together these multiple biochemical elements (bacterial dormancy and resuscitation, LPS-induced inflammatory cytokine production, cytokine-induced cell death, cell-death-induced translocation, and localised cell death induced by LPS), and (ii) by showing their commonality, and the centrality of LPS, across a range of chronic, inflammatory diseases normally considered to lack a microbial component. |
Introduction
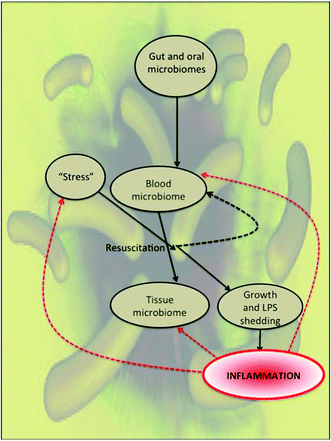
Blood is normally considered a sterile environment in the sense of lacking active microbes, since any bacteraemia or sepsis is potentially extremely life-threatening.1 However, this does not exclude the presence in blood of dormant bacteria, that by definition2,3 are not growing but resist detection by standard culture techniques, yet are not ‘dead’ as they may be resuscitated and then proliferate. We have recently summarised the considerable evidence4,5 to the effect that human blood contains an authentic but dormant microbiome that can contribute significantly to a large variety of chronic inflammatory diseases, a set of diseases that is strikingly similar to those for which we had previously noted the presence of iron dysregulation6–10 and hypercoagulability.9Given the well-established facts (i) that microbial growth in vivo is normally strongly limited by the (non-) availability of free iron (e.g.ref. 11–21), and (ii) that bacterial components such as lipopolysaccharide (LPS) are strongly inflammatory (e.g.ref. 22 and 23), such an analysis leads to the recognition that the iron-related inflammatory diseases also have a major microbial component involving the resuscitation of dormant organisms and their shedding of inflammatory molecules, and especially of cell wall components such as LPS (Fig. 1). LPS is commonly known as endotoxin, albeit that it is frequently shed, and we shall use this name interchangeably unless otherwise specified. Most work has been done with LPS from Gram-negative bacteria, but unless specified, we recognise that much of what we have to say should be taken to apply to inflammatory processes catalysed by cell wall components (such as lipoteichoic acids24) from Gram-positive organisms, ultramicrobacteria,25 and potentially (though there seems to be relatively little work on this26–29) from the cell envelopes of archaea. Also, though many of the ideas developed here very likely apply to them too, and there is a considerable literature, we shall not discuss viruses,30 nor mycoplasmas31–33 in much detail.
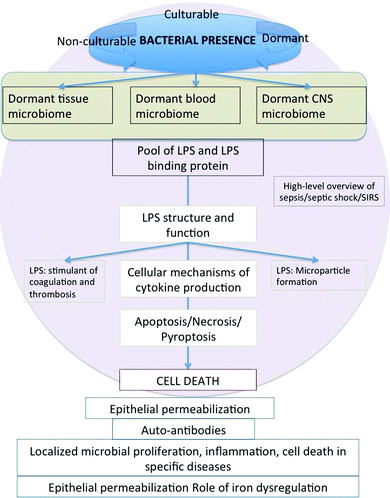
The earlier overviews4,5 recognised that the chief sources of the blood microbiome were likely to be via translocations of microbes from the gut and oral cavities, and although a number of the diseases discussed were neurodegenerative in nature, we did not look at the evidence (and mechanism) for the transport of cells from blood into tissues such as the CNS. A chief purpose of the present review is thus to take a systems approach, designed to bring together the evidence for the strong involvement of microbes and their inflammatory bacterial cell wall components in both the diseases themselves and their dynamics, and relating the known ability of LPS and related component to induce (mainly apoptotic) cell death. This turns out to be sufficient to explain many of the acute and chronic sequelae of the presence of microbes and their cell envelope products in mammals. An overview of the present article is given in the form of a mind map (Fig. 2). As background, we first discuss microbial dormancy, culturability and non-culturability.
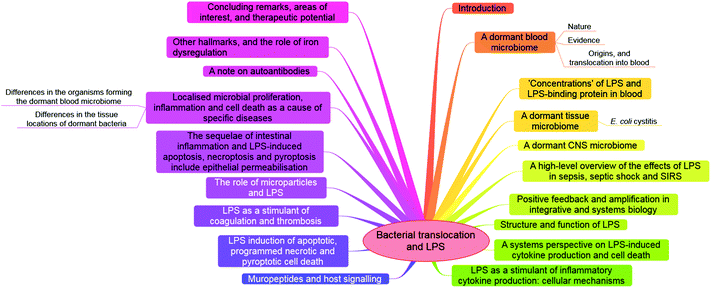 | ||
| Fig. 2 A ‘mind map’34 summarising the article. | ||
A dormant blood microbiome
Nature
As foreshadowed in the introduction, we4,5 and others35,36 have summarised the rather extensive evidence that the presence of bacteria in blood is – perhaps unsurprisingly, given the assumption that blood is normally sterile – much commoner than is usually supposed, and we have pointed out4,5 that, in the usual absence of overt bacteraemia, such organisms are best considered as being in a dormant physiological state.Dormancy is defined as a reversible non-replicating state, often of low metabolic activity. Leaving aside obviously specialised forms such as spores and seeds (‘constitutive dormancy’2), in non-sporulating bacteria it manifests typically as an inability of an individual cell to proliferate (e.g. to form a colony) under conditions normally considered adequate for cultivation, but where the cell is not operationally ‘dead’3 in that it can revert to a state of ‘aliveness’ or ‘culturability’ via processes referred to as resuscitation. Thus, by definition, dormant cells are resuscitable, but this necessarily operational definition means that we can only tell that they were dormant, not that they are dormant.2,3 Indeed, the ability to undergo dormancy (as with pheromone production37) is increasingly being recognised as an adaptive phenotypic trait (e.g.ref. 38–41). Ewald in particular (e.g.ref. 42–44) has stressed the evolutionary aspects of infectious diseases.
Evidence
The evidence for a dormant blood microbiome comes mainly4,5 from its direct assessment using culture-independent methods (although we know of Domingue's resuscitation papers45–47). Culture-independent methods include the detection of blood (or tissue) microbial macromolecules such as rDNA48–59 and the direct visualisation of cells using ultramicroscopic methods (e.g.ref. 5, 43, 45, 46, 60–64). In particular, it is recognised that dormant blood bacteria could ‘hide’ (or at least survive65–68) not only in white cells (e.g.ref. 69) but also within the (more than 1000-fold more numerous) erythrocytes. The significance of this, of course, is the sheer numbers that may be involved. If only one in 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 erythrocytes (that are present in blood at ca. 5 × 109 mL−1) each harboured just a single dormant bacterium, there would still be more than 105 mL−1 (a number equivalent in urine to the threshold normally given for culturable cells in defining clinical bacteriuria).
000 erythrocytes (that are present in blood at ca. 5 × 109 mL−1) each harboured just a single dormant bacterium, there would still be more than 105 mL−1 (a number equivalent in urine to the threshold normally given for culturable cells in defining clinical bacteriuria).
Although there is evidence that a surprisingly large variety of bacteria can invade erythrocytes,5 we know next to nothing about how they enter and egress from such cells. Even whether the latter involves pore-forming toxins70,71 or haemolysins72 that may effect membrane destruction, is unclear.
Origins, and translocation into blood
We also rehearsed5 the considerable evidence that minor leakages from the gut microbiome (e.g.ref. 73–75), even if only a tiny fraction of the 10–100 trillion76 cells involved, whether via specialised structures such as M cells or more significant breaches in the gut epithelium (as can also occur in some cancers77 and stroke78), are more than sufficient to provide a continuing inoculum to the bloodstream. Clearly the innate and adaptive immune system will normally remove those organisms threatening growth and noticeable bacteraemia, but this statement does not account for the fraction that become dormant and hide therefrom (whether geographically in cells and/or by losing their immunogenic potential, for instance by creating L-forms64). While the dormant bacteria do seem mainly to be hidden inside cells, their inflammatory products may not be. In the blood, LPS is typically bound either to an LPS binding protein (LBP)79–82 which is a glycoprotein with a molecular weight of some 58 kDa83 (452 amino acids84) or to the lipoprotein ApoE that is protective against LPS.85–87 The ApoE4 polymorphism is of course well known as a genetic locus favouring the development of Alzheimer's disease (e.g.ref. 87–92).‘Concentrations’ of LPS and LPS-binding protein in blood
Our central argument is that low grade inflammation is mainly effected via the continuing production and shedding of LPS (and similar molecules) as dormant bacteria periodically awaken, proliferate and produce LPS before returning to dormancy. If this is going to be true, it is instructive to determine how much LPS and related molecules are typically found in human blood under various conditions. The potential load of LPS in the alimentary canal is ∼1 g.93 We note, of course, that (as with serum ferritin8) the basis of these assays used to estimate concentration is typically a binding reaction, whether to an antibody or (in the case of LPS) based on a Limulus amoebocyte lysate (or its recombinant factor C94). Thus these and other (e.g.ref. 95–97) assays typically measure the (thermodynamically active) free forms, while the total amounts may be very much greater if (as with LPS) they are mainly bound to LBP or ApoE of HDL/cholesterol, or even monocyte surfaces.98 Indeed, given that HDL–cholesterol is capable of sequestering LPS99 (and lipoteichoic acid100), it is not surprising that there is considerable evidence that HDL–cholesterol is protective against sepsis and sepsis-related death,83,101–107 showing further the importance of free LPS levels in disease prognosis.This said, it is important to point out that if a substance is hydrophobic, i.e. poorly water-soluble, and its targets are hydrophobic (i.e. insoluble) as well, its measured potency also depends on the concentration of the hydrophobic elements containing the target (or otherwise).108,109 In such circumstances, it is arguably better to speak of functional concentrations in terms of nmol per nmol target or similar, rather than in concentration terms (e.g. nM). In a similar vein, when considering properties such as cell death, what matters is the distribution of ligands between targets and the fraction of cells that die. In other words, if an added toxic molecule kills a cell (i.e. irreversibly) then this is a quantised property of the molecule, and again ‘concentrations’ are not an entirely meaningful manner with which to describe the toxic stimulus.3,110,111
Our main purpose here, though, is comparative, and aimed at obtaining a feel for the typical concentrations in health and disease, and those that are used in research studies. Note that as well as coming from infections, LPS is a common component of dust.112 Thus, reported LPS ‘concentrations’ in healthy subjects seem to be of the order of 10–15 ng L−1, while those of LBP are roughly 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times greater at 5–15 mg L−1 (with both values increasing during sepsis) (Tables 1 and 2). LPS challenges of 5–100 μg per patient are commonly administered as experimental challenges and seen as ‘safe’.112 The larger volume, if distributed in 5 L of blood (a typical human value) equates to 20
000 times greater at 5–15 mg L−1 (with both values increasing during sepsis) (Tables 1 and 2). LPS challenges of 5–100 μg per patient are commonly administered as experimental challenges and seen as ‘safe’.112 The larger volume, if distributed in 5 L of blood (a typical human value) equates to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ng L−1, which is obviously much higher than those free amounts typically measured even in sepsis. In terms of relating LPS to microbial biomass (see ref. 113), Watson and colleagues114 showed in laboratory cultures that LPS amounted to some 50 fg cell−1 in a logarithmic growth phase, falling to 29 fg cell−1 in stationary phase, but in the oligotrophic conditions of seawater was just some 2.8 fg cell−1. This shows at once that LPS contents per cell can be quite variable, and that bacteria can shed a considerable amount of LPS at no major harm to themselves. On the basis that 1 mg dry weight of bacteria is about 109 cells, each cell is about 1 pg, so 50 fg LPS per cell equates to about 5% of its dry weight, a reasonable and self-consistent figure for approximate calculations. To deal with the fact that LPS is typically not a molecularly defined substance, its activity is sometimes reported in ‘endotoxin units’ (EU) based on a standard taken115 from an E. coli O55:B5 strain; an approximate relationship is that 1 ng endotoxin ∼10 EU. While the Limulus amoebocyte lysate assay is widely and effectively used as a test for pyrogens in parenteral solutions, its use in the estimation of LPS in blood is not considered especially reliable,116–120 and it may be better to look more closely at LBP. This said, it is LPS that is the stimulus, and thus knowing its effective concentration is important. Unfortunately (Table 2), although the values in sepsis are considerably greater than are those in controls, there is a rather substantial variation between different studies, likely reflecting the rather different qualities of the assays used, the variation in the nature of the LPS (which is not a molecular entity), and the fact that much of the (rather hydrophobic) LPS in vivo is bound to other substances such that the result of the assay depends in significant measure on the extent and nature of any pre-extraction methods employed. Indeed, there are surprisingly few measurements of LPS in non-infectious low-grade inflammation, and very little evidence that plasma or serum LPS might be a particularly useful marker of it. The situation is a little clearer with LPS-binding protein (LBP), with a much more obvious distinction between controls and those with sepsis. Less severe instances of infection include a median of 16 mg L−1 for cirrhosis (interestingly reversed by the antibiotic norfloxacin).121 Thus on the basis of present assay methods, there seems little benefit of seeking to follow the behaviour of a dormant blood microbiome with LPS measurements.
000 ng L−1, which is obviously much higher than those free amounts typically measured even in sepsis. In terms of relating LPS to microbial biomass (see ref. 113), Watson and colleagues114 showed in laboratory cultures that LPS amounted to some 50 fg cell−1 in a logarithmic growth phase, falling to 29 fg cell−1 in stationary phase, but in the oligotrophic conditions of seawater was just some 2.8 fg cell−1. This shows at once that LPS contents per cell can be quite variable, and that bacteria can shed a considerable amount of LPS at no major harm to themselves. On the basis that 1 mg dry weight of bacteria is about 109 cells, each cell is about 1 pg, so 50 fg LPS per cell equates to about 5% of its dry weight, a reasonable and self-consistent figure for approximate calculations. To deal with the fact that LPS is typically not a molecularly defined substance, its activity is sometimes reported in ‘endotoxin units’ (EU) based on a standard taken115 from an E. coli O55:B5 strain; an approximate relationship is that 1 ng endotoxin ∼10 EU. While the Limulus amoebocyte lysate assay is widely and effectively used as a test for pyrogens in parenteral solutions, its use in the estimation of LPS in blood is not considered especially reliable,116–120 and it may be better to look more closely at LBP. This said, it is LPS that is the stimulus, and thus knowing its effective concentration is important. Unfortunately (Table 2), although the values in sepsis are considerably greater than are those in controls, there is a rather substantial variation between different studies, likely reflecting the rather different qualities of the assays used, the variation in the nature of the LPS (which is not a molecular entity), and the fact that much of the (rather hydrophobic) LPS in vivo is bound to other substances such that the result of the assay depends in significant measure on the extent and nature of any pre-extraction methods employed. Indeed, there are surprisingly few measurements of LPS in non-infectious low-grade inflammation, and very little evidence that plasma or serum LPS might be a particularly useful marker of it. The situation is a little clearer with LPS-binding protein (LBP), with a much more obvious distinction between controls and those with sepsis. Less severe instances of infection include a median of 16 mg L−1 for cirrhosis (interestingly reversed by the antibiotic norfloxacin).121 Thus on the basis of present assay methods, there seems little benefit of seeking to follow the behaviour of a dormant blood microbiome with LPS measurements.
| Tissue type | LBP in disease (mg L−1) | LBP in control (mg L−1) | Place | Ref. |
|---|---|---|---|---|
| Bacterial gastrointestinal infections | 28.5 ± 16.5 | — | Serum | 122 |
| Crohn's disease (CD) and ulcerative colitis (UC) | 57.11 (49.4–65.8) | 50.01 (37.1–63.9) | Plasma | 123 |
| Diabetes type 2 | 19.78 ± 6.40 | 20.53 ± 6.99 | Serum | 124 |
| Endocarditis | ||||
| Infectious endocarditis | Median 33.41 | Median 5.61 | Serum | 125 |
| Noninfectious heart valve diseases | Median 6.67 | |||
| Inflammatory bowel disease | 52.7 (45.4–64.6) | 39.1 (32.1–43.7) | Serum | 123 |
| Lifestyle factors | ||||
| Smoking | 7.11 (5.85–8.74) | 7.18 (5.42–9.15) | Serum | 83 |
| Obese | 5.90 (5.09–7.67) | 7.75 (6.35–9.47) | Serum | |
| Overweight | 5.90 (5.09–7.67) | 7.29 (5.96–8.78) | Serum | |
| Metabolic syndrome | 6.82 (5.48–8.40) | 8.02 (6.63–9.82) | Serum | |
| Obesity, T2D and metabolic syndrome | 27 | 10 | Plasma | 126 |
| Liver | ||||
| Hepatocytes | 5 to 15 | — | Cells | 127 |
| Hepatic macrophages | LPS concentration were 10-fold higher than in the healthy controls | Plasma | 128 | |
| Urinary tract infection in childhood | >43.23 | — | Serum | 129 |
| Sepsis | ||||
| Sepsis/septic shock LBP concentration at onset of severe sepsis | 46.2 (3.74–155) | 7.94 | Serum | 127 |
| Sepsis in neonates | Median 36.6 | Median 7.8 | Plasma | 130 |
| Late-onset neonatal sepsis (LONS) | 17.5 | Unstated | Plasma | 131 |
| Gram +ve or Gram −ve sepsis | 216 | 16 | Plasma | 132 |
| (Higher in survivors) | 31 | 4 | Plasma | 133 |
| Septic shock tests | 200 | 5–15 | Unstated | 134 |
| Non-survivors 121 vs. 77 at 48 h | 116–132 baseline | Serum | 135 | |
| Remained much higher in non-survivors | 34–55 | 8–15 | Serum | 136 |
| Disease | LPS in disease (ng L−1 unless indicated as EU mL−1) | LPS in controls (ng L−1 unless indicated as EU mL−1) | Tissue type | Ref. |
|---|---|---|---|---|
| Healthy individuals | — | 0.15 to 0.35 EU mL−1 | Plasma | 137 |
| Non-obese, post-menopausal women | 10–20 | Serum | 93 | |
| Healthy controls | 5 | Plasma | 133 | |
| HIV infection | 60 | 138 | ||
| Inflammatory bowel disease | 12.6 (5.9–16.2) | 12.2 (3.8–26.3) | Serum | 123 |
| Non-alcoholic fatty liver disease | 7.8–14.8 EU mL−1 | 3.2–5.2 EU mL−1 | Serum | 139 |
| Sepsis | ||||
| 300 | 7.3 | Plasma | 133 | |
| 470 | Not noted | Whole blood | 140 | |
| Type 1 diabetes | ||||
| Microalbuminuria group | 31–60 EU mL−1 | Plasma LAL assay | 141 | |
| Normoalbuminuric group | 38–74 EU mL−1 | Plasma LAL assay | ||
| Type 2 diabetes | ||||
| Non-obese postmemoposal women | — | 0.37 ± 0.02 EU mL−1 | Plasma LAL assay | 93 |
| Diabetic non-obese postmenoposal women | 0.39 ± 0.03 EU mL−1 | Plasma LAL assay | ||
| Insulin-treated diabetes | 6.6–10.7 EU mL−1 | 3.1–5.1 EU mL−1 | Serum | 142 |
| Atherosclerosis | Above 50 gave 3× greater chance of atherosclerosis | 14 | 143 | |
A dormant tissue microbiome
Our previous reviews4,5 (and many other works, e.g. those summarised in ref. 43, 62, 64 and 144) outlined in some detail the fact that many known infectious agents can enter cells and persist intracellularly, and those discussions are not repeated in detail here. Indeed, the very existence of eukaryotes is considered to be based on the intracellular uptake of prokaryotes to form structures such as mitochondria,145 and there is increasing evidence for dinitrogen fixation by endosymbionts in plant leaf cells (e.g.ref. 146 and 147). Regarding human tissue, as Nash and colleagues put it,144 “the blood is the most effective vehicle of all for the spread of microbes through the body. After entering the blood they can be transported within a minute or two to a vascular bed in any part of the body. In small vessel such as capillaries and sinusoids where blood flows slowly, there is an opportunity for the microorganism to be arrested and to establish infection in neighbouring tissues.” (The same holds true, of course, for circulating tumour cells and their role in metastasis.) Later we shall look at this translocation from blood to tissues in more detail. However, we first mention an example that we did not deal with previously in much detail, viz. E. coli-based cystitis.E. coli-based cystitis
Cystitis (inflammation of the bladder) is commonly caused by urinary tract infection, typically by E. coli,148,149 and especially in women. It can also lead to bacteraemia.150 A particular point of present interest regarding dormancy151 is the fact that a high percentage of cystitis patients suffer reinfection,152–159 that is often clearly from the same strains that caused the original infection.160–164 This has led to the recognition in bladder epithelial cells of so-called ‘quiescent intracellular reservoirs’156,158,165–170 of dormant cells that can resuscitate. Because one cannot determine these things in humans in vivo, it is not known precisely how they enter such cells after binding to appropriate receptors such as uroplakins,171 but it is presumed that as with many other cells where it is better understood this occurs via endocytosis of some kind.Separating the blood from certain tissues are physical barriers such as the blood–brain, blood–retina and blood–testis barriers, consisting of layers of epithelial cells with especially tight junction. They are of notable significance to drug transport(ers) as well.172–175 It is of particular interest that even here we can find that these barriers are (or must be) breached from time to time, as dormant microbes can be found even in the CNS. Possible means of resuscitation are discussed below and elsewhere.4
A dormant CNS microbiome
As one might suppose, the CNS differs little from other tissues with regard to the possibility that dormant microbes may persist there, occasionally ‘waking up’ to cause trouble. Three examples, as they pertain to the aetiology of Alzheimer's disease (and presumably other dementias) are represented by Chlamydia pneumoniae (as stressed by Balin and colleagues, e.g.ref. 176–185), by herpes simplex virus (as highlighted by Itzhaki and colleagues30,88,182,186–195) and by a variety of spirochetes (as championed by Miklossy and colleagues196–203). The latter is consistent with the well-established dementia in the terminal stages of another spirochetal disease in the form of syphilis, and also with Lyme disease.199,204–206 Of course multiple classes of microorganisms may contribute. There is also evidence for a CNS involvement of the parasitic protozoan Toxoplasma gondii in a numbers of neurodegenerative diseases.207–209The ‘gut-brain axis’ describes the well-established observations of a bidirectional neurohumoral communication system in the human body. Given the above, it is not surprising that there has been increasing recognition over the last couple of years of quite overt (and bidirectional) communication between the gut microbiome and the CNS via the gut-brain axis, in particular, the idea that bacteria in the gastrointestinal (GI) tract can activate neural pathways and CNS signalling systems, from the earliest moments in life. Dysfunctions of this axis can lead to all kinds of neurological problems, including anxiety, depression and other CNS disorders (e.g.ref. 210–229). Most studies have focussed on the endocrine and immune systems. As yet, however, we have found no literature that has sought a role for LPS here, although given that LPS is actually used in a variety of experimental models (e.g. for Parkinson's4,230–237 and even obesity238) to initiate CNS disorders, it is an easy prediction that it is likely to play a significant role in the gut microbiota–brain interaction.
A high-level overview of the effects of LPS in sepsis, septic shock and SIRS
Although our focus here is more on chronic inflammatory states induced by dormant and resuscitating bacteria, it is instructive first to consider events that occur in the more extreme and life-threatening cases of sepsis, septic shock, and the systemic inflammatory response syndrome (SIRS). While these are commonly observed in the Intensive Therapy Unit as a result of an initial infection (hence the term ‘sepsis’), their typical treatment there with broad spectrum antibiotics means that proliferating microbes are rare or absent, and it is their products such as LPS that are then the main problem. Specifically, although these are responsible for invoking the innate immune response that triggers cells to attack and dispose of the invading microbes, an overstimulation of these activities leads to the life-threatening ‘cytokine storms’ that are the proximate causes of, and reflect, endotoxic or septic shock or SIRS (e.g.ref. 7 and 239–247).It is worth rehearsing the definitions248,249 to help discriminate sepsis250,251 from its sequelae. Thus, sepsis has been defined as “the presence (probable or documented) of infection together with systemic manifestations of infection”,252 while severe sepsis is defined as sepsis plus sepsis-induced organ dysfunction or tissue hypoperfusion252 Septic shock is “sepsis-induced hypotension persisting despite adequate fluid resuscitation”.252 SIRS refers to “the systemic inflammatory response to a variety of severe clinical insults” (infectious or otherwise). It usually involves two or more of the following criteria: (i) temperature >38 °C or <36 °C; (ii) heart rate >90 beats per m; (iii) respiratory rate >20 breaths per m or PaCO2 <32 mm Hg; (iv) WBC count >12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μL−1 or <4000 μL−1 or (v) >10% immature neutrophil forms (i.e., “bands”).253,254 The chief point about recognising and using SIRS instead of ‘sepsis’ is, of course, that it does not rely on the presence of observable (or culturable) microbes (and it can anyway be caused by traumas lacking an immediate microbial component). A Venn diagram (redrawn from ref. 254) illustrates the main ideas (Fig. 3).
000 μL−1 or <4000 μL−1 or (v) >10% immature neutrophil forms (i.e., “bands”).253,254 The chief point about recognising and using SIRS instead of ‘sepsis’ is, of course, that it does not rely on the presence of observable (or culturable) microbes (and it can anyway be caused by traumas lacking an immediate microbial component). A Venn diagram (redrawn from ref. 254) illustrates the main ideas (Fig. 3).
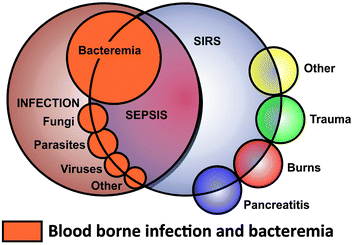 | ||
| Fig. 3 Relationships and overlaps between bacteraemia, sepsis and systemic inflammatory response syndrome. Redrawn from ref. 254. | ||
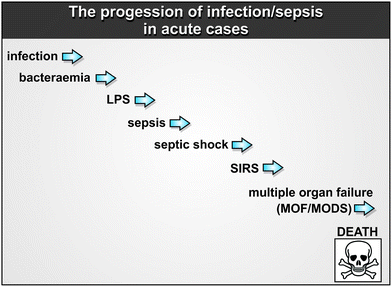
Clearly any increases in microbial cell numbers increase the likelihood of LPS production and shedding that leads to the cytokine storm. Thus, the progression of the microbial variant in unfavourable cases goes roughly from left to right in Fig. 4, as infection → bacteraemia → LPS → sepsis → septic shock → SIRS → multiple organ failure (MOF/MODS) → death. Mortality rates from sepsis/SIRS are extremely high (30–70% in intensive care units),241,242 and dependent on age.255 Note too that antibiotics can themselves promote shedding of LPS from dying bacteria (e.g.ref. 118 and 256–265), especially from spirochetes such as Borrelia burgdorferi, leading to a Jarisch–Herxheimer (JH) reaction.266,267 The JH reaction can be mitigated by antibodies to TNF-α.268 Importantly, this continual shedding of LPS is a normal property of growing Gram-negative bacteria, especially in media such as those containing serum that are inimical to their growth.257,269 This has obvious implications.
Even if one survives septic shock, there are other sequelae, such as long-term cognitive impairment,270 cardiovascular271 and other272 complications that may reflect or contribute to symptoms seen under more chronic conditions.273 Indeed, there are a great many comorbidities between various diseases with a microbial component.36
Clearly, LPS plays a central role in the development of inflammation. We therefore spend time in the next paragraphs to discuss the role of LPS in the cellular inflammatory processes.
Positive feedback and amplification in integrative and systems biology
The first point to make here is a general one about how various kinds of kinetic schemes or network topologies can amplify biochemical signals. For a single enzyme, if its product or any other molecule is an uncompetitive inhibitor that binds only to the enzyme–substrate complex, a small amount of this can lead to a very large increase in a substrate concentration. This serves to explain both the peculiar effectiveness of glyphosate as a herbicide,274–277 and the extreme rarity of uncompetitive inhibition in natural systems.278 One type of network-based amplification, that is familiar in signalling cascades, is one in which a signalling activity such as a kinase changes the activity of another kinase, and so on. Here, as well as amplification, the cascade is partly about serving as a suitable delay loop,279 as clearly variations in amino acid sequence can and do280 have major effects on the activities of individual proteins such that the cascade would otherwise seem unnecessary. A second kind of amplification, known as ultrasensitivity, comes from a structure in which an effector stimulates by covalent modification (e.g. phosphorylation) of an enzyme catalysing a particular reaction, while simultaneously inhibiting a second enzyme (e.g. a phosphatase) catalysing the removal of the covalent modification. This leads to very large changes in flux and network behaviour as the concentration of the effector passes a threshold.281–285 Similarly, pulsatile or oscillatory signals can be much more effective for the same ‘average’ concentration.286,287 Thus, a variety of network motifs can provide “sniffers, buzzers, toggles and blinkers”.288 And most of all, although other behaviours are possible,289 a variety of simple systems with positive feedback can amplify a very small signal into a much larger one (Fig. 5). This can typically occur in inflammatory systems7 where molecules whose production is induced by LPS, such as IL-1β290–293 (Fig. 6) or TNFα (see later), can stimulate their own synthesis or effect crosstalk (e.g. IL-1 induced TNF-α294).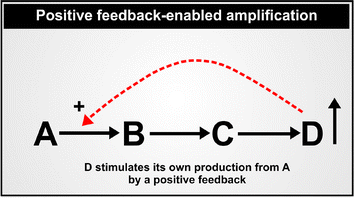 | ||
| Fig. 5 A system exhibiting positive feedback in which a product stimulates its own synthesis. In a biochemical context, A to D represent metabolites, while the arrows represent enzymatic steps. | ||
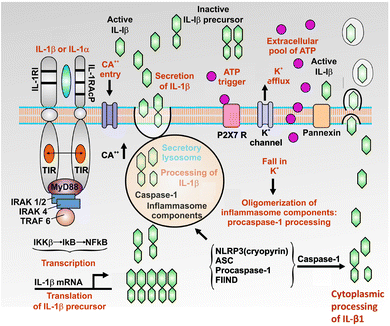 | ||
| Fig. 6 IL-1β is an example of a cytokine that can stimulate its own synthesis (figure redrawn from and based on one in ref. 291). | ||
A second major area where an effector can appear to amplify a small stimulus, or have a large effect, is when it interacts with multiple targets simultaneously (in drug discovery this is known as polypharmacology (e.g.ref. 295–297)). This need actually follows from the principles of systems biology as encapsulated in metabolic control analysis,298–301 where mediators that modify only one target can rarely be expected to have much effect. As we shall see, LPS qualifies here too, as it stimulates a great many proinflammatory and proapoptotic pathways, including those necessary for its translocation in both free and bacterial cell-associated forms.
Thus we shall see that LPS is likely to serve as a major nexus in inflammation in general and that induced by microbes in particular.
Structure and function of LPS
An overall ‘cartoon’-type structure of typical LPS molecules is given in Fig. 7A based on.302 They consist of an inner lipid A core and a number of branched polysaccharide chains (e.g.ref. 302–307), terminating in those that determine the strain's serology via the O-antigen.308 The biosynthesis is discussed by Wang and Quinn.309 According to the comprehensive LIPIDMAPS classification,310 LPS is a saccharolipid glycan. The lipid A core is significantly the most inflammatory part of the molecule,311 with typically two N-acetylglucosamine residues attached to a 2-keto-3-deoxy-D-manno-octulosonic acid (Kdo) disaccharide (Fig. 7B). Bacteria lacking the outer O-antigen chains are known as ‘rough’ and are significantly more immunostimulatory than are their ‘smooth’ equivalents that contain them.312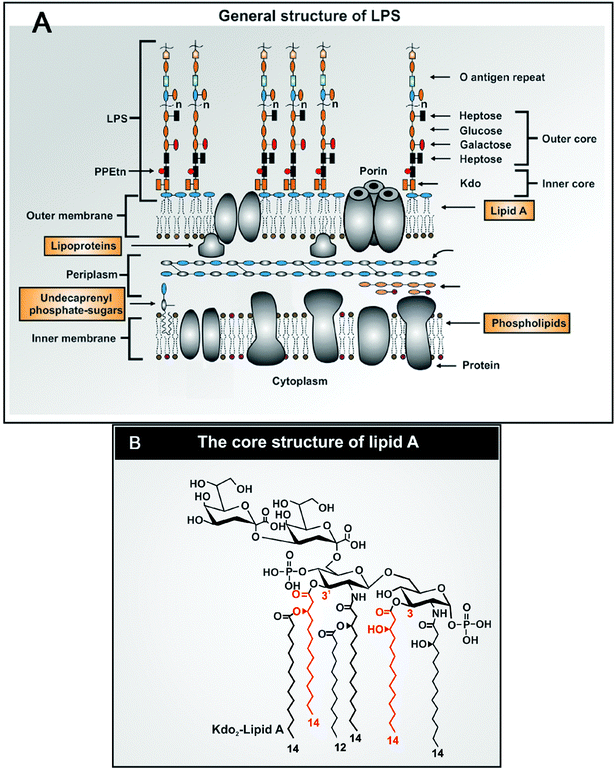 | ||
| Fig. 7 The general structure of (A) LPS and (B) its core lipid A component. Redrawn from ref. 302. | ||
A systems perspective on LPS-induced cytokine production and cell death
A standard approach to systems biology modelling (e.g.ref. 279 and 289) has four main stages. The first two are qualitative, and involve determining the players and how they interact, whether as substrates, products or effectors – this establishes the topology of the network. The second two are more quantitative, involving the mathematical form of the equations describing each step, and their parametrisation, also involving the running of the model, typically as a set of coupled ordinary differential equations, using suitable software (e.g.ref. 313–315). Note too that in a typical network of this type, a systems approach typically discriminates parameters from variables. Parameters are either fixed or outside the control of the experimenter; they typically include the unchanging concentrations of substances involved in flux-generating steps, as well as kinetic constants such as Km and kcat. By contrast, variables are those things that vary during an experiment, typically involving concentrations of intermediary substances and fluxes through pathways towards ‘exit’ variables. Thus, figures such as those in Fig. 1, while accurate in the sense of illustrating flows of information, are misleading because they are at once both static and qualitative. While we can and shall point to many papers that show clearly that “LPS can cause inflammation (or apoptosis)”, such a statement too is less than complete. This is because individual papers rarely if ever state, use or vary systematically a number of parameters that are known to have a huge impact on cell fate. These include• The exact type of LPS (even though some, especially their lipid A component, are known to be much more immunogenic or inflammatory than are others – see e.g.ref. 306, 309 and 316–324 and later)
• The rather variable amount of LPS added, whether the assay is for cytokine production (Table 3) or ‘viability’ (Table 4)
| Cell type/animal origin | Cell name | LPS concn administered to cells/animals | LPS type | Measurement: interleukin and TNF-α production | Ref. |
|---|---|---|---|---|---|
| Mouse origin | |||||
| Peritoneal mouse macrophages | Raw 264.7 mouse macrophages | 100 ng mL−1 | Escherichia coli O55:B5 | Increased IL-6 production | 338 |
| C57BL/6 mice | Primary microglial cultures | 100 ng mL−1 | E. coli O26:B6 | Massive increase in IL-1β when purinergic receptor activated | 335 |
| IL-1Ra knockout (KO) and wild-type (WT) mice | Peritoneal macrophages (primary cultures) | 10 μg mL−1 | Aggregatibacter actinomycetemcomitans LPS | Increased production of IL-1α, IL-1β, TNF-α and IL-6 in IL-1Ra KO mice, compared with the levels in WT mice | 339 |
| Mouse alveolar macrophages | MH-S cell lines | 100 ng mL−1 | Sigma LPS type not stated | LPS stimulated overproduction of TNF-α and IL-10 | 340 |
| C57BL/6 mouse | Primary macrophages | 1 μg mL−1 | E. coli O26:B6 | Massive increase in IL-1β but not IL-6 | 341 |
| Mouse microglia | BV2 microglia | 100 ng mL−1 | E. coli O55:B5 | LPS-stimulation induces the expression of inducible NOS (iNOS) and COX-2 | 342 |
| Intact mice (ob/ob), or C57BL/6 with diet-induced obesity | 5 μg or 100 μg kg−1 | E. coli O127:B8 | Massive increases in IL-1β, IL-6 and IL-RA | 238 | |
| J774.A1 macrophages | Primary cultures | 10 μg mL−1 | Not stated, but was E. coli O26:B6 | Huge increase in IL-1β production | 343 |
| Rat origin | |||||
| Rats | Serum level measurement | 100 μg per rat, ip | Not stated | Increased IL-6 and TNF-α production | 344 |
| Sprague-Dawley rats hippocampal brain slices | Primary culture | 10 μg mL−1 | E. coli O55:B5 | Significant increased concentration of TNF-α and IL-1β | 345 |
| Human origin | |||||
| Human bone marrow aspirates | Bone marrow mesenchymal stem cell primary cultures | 0 to 10 μg mL−1 | Porphyromonas gingivalis LPS | IL-6 production independent of the LPS dosage | 346 |
| Peripheral blood mononuclear cells | Primary culture white blood cells | 0.1, 1 or 100 ng mL−1 | Not stated | IL-6 and TNF-α were both strongly upregulated | 347 |
| HepG2 | Hepatoma cell line | 0.1, 1 or 100 ng mL−1 | Not stated | Slight inhibition of TNF-α | 347 |
| Peripheral blood mononuclear cells (PBMC) (from healthy and type 2 diabetes individuals (T2D)) | Primary culture white blood cells | 2 and 0.2 ng mL−1 | TLR4 ligands LPS | With low LPS dose the T2D cohort exhibited enhanced IL-1β relative to healthy cells | 348 |
| Nasal polypfibroblasts | Primary cultures | 10 μg mL−1 | E. coli O111:B4 | LPS enhanced the secretion of IL-6 | 349 |
| Other animal origin | |||||
| Airway neutrophilia was induced in horses by inhalation of LPS | Animal model | 1 mg mL−1 per horse | Sigma LPS type not stated | Significant increased concentration of IL-6 and TNF-α | 350 |
| Cell type | Cell name | LPS conc (ng mL−1) | LPS type | Viability assay | Viability % | Ref. |
|---|---|---|---|---|---|---|
| Rat origin | ||||||
| Rat duodenum | Epithelial cells | 0.75–3 mg kg−1 i.v. or 3–12 mg kg−1 p.o | H. pylori LPS | MTT | 60% | 351 |
| Rat myocytes | 25–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (most 100) 000 (most 100) |
Unstated | Apoptosis | 80% | 352 | |
| Alveolar macrophages | NR8383 | 10 | Unstated | LDH and Hoechst/PI | 90% without particulates | 353 |
| Myocardial myocytes | H9c2 | 1000 | MTT, LDH, TUNEL, JC-1 | 85% | 354 | |
| Myocardial myocytes | H9c2 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
MTT | 65% | 355 | |
| Mouse origin | ||||||
| C57BL/6 mice | Primary microglial cultures | 100 | E. coli O26:B6 | LDH release | ∼10% in presence of ATP | 335 |
| Osteoblast | MC3T3-E1 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
E. coli O55:B5 | MTT | 70% | 356 |
| Mouse macrophages | RAW 264.7 | 100 | Unstated | MTT | Not stated; increased apoptosis | 357 |
| Macrophages | BMDM | 100 (6 h) | Unstated | DNA fragmentation | ∼60% | 358 |
| Human origin | ||||||
| Pulmonary epithelia | A549 | 1000 | Unstated Sigma | Unstated kit | 60–70% | 359 |
| Human PBMC | PBMC | 0.01–3 | E. coli unstated | None – only IL-8 and ROS | 360 | |
| Microgial | BV2 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–30 000–30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Unstated | MTT and trypan blue | ∼60% | 361 |
| Vascular human endothelia | HUVECs | 500 | Unstated | Annexin | 80% | 362 |
| Dopaminergic | SH-SY5Y | 100 | E. coli O127:B8 | MTT | Unclear | 363 |
• The nature of the host (human or rodent, that respond differently,325–327 or in vitro)
• Whether the LPS is added in a bolus or a dynamic manner (this matters a lot287)
• Whether measurements are done in single cells or as an ensemble, and or quasi-continuously; this matters because much evidence in the NF-κB system287,328–330 and other related systems331–334 shows that it is the nature and dynamics of the oscillations that determines which genes are transcribed and with which kinetics, as well as cell fate, and not just say an NF-κB concentration at a particular time. Thus knowledge of single-cell behaviour is vital3,110,111 (also in pharmaceutical drug uptake174,175).
• Which other substances, conditions or parameters have been co-varied or even recorded, and whether they themselves are known to modify the effect of LPS alone. One example is ATP, which activates the purinergic PX27 receptors and increases massively the extent of cell death.335,336 Another is acidosis.337
LPS as a stimulant of inflammatory cytokine production: cellular mechanisms
While only partly consistent with the ‘danger theory’ of the immune response,364,365 and more obviously stemming from the ideas of Janeway,366 LPS is recognised as a major ‘pathogen-associated molecular pattern’ or PAMP that triggers the body's innate immune response to pathogens (e.g.ref. 367). In addition, cells release damage-associated molecular pattern molecules (DAMPs) as signals that alert the innate immune system to unexpected cell death and to microbial invasion. It is now very well established that a chief means by which LPS excites an inflammatory innate immune response is by binding to the toll-like receptor 4 (TLR4).368–373 Typically, the LPS is bound in blood to LBP, and the LPS is ferried to the TLR4 receptor via a CD14 co-receptor (which also links innate immunity with Alzheimer's disease374–377). This binding of LPS to TLR4 in turn activates the production of a variety of pro-inflammatory cytokines.378–381 The extent of cytokine activation reflects in part the strength of binding to CD14/TLR4.320 These inflammatory cytokines are induced via a set of canonical pathways illustrated in Fig. 8, with the transcription factor NF-κB playing a prominent role.382–385 As is also well known (and see below), NF-κB is normally held inactive in the cytoplasm by being bound to an inhibitor IκB protein, and the means by which extracellular signals such as LPS are transduced involve a series of kinases, one of which (IKK) in particular phosphorylates the IκB and thereby releases the NF-κB that can translocate to the nucleus to turn on a large variety of other genes, including in particular TNF-α and IL-6.386 There is also a ‘non-canonical’ inflammasome LPS activation pathway independent of TLR4,387–389 that occurs at higher external concentrations of LPS,23,390 comes into play when the LPS is internalised, and involves (via p38 MAP kinase and intracellular LPS) the activation and secretion of cytokines such as IL-1β (Fig. 9) and also TNF-α).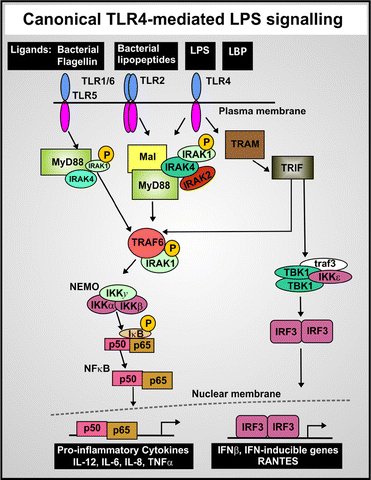 | ||
| Fig. 8 The LPS-mediated cellular production of inflammatory cytokines. Canonical pathway of LPS-mediated release and nuclear translocation of NF-κB (based on ref. 379). | ||
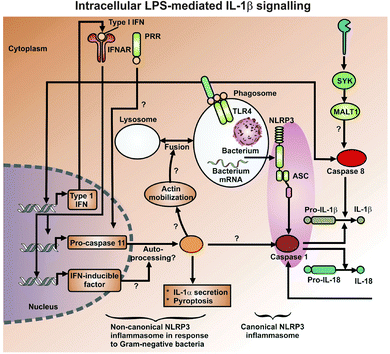 | ||
| Fig. 9 The intracellular LPS-mediated activation of caspase-1 leading to IL-1β production (after ref. 391). | ||
Muropeptides and host signalling
A more recent recognition is that as well as lipid A, shorter bacterial cell-wall derived muropeptides also have a significant role in innate immunity (e.g.ref. 392–405; they act synergistically with LPS,396 probably because they also interact with the NF-κB pathway, via the RICK/Rip2/CARDIAK kinase406,407). It is of particular interest in the context of bacterial dormancy that such muropeptides seem to be part of the ‘wake-up’ activity of the bacterial Rpf408 and other bacterial resuscitation systems.409 See Fig. 10 for a visual representation of the host signalling pathway of MDPs.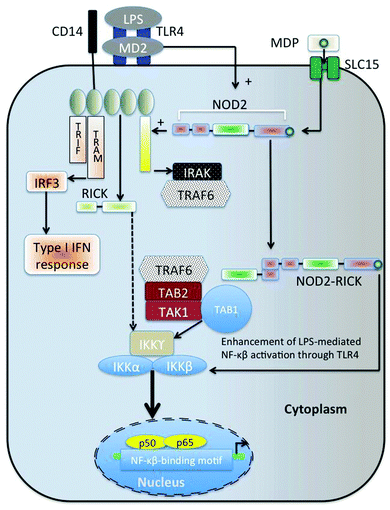 | ||
| Fig. 10 Host signalling pathways of muramyl peptides (MDP). Based on and redrawn from ref. 405. MDP are taken up via the pepT1 oligopeptide transport system (SLC15 – see ref. 410). | ||
LPS induction of apoptotic, programmed necrotic and pyroptotic cell death
A particularly important inflammatory cytokine whose secretion is induced by NF-κB (and also by p38 MAP kinase) is TNF-α (e.g.ref. 411–414). This too is inflammatory and itself induces further changes in NF-kB expression. The same is true for IL-1β.343,378 In particular, these inflammatory cytokines can lead to apoptotic cell death (e.g.ref. 343 and 358). As we shall see, however, and as presaged in Tables 3 and 4, it is quite difficult to establish precisely what is going on in many cases, as individual studies tend to study a restricted set of pathways and individual players, be they NF-κB, p38, IL-1β or IL-6. It would not in our view be completely unfair to describe a lot of these studies collectively as ‘a bit of a mess’. Taking NF-κB as a canonical example (though the same is true for p38415), what are ostensibly the same kinds of signal can lead to dramatically different cell fates, e.g. proliferation vs. apoptosis416–424 depending on the conditions. What this is telling us, of course, since these processes are considered largely deterministic, is that we are not measuring or controlling all of the relevant factors (see also Table 3), and in great measure these studies are largely qualitative in nature. Here our purposes are thus simply to recognise that the genes induced or repressed via transcription factors such as NF-κB can be pro- or anti-apoptotic, and which ones are activated depend on all prevailing conditions.As well as apoptosis, there is a less tightly (but partly) regulated form of cell death known as ‘programmed necrosis’ or ‘necroptosis’,425–437 that may be induced by inflammatory ligands such as TNF, especially during infection, and that sometimes also involve NF-κB. Another important mode of cell death induced by related stimuli is pyroptosis438–450 (that involves the caspase 1-dependent production of IL-1β). Ferroptosis451,452 is a cell death mechanism that stresses the importance of unliganded iron (see also ref. 6, 7, 453 and 454). Although the stimulus in each of these cases is nominally the same (LPS of some kind) there are presumably pre-existing conditions that differ and thereby determine precisely the kind of cell death that ensues. However, we do not discuss the emerging differences in their molecular details nor taxonomy here, since the important thing for the present arguments is simply that the cells die, disappear, and thereby leave gaps where once they lived.
LPS as a stimulant of coagulation and thrombosis
As we recently reviewed,9 a hallmark of many chronic, inflammatory diseases is the fact that they simultaneously exhibit both hypercoagulability and hypofibrinolysis. While a great many biochemicals can influence both the kinetics and endproduct structures of the clotting process, and we previously highlighted unliganded iron8,10,455–458 and the fibrin concentration itself,9 we can hardly avoid noting that LPS itself is a strong procoagulant.459–463 How direct some of these mechanisms are seems not to have been established, though certainly LPS can bind to erythrocyte membranes.464,465 Given that promiscuity correlates with hydrophobicity (e.g.ref. 173 and 466–469), it is not surprising that the very hydrophobic LPS can potentially interact with a great many (lipo)proteins; its ability to convert prions to their more toxic PrPSc form470 is a pertinent case in point.The role of microparticles and LPS
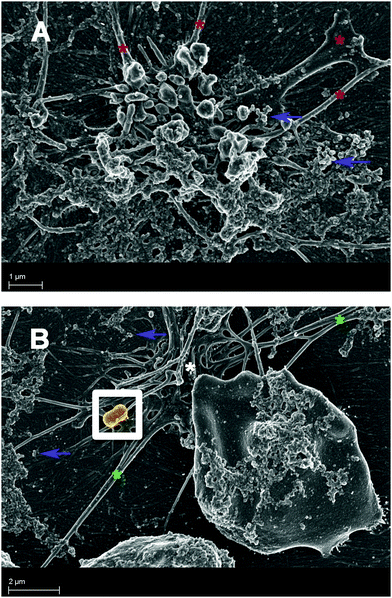
We wish, however, to spend some time on the possible involvement of LPS in microparticle formation. Microparticle formation is typically via apoptosis and the related pathways described above.471–474 Such microparticle formation is prominently associated with inflammatory conditions. Microparticles have also been associated with proinflammatory effects and also with autoimmune processes, and they are thought to be a source of autoantigenic nuclear material, which can form immune complexes.475 Various cells such as platelets, lymphocytes, endothelial cells, erythrocytes and monocytes do release surface-derived microparticles476 and these microparticles are seen as multi-purpose carriers.477 They carry proteins, lipids and nucleic acids, and play a fundamental role in the pathogenesis of thrombosis and are known to modulate the properties of target cells.478 Microparticles from erythrocytes also carry heme and these heme-laden microparticles have a physiopathological impact on the rest of the haematological system.479 Microparticles also frequently elicit an immune response.480–483As microparticles are known to be present in many inflammatory diseases,8,478,484–487 they might therefore develop via an external or internal stimulus on cells (e.g. erythrocytes and platelets), and here we suggest that the stimulus might be LPS. While pure lipid systems are not always good membrane mimics,172–174 LPS has been shown to insert spontaneously into lipid bilayers, and this insertion can lead to membrane breakdown.480,488,489 This insertion capability has also been demonstrated in lipid raft models.490 Given that we have recently shown that bacteria can hide inside erythrocytes, shedding of LPS may thus occur within the cells (as well as obviously outside the cells, where “free” bacteria may shed LPS). There is considerable literature that suggests that LPS can be a cause of apoptosis etc, so LPS shed from internalized cellular bacteria may also trigger apoptotic pathways from within the cells (via caspase-11, caspase-4 and IL-1β pathways), resulting in microparticle formation. This eventually stimulates the processes of coagulation and thrombosis already known to be associated with microparticle presence in inflammatory conditions.491,492 An example of this is shown in Fig. 11A and B, where microparticle formation in thrombo-embolic ischemic stroke is seen associated with both hyperactivated platelets and damaged erythrocytes, together with the presence of bacteria.
The sequelae of intestinal inflammation and LPS-induced apoptosis, necroptosis and pyroptosis include epithelial permeabilisation
It is clear that as the concentration(s) or activities of LPS, inflammatory cytokines and other mediating factors increase, cell death is an inevitable consequence.425,493–495 While, as mentioned above, there is almost certainly a continuing small leakage of microbes from the gut496 (known as the ‘leaky gut’ hypothesis), it is evident that a variety of conditions, that we may loosely refer to as ‘stress’ can increase this considerably (e.g.ref. 73, 78 and 497–505). Bacterial sepsis itself is one such stress,506 and interestingly there is now a burgeoning literature to the effect that the immunodeficiency seen in HIV/AIDS patients may actually be caused by gut-derived LPS causing hyperactivation (then death) of CD4+ cells (e.g.ref. 138 and 507–509). There does not seem to be a major genetic contribution to leaky gut.510 Although many of the same signalling pathways are involved, the extent to which this is mediated via LPS is not yet clear, albeit LPS itself can indeed disrupt tight junctions and increase intestinal permeability.511–516 Overall, these kinds of endothelial dysfunction clearly lead to increased leakiness or permeabilisation430 (Fig. 12).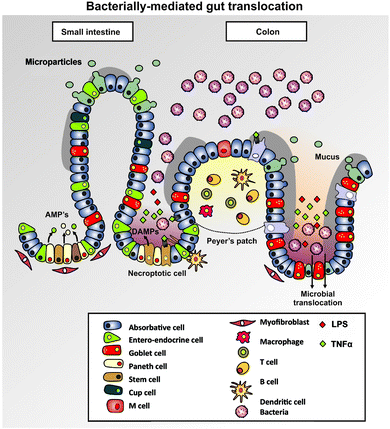 | ||
| Fig. 12 Some of the mechanisms of bacterial translocation from the intestine in a ‘leaky gut’, based on ref. 430. Some cells (and LPS) may pass through tight junctions, while others pass through Peyer's patches or through spaces vacated by dead cells whose death may be initiated by LPS. Diagram is not to scale. | ||
Localised microbial proliferation, inflammation and cell death as a cause of specific diseases
We have here sought to provide a rather general explanation for the role of LPS-induced chronic inflammation in a variety of different diseases, with the ‘continuing’ element driven by the resuscitation of a resident blood and tissue microbiome. However, it is obvious that while all these diseases are inflammatory (and have been linked to microbes4,5 and/or LPS (Fig. 2), diseases such as Alzheimer's, Parkinson's, atherosclerosis and rheumatoid arthritis obviously occur – or one might better say manifest – in largely different tissue or tissue subtypes. Thus, the disappearance of cells from the CNS can manifest as Parkinson's disease if in the dopaminergic neurons of the substantia nigra pars compacta,517 or to more widespread cell disappearance is diseases such as those mediated by prions (e.g.ref. 7 and 518). Note of course that LPS is actually used to induce a form of Parkinson's disease in experimental animals.4,230–237Within the present framework, there are at least three general and straightforward explanations for this differentiation of diseases despite a broad common cause. The first is that (as with infectious diseases), the nature of the microorganisms differs, with some of these inflammatory diseases clearly being more associated with some microbes than are others (e.g.ref. 5, 43 and 64). The second is simply that the tissue location of the dormant bacteria differs. The third follows as much from our ignorance of the molecular details, as discussed above, as from what we know – namely the fact that we know little of the different effects of LPS type and concentration, and the effects of different levels of cytokines and other molecules that may themselves vary this. Sharpening these kinds of analyses will obviously feature as an important area for future research, and we rehearse the first two points briefly.
Differences in the organisms forming the dormant blood microbiome
We5 and others (e.g.ref. 36, 43, 48, 49, 52, 62, 64 and 519–522) have provided both (ultra)microsocopic and molecular (sequence-based) evidence for a very great variety of culture-negative organisms that have been noted to be present in the blood, despite the fact that it is normally considered to be sterile.Differences in the tissue locations of dormant bacteria
In a similar vein, microbes – presumably persistent, dormant organisms – are regularly detected in other tissues in which any degree of proliferation would be highly inimical to the host. Some recent examples involving just reproductive disorders include the vagina,523–525 the placenta,526–530 and the amniotic fluid,531–540 while recent evidence has also been provided for a sub-epidermal microbiome.541 The considerable evidence for a dormant CNS microbiome was discussed earlier, and we here note the presence in the brain of TLR4 and its major role in neurodegeneration,542 consistent with the idea that waking up dormant microbes can stimulate overt neurodegenerative disease.A note on autoantibodies
A number of the diseases (e.g. rheumatoid arthritis, multiple sclerosis, psoriasis) for which we are invoking a microbial component involving LPS are usually considered to be autoimmune diseases. The question then arises as to the origins of this autoimmunity. If LPS were a protein with a defined structure it would be relatively easy to compare its epitope sequences with those of the targets of host antibodies in different circumstances (e.g.ref. 543–546), but of course it is not. This said, there is plenty of evidence that host autoantibodies are elicited by LPS that have less than perfect specificity for the immunogen (e.g.ref. 547–558), so while most of this work is not very recent, this question of LPS-induced antibody non-specificity seems an avenue well worth exploring. We note too the potential toxicity of exogenously administered anti-LPS antibodies.559,560 Regarding an autoimmune hypothesis, Marshall, Proal and colleagues highlight precisely this, along with a role for the vitamin D receptor.35,36,561–565Other hallmarks, and the role of iron dysregulation
While LPS itself as commonly measured seems to be a rather inadequate biomarker for chronic, inflammatory disease (Table 2), LBP (Table 1) and longer-lived markers of LPS exposure like IgM and IgA antibodies4,543–545,566–570 may be more promising.What serves to wake up the dormant microbes is not yet clear, and it is unlikely to be a single element. One possibility is certainly host stress as reflected in noradrenaline levels, a mechanism championed with considerable evidence (at least in terms of stimulating the growth of Gram-negative organisms) by Lyte, Freestone and colleagues.215,222,571–579 We note that catecholamine synthesis may be induced in macrophages by LPS,580,581 and that catecholamines increase inflammation582,583 (another positive feedback loop). It is of special interest here that noradrenaline can act as an iron chelator,584–586 since iron is normally seen as the nutrient most limiting to bacterial growth in vivo (e.g.ref. 16, 19–21 and 587–591). As we have pointed out before,6–10 the diseases highlighted here are precisely those where iron dysregulation, inflammation, coagulopathies, microparticles,592,593 and now a microbial component involving LPS sensu lato, are seen to coexist.
Concluding remarks, areas of ignorance, and therapeutic potential
It is undoubtedly the case that despite our greatly improved understanding of many of the pathways involved in inflammation and sepsis (sensu lato), strategies for drug development have been noted mainly by their relative failure,411,594–597 probably in significant measure because they aim at individual targets (see ref. 297). As a systems problem, the way to improve our therapeutic strategies is to recognise that there are multiple targets or steps. We have here stressed that LPS (sensu lato) created by the resuscitation of dormant bacteria is likely heavily involved, and that the latter can be affected greatly by the concentrations of free iron (that may also be a target for therapy). Fig. 13 shows some of the various translocations of dormant and proliferating bacteria between the different tissues, and the various steps that might thus be targeted. Improving methods for quantitative lipidomics (e.g.ref. 598–603) are likely to be of considerable value here.Here we have stressed the significance of LPS, but initially promising trials of an anti-LPS antibody (HA-1A, Centoxin)604 were not sustained (as is common605,606) because of ineffectiveness607,608 or toxicity.559,560 This was probably because of its lack of specificity between lipid A and other hydrophobic ligands,609 and drug promiscuity is a function of hydrophobicity more generally (e.g.ref. 173, 468 and 610–612). Recognising that factor C, the active component of the Limulus amoebocyte lysate,94 necessarily binds to LPS, Ding, Wohland and colleagues have isolated so-called sushi peptides therefrom,613–620 in particular sushi peptides S1 and S3. These are 34mers that that can bind to and inactivate endotoxin molecules (and are in some cases directly antibacterial), in concert with bacterial phospholipids.621 They are said to have low human cell toxicity, and these peptides (or variants622,623) would seem to have significant therapeutic potential. As a hydrophobic target, variant lipocalins binding LPS (‘anticalins’624–628) might also be of value here. The evidence that HDL can bind to and sequester LPS was given above.
It is hard to gauge whether a better treatment than an anti-LPS antibody might involve recombinant LBP because while LBP can sequester LPS it can also transfer it to TLR4.127,134,629–631 Probably the best strategy is to avoid letting the bacteria proliferate in an uncontrolled manner at all, and to this end an iron-withholding strategy632 seems most suitable (e.g.ref. 6, 7 and 633–636) (and not a fortification one637,638). In a similar vein, many of the substances found to be anti-inflammatory are so because of their iron-chelating properties (e.g.ref. 7 and 639–641), though clearly there are a vast number of possible steps (plural) at which anti-inflammatory substances may act. Those involving downstream cytokines (e.g.ref. 539) are outwith our scope here.
In addition, provided one can avoid excessive LPS shedding and a Jarisch–Herxheimer reaction,266,268 suitable antibiotics (e.g. minocycline642–650) that have polypharmacological properties651,652 must certainly have a role in treating purportedly non-communicable inflammatory diseases, as well as those established as infectious. It is at least worthy of mention,653 albeit drugs can be quite promiscuous with regard to hitting multiple targets,173,466,651,652,654 that certain antibiotics are both used and effective in the treatment of diseases such as multiple sclerosis,655–661 rheumatoid arthritis,544,642,651,652,662–665 and psoriasis,666–668 while vaccination can also work to protect against supposedly non-communicable disease.669,670
A modern trend is to use patients as their own controls (so-called n = 1671 or n-of-1672,673 methods), in suitable crossover designs. These seem ideally suited to chronic diseases of the type discussed here, especially for those in which disease severity can itself change significantly (‘flare’674–677) on quite rapid timescales (for the large and very numerous circadian changes in a mammalian transcriptome, see ref. 678). Indeed, we predict that such flares will be accompanied by major changes in the relevant microbiomes. But to understand the precise mechanisms involved we shall need much better, more quantitative, and above all reliable methods for measuring and manipulating the microbes, pathways and pathway elements involved. This is the most urgent task for the future.
Note added in proof
A recent and complementary article highlights the role of LPS translocation directly from the gut.679Acknowledgements
We thank Dr David Brough for commenting on the manuscript and Prof Ian Roberts for drawing our attention to the literature on quiescent intracellular reservoirs. We thank the Biotechnology and Biological Sciences Research Council (grant BB/L025752/1) as well as the National Research Foundation (NRF) of South Africa for supporting this collaboration. This is also a contribution from the Manchester Centre for Synthetic Biology of Fine and Speciality Chemicals (SYNBIOCHEM) (BBSRC grant BB/M017702/1).References
- Mandell, Douglas and Bennett's Principles and Practice of Infectious Diseases, ed. J. E. Bennett, R. Dolin and M. J. Blaser, Saunders Elsevier, Philadelphia, 8th edn, 2015 Search PubMed.
- A. S. Kaprelyants, J. C. Gottschal and D. B. Kell, Dormancy in non-sporulating bacteria, FEMS Microbiol. Rev., 1993, 10, 271–286 CrossRef CAS PubMed.
- D. B. Kell, A. S. Kaprelyants, D. H. Weichart, C. L. Harwood and M. R. Barer, Viability and activity in readily culturable bacteria: a review and discussion of the practical issues, Antonie van Leeuwenhoek, 1998, 73, 169–187 CrossRef CAS.
- D. B. Kell, M. Potgieter and E. Pretorius, Individuality, phenotypic differentiation, dormancy and ‘persistence’ in culturable bacterial systems: commonalities shared by environmental, laboratory, and clinical microbiology, F1000Research, 2015, 4, 179 Search PubMed.
- M. Potgieter, J. Bester, D. B. Kell and E. Pretorius, The dormant blood microbiome in chronic, inflammatory diseases, FEMS Microbiol. Rev., 2015, 39, 567–591 CrossRef PubMed.
- D. B. Kell, Iron behaving badly: inappropriate iron chelation as a major contributor to the aetiology of vascular and other progressive inflammatory and degenerative diseases, BMC Med. Genomics, 2009, 2, 2 CrossRef PubMed.
- D. B. Kell, Towards a unifying, systems biology understanding of large-scale cellular death and destruction caused by poorly liganded iron: Parkinson's, Huntington's, Alzheimer's, prions, bactericides, chemical toxicology and others as examples, Arch. Toxicol., 2010, 577, 825–889 CrossRef PubMed.
- D. B. Kell and E. Pretorius, Serum ferritin is an important disease marker, and is mainly a leakage product from damaged cells, Metallomics, 2014, 6, 748–773 RSC.
- D. B. Kell and E. Pretorius, The simultaneous occurrence of both hypercoagulability and hypofibrinolysis in blood and serum during systemic inflammation, and the roles of iron and fibrin(ogen), Integr. Biol., 2015, 7, 24–52 RSC.
- E. Pretorius and D. B. Kell, Diagnostic morphology: biophysical indicators for iron-driven inflammatory diseases, Integr. Biol., 2014, 6, 486–510 RSC.
- R. L. Jurado, Iron, infections, and anemia of inflammation, Clin. Infect. Dis., 1997, 25, 888–895 CAS.
- M. A. Fischbach, H. N. Lin, D. R. Liu and C. T. Walsh, How pathogenic bacteria evade mammalian sabotage in the battle for iron, Nat. Chem. Biol., 2006, 2, 132–138 CrossRef CAS PubMed.
- S. T. Ong, J. Z. S. Ho, B. Ho and J. L. Ding, Iron-withholding strategy in innate immunity, Immunobiology, 2006, 211, 295–314 CrossRef CAS PubMed.
- M. Miethke and M. A. Marahiel, Siderophore-based iron acquisition and pathogen control, Microbiol. Mol. Biol. Rev., 2007, 71, 413–451 CrossRef CAS PubMed.
- L. J. Wang and B. J. Cherayil, Ironing out the wrinkles in host defense: interactions between iron homeostasis and innate immunity, J. Innate Immun., 2009, 1, 455–464 CrossRef CAS PubMed.
- B. C. Chu, A. Garcia-Herrero, T. H. Johanson, K. D. Krewulak, C. K. Lau, R. S. Peacock, Z. Slavinskaya and H. J. Vogel, Siderophore uptake in bacteria and the battle for iron with the host; a bird's eye view, BioMetals, 2010, 23, 601–611 CrossRef CAS PubMed.
- K. E. Sivick and H. L. T. Mobley, Waging war against uropathogenic Escherichia coli: winning back the urinary tract, Infect. Immun., 2010, 78, 568–585 CrossRef CAS PubMed.
- M. Wessling-Resnick, Iron homeostasis and the inflammatory response, Annu. Rev. Nutr., 2010, 30, 105–122 CrossRef CAS PubMed.
- K. P. Haley and E. P. Skaar, A battle for iron: host sequestration and Staphylococcus aureus acquisition, Microbes Infect., 2012, 14, 217–227 CrossRef CAS PubMed.
- A. E. Armitage and H. Drakesmith, The battle for iron, Science, 2014, 346, 1299–1300 CrossRef CAS PubMed.
- S. Subashchandrabose and H. L. T. Mobley, Back to the metal age: battle for metals at the host–pathogen interface during urinary tract infection, Metallomics, 2015, 7, 935–942 RSC.
- L. Diacovich and J. P. Gorvel, Bacterial manipulation of innate immunity to promote infection, Nat. Rev. Microbiol., 2010, 8, 117–128 CrossRef CAS PubMed.
- A. Płóciennikowska, A. Hromada-Judycka, K. Borzcka and K. Kwiatkowska, Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling, Cell. Mol. Life Sci., 2015, 72, 557–581 CrossRef PubMed.
- S. Morath, A. Geyer and T. Hartung, Structure–function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus, J. Exp. Med., 2001, 193, 393–397 CrossRef CAS.
- B. Luef, K. R. Frischkorn, K. C. Wrighton, H. Y. Holman, G. Birarda, B. C. Thomas, A. Singh, K. H. Williams, C. E. Siegerist, S. G. Tringe, K. H. Downing, L. R. Comolli and J. F. Banfield, Diverse uncultivated ultra-small bacterial cells in groundwater, Nat. Commun., 2015, 6, 6372 CrossRef CAS PubMed.
- P. B. Eckburg, P. W. Lepp and D. A. Relman, Archaea and their potential role in human disease, Infect. Immun., 2003, 71, 591–596 CrossRef CAS.
- P. Blais Lecours, C. Duchaine, M. Taillefer, C. Tremblay, M. Veillette, Y. Cormier and D. Marsolais, Immunogenic properties of archaeal species found in bioaerosols, PLoS One, 2011, 6, e23326 Search PubMed.
- B. Dridi, Laboratory tools for detection of archaea in humans, Clin. Microbiol. Infect., 2012, 18, 825–833 CrossRef CAS PubMed.
- R. I. Aminov, Role of archaea in human disease, Front. Cell. Infect. Microbiol., 2013, 3, 42 CrossRef PubMed.
- R. F. Itzhaki, Herpes simplex virus type 1 and Alzheimer's disease: increasing evidence for a major role of the virus, Front. Aging Neurosci., 2014, 6, 202 CrossRef PubMed.
- C. H. Chiang, C. C. Huang, W. L. Chan, Y. C. Chen, T. J. Chen, S. J. Lin, J. W. Chen and H. B. Leu, Association between Mycoplasma pneumonia and increased risk of ischemic stroke: a nationwide study, Stroke, 2011, 42, 2940–2943 CrossRef PubMed.
- C. Gil, A. A. S. González, I. L. León, A. Rivera, R. S. Olea and L. Cedillo, Detection of Mycoplasmas in Patients with Amyotrophic Lateral Sclerosis, Adv. Microbiol., 2014, 4, 712–719 CrossRef.
- G. L. Nicolson, R. Gan, N. L. Nicolson and J. Haier, Evidence for mycoplasma ssp., Chlamydia pneumoniae, and human herpes virus-6 coinfections in the blood of patients with autistic spectrum disorders, J. Neurosci. Res., 2007, 85, 1143–1148 CrossRef CAS PubMed.
- T. Buzan, How to mind map, Thorsons, London, 2002 Search PubMed.
- A. D. Proal, P. J. Albert and T. G. Marshall, The human microbiome and autoimmunity, Curr. Opin. Rheumatol., 2013, 25, 234–240 CrossRef CAS PubMed.
- A. D. Proal, P. J. Albert and T. G. Marshall, Inflammatory disease and the human microbiome, Discov. Med., 2014, 17, 257–265 Search PubMed.
- D. B. Kell, A. S. Kaprelyants and A. Grafen, On pheromones, social behaviour and the functions of secondary metabolism in bacteria, Trends Ecol. Evolution, 1995, 10, 126–129 CrossRef CAS.
- G. V. Mukamolova, A. S. Kaprelyants, D. B. Kell and M. Young, Adoption of the transiently non-culturable state – a bacterial survival strategy?, Adv. Micr. Physiol., 2003, 47, 65–129 CAS.
- S. V. Avery, Microbial cell individuality and the underlying sources of heterogeneity, Nat. Rev. Microbiol., 2006, 4, 577–587 CrossRef CAS PubMed.
- N. Q. Balaban, Persistence: mechanisms for triggering and enhancing phenotypic variability, Curr. Opin. Genet. Dev., 2011, 21, 768–775 CrossRef CAS PubMed.
- S. S. Epstein, The phenomenon of microbial uncultivability, Curr. Opin. Microbiol., 2013, 16, 636–642 CrossRef CAS PubMed.
- P. W. Ewald, Evolution of infectious disease, Oxford University Press, New York, 1994 Search PubMed.
- P. W. Ewald, Plague time: the new germ theory of disease, Anchor Books, New York, 2002 Search PubMed.
- P. W. Ewald, Evolution of virulence, Infectious disease clinics of North America, 2004, 18, 1–15 CrossRef.
- M. T. Green, P. M. Heidger Jr. and G. Domingue, Proposed reproductive cycle for a relatively stable L-phase variant of Streptococcus faecalis, Infect. Immun., 1974, 10, 915–927 CAS.
- M. T. Green, P. M. Heidger Jr. and G. Domingue, Demonstration of the phenomena of microbial persistence and reversion with bacterial L-forms in human embryonic kidney cells, Infect. Immun., 1974, 10, 889–914 CAS.
- G. J. Domingue and J. U. Schlegel, Novel bacterial structures in human blood: cultural isolation, Infect. Immun., 1977, 15, 621–627 CAS.
- S. Nikkari, I. J. McLaughlin, W. Bi, D. E. Dodge and D. A. Relman, Does blood of healthy subjects contain bacterial ribosomal DNA?, J. Clin. Microbiol., 2001, 39, 1956–1959 CrossRef CAS PubMed.
- K. Moriyama, C. Ando, K. Tashiro, S. Kuhara, S. Okamura, S. Nakano, Y. Takagi, T. Miki, Y. Nakashima and H. Hirakawa, Polymerase chain reaction detection of bacterial 16S rRNA gene in human blood, Microbiol. Immunol., 2008, 52, 375–382 CrossRef CAS PubMed.
- A. Cherkaoui, S. Emonet, D. Ceroni, B. Candolfi, J. Hibbs, P. Francois and J. Schrenzel, Development and validation of a modified broad-range 16S rDNA PCR for diagnostic purposes in clinical microbiology, J. Microbiol. Methods, 2009, 79, 227–231 CrossRef CAS PubMed.
- S. G. Sakka, A. J. Kochem, C. Disqué and N. Wellinghausen, Blood infection diagnosis by 16S rDNA broad-spectrum polymerase chain reaction: the relationship between antibiotic treatment and bacterial DNA load, Anesth. Analg., 2009, 109, 1707–1708 Search PubMed.
- J. Amar, M. Serino, C. Lange, C. Chabo, J. Iacovoni, S. Mondot, P. Lepage, C. Klopp, J. Mariette, O. Bouchez, L. Perez, M. Courtney, M. Marre, P. Klopp, O. Lantieri, J. Doré, M. A. Charles, B. Balkau, R. Burcelin and D. S. Grp, Involvement of tissue bacteria in the onset of diabetes in humans: evidence for a concept, Diabetologia, 2011, 54, 3055–3061 CrossRef CAS PubMed.
- K. Grif, I. Heller, W. M. Prodinger, K. Lechleitner, C. Lass-Florl and D. Orth, Improvement of detection of bacterial pathogens in normally sterile body sites with a focus on orthopedic samples by use of a commercial 16S rRNA broad-range PCR and sequence analysis, J. Clin. Microbiol., 2012, 50, 2250–2254 CrossRef CAS PubMed.
- K. Grif, M. Fille, R. Wurzner, G. Weiss, I. Lorenz, G. Gruber, S. Eschertzhuber, D. Nachbaur, C. Lass-Flörl and D. Orth, Rapid detection of bloodstream pathogens by real-time PCR in patients with sepsis, Wien. Klin. Wochenschr., 2012, 124, 266–270 CrossRef CAS PubMed.
- A. Fernández-Cruz, M. Marin, M. Kestler, L. Alcala, M. Rodriguez-Créixems and E. Bouza, The value of combining blood culture and SeptiFast data for predicting complicated bloodstream infections caused by Gram-positive bacteria or Candida species, J. Clin. Microbiol., 2013, 51, 1130–1136 CrossRef PubMed.
- P. Gaibani, M. Mariconti, G. Bua, S. Bonora, D. Sassera, M. P. Landini, P. Mulatto, S. Novati, C. Bandi and V. Sambri, Development of a broad-range 23S rDNA real-time PCR assay for the detection and quantification of pathogenic bacteria in human whole blood and plasma specimens, BioMed Res. Int., 2013, 2013, 264651 Search PubMed.
- J. Sato, A. Kanazawa, F. Ikeda, T. Yoshihara, H. Goto, H. Abe, K. Komiya, M. Kawaguchi, T. Shimizu, T. Ogihara, Y. Tamura, Y. Sakurai, R. Yamamoto, T. Mita, Y. Fujitani, H. Fukuda, K. Nomoto, T. Takahashi, T. Asahara, T. Hirose, S. Nagata, Y. Yamashiro and H. Watada, Gut dysbiosis and detection of “live gut bacteria” in blood of Japanese patients with type 2 diabetes, Diabetes Care, 2014, 37, 2343–2350 CrossRef CAS PubMed.
- C. L. Liu, H. W. Ai, W. P. Wang, L. Chen, H. B. Hu, T. Ye, X. H. Zhu, F. Wang, Y. L. Liao, Y. Wang, G. Ou, L. Xu, M. Sun, C. Jian, Z. J. Chen, L. Li, B. Zhang, L. Tian, B. Wang, S. Yan and Z. Y. Sun, Comparison of 16S rRNA gene PCR and blood culture for diagnosis of neonatal sepsis, Arch. Pediatr., 2014, 21, 162–169 CrossRef CAS PubMed.
- F. Valencia-Shelton and M. Loeffelholz, Nonculture techniques for the detection of bacteremia and fungemia, Future Microbiol., 2014, 9, 543–559 CrossRef CAS PubMed.
- G. Domingue, B. Turner and J. U. Schlegel, Cell-wall deficient bacterial variants in kidney tissue. Detection by immunofluorescence, Urology, 1974, 3, 288–292 CrossRef CAS.
- G. J. Domingue, Electron dense cytoplasmic particles and chronic infection – a bacterial pleomorphy hypothesis, Endocytobiosis Cell Res., 1995, 11, 19–40 Search PubMed.
- G. J. Domingue and H. B. Woody, Bacterial persistence and expression of disease, Clin. Microbiol. Rev., 1997, 10, 320–344 Search PubMed.
- G. J. Domingue, Demystifying pleomorphic forms in persistence and expression of disease: are they bacteria and is peptidoglycan the solution?, Discov. Med., 2010, 10, 234–246 Search PubMed.
- L. Mattman, Cell wall deficient forms: stealth pathogens, CRC Press, Boca Raton, 3rd edn, 2001 Search PubMed.
- M. Miskinyte, A. Sousa, R. S. Ramiro, J. A. de Sousa, J. Kotlinowski, I. Caramalho, S. Magalhães, M. P. Soares and I. Gordo, The genetic basis of Escherichia coli pathoadaptation to macrophages, PLoS Pathog., 2013, 9, e1003802 Search PubMed.
- M. Miskinyte and I. Gordo, Increased survival of antibiotic-resistant Escherichia coli inside macrophages, Antimicrob. Agents Chemother., 2013, 57, 189–195 CrossRef CAS PubMed.
- P. Liehl, V. Zuzarte-Luis and M. M. Mota, Unveiling the pathogen behind the vacuole, Nat. Rev. Microbiol., 2015, 13, 589–598 CrossRef CAS PubMed.
- D. Ribet and P. Cossart, How bacterial pathogens colonize their hosts and invade deeper tissues, Microbes Infect., 2015, 17, 173–183 CrossRef CAS PubMed.
- G. E. Thwaites and V. Gant, Are bloodstream leukocytes Trojan Horses for the metastasis of Staphylococcus aureus?, Nat. Rev. Microbiol., 2011, 9, 215–222 CrossRef CAS PubMed.
- F. C. O. Los, T. M. Randis, R. V. Aroian and A. J. Ratner, Role of pore-forming toxins in bacterial infectious diseases, Microbiol. Mol. Biol. Rev., 2013, 77, 173–207 CrossRef CAS PubMed.
- T. J. LaRocca, E. A. Stivison, E. A. Hod, S. L. Spitalnik, P. J. Cowan, T. M. Randis and A. J. Ratner, Human-specific bacterial pore-forming toxins induce programmed necrosis in erythrocytes, mBio, 2014, 5, e01251–01214 CrossRef CAS PubMed.
- A. K. May, T. G. Gleason, R. G. Sawyer and T. L. Pruett, Contribution of Escherichia coli alpha-hemolysin to bacterial virulence and to intraperitoneal alterations in peritonitis, Infect. Immun., 2000, 68, 176–183 CrossRef CAS.
- L. Ferrier, L. Mazelin, N. Cenac, P. Desreumaux, A. Janin, D. Emilie, J. F. Colombel, R. Garcia-Villar, J. Fioramonti and L. Bueno, Stress-induced disruption of colonic epithelial barrier: role of interferon-gamma and myosin light chain kinase in mice, Gastroenterology, 2003, 125, 795–804 CrossRef CAS.
- K. Honda and D. R. Littman, The microbiome in infectious disease and inflammation, Annu. Rev. Immunol., 2012, 30, 759–795 CrossRef CAS PubMed.
- N. R. Klatt, N. T. Funderburg and J. M. Brenchley, Microbial translocation, immune activation, and HIV disease, Trends Microbiol., 2013, 21, 6–13 CrossRef CAS PubMed.
- P. J. Turnbaugh, R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight and J. I. Gordon, The human microbiome project, Nature, 2007, 449, 804–810 CrossRef CAS PubMed.
- W. S. Garrett, Cancer and the microbiota, Science, 2015, 348, 80–86 CrossRef CAS PubMed.
- J. R. Caso, O. Hurtado, M. P. Pereira, B. Garcia-Bueno, L. Menchen, L. Alou, M. L. Gomez-Lus, M. A. Moro, I. Lizasoain and J. C. Leza, Colonic bacterial translocation as a possible factor in stress-worsening experimental stroke outcome, Am. J. Physiol.: Regul., Integr. Comp. Physiol., 2009, 296, R979–R985 CrossRef CAS PubMed.
- R. R. Schumann and J. Zweigner, A novel acute-phase marker: lipopolysaccharide binding protein (LBP), Clin. Chem. Lab. Med., 1999, 37, 271–274 CrossRef CAS PubMed.
- R. L. Kitchens and P. A. Thompson, Impact of sepsis-induced changes in plasma on LPS interactions with monocytes and plasma lipoproteins: roles of soluble CD14, LBP, and acute phase lipoproteins, J. Endotoxin Res., 2003, 9, 113–118 CrossRef CAS PubMed.
- R. R. Schumann, Old and new findings on lipopolysaccharide-binding protein: a soluble pattern-recognition molecule, Biochem. Soc. Trans., 2011, 39, 989–993 CrossRef CAS PubMed.
- P. H. Ding and L. J. Jin, The role of lipopolysaccharide-binding protein in innate immunity: a revisit and its relevance to oral/periodontal health, J. Periodontal Res., 2014, 49, 1–9 CrossRef CAS PubMed.
- A. Gonzalez-Quintela, M. Alonso, J. Campos, L. Vizcaino, L. Loidi and F. Gude, Determinants of serum concentrations of Lipopolysaccharide-Binding Protein (LBP) in the adult population: the role of obesity, PLoS One, 2013, 8, e54600 CAS.
- J. Zweigner, R. R. Schumann and J. R. Weber, The role of lipopolysaccharide-binding protein in modulating the innate immune response, Microbes Infect., 2006, 8, 946–952 CrossRef CAS PubMed.
- M. Van Oosten, P. C. Rensen, E. S. Van Amersfoort, M. Van Eck, A. M. Van Dam, J. J. Breve, T. Vogel, A. Panet, T. J. Van Berkel and J. Kuiper, Apolipoprotein E protects against bacterial lipopolysaccharide-induced lethality. A new therapeutic approach to treat Gram-negative sepsis, J. Biol. Chem., 2001, 276, 8820–8824 CrossRef CAS PubMed.
- C. Cuaz-Pérolin, L. Billiet, E. Baugé, C. Copin, D. Scott-Algara, F. Genze, B. Buchele, T. Syrovets, T. Simmet and M. Rouis, Antiinflammatory and antiatherogenic effects of the NF-kappaB inhibitor acetyl-11-keto-beta-boswellic acid in LPS-challenged ApoE-/- mice, Arterioscler., Thromb., Vasc. Biol., 2008, 28, 272–277 CrossRef PubMed.
- N. Urosevic and R. N. Martins, Infection and Alzheimer's disease: the APOE epsilon4 connection and lipid metabolism, J. Alzheimer's Dis., 2008, 13, 421–435 CAS.
- R. F. Itzhaki and M. A. Wozniak, Herpes simplex virus type 1, apolipoprotein E, and cholesterol: a dangerous liaison in Alzheimer's disease and other disorders, Prog. Lipid Res., 2006, 45, 73–90 CrossRef CAS PubMed.
- V. Leoni, The effect of apolipoprotein E (ApoE) genotype on biomarkers of amyloidogenesis, tau pathology and neurodegeneration in Alzheimer's disease, Clin. Chem. Lab. Med., 2011, 49, 375–383 CrossRef CAS PubMed.
- C. C. Liu, T. Kanekiyo, H. Xu and G. Bu, Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy, Nat. Rev. Neurol., 2013, 9, 106–118 CrossRef CAS PubMed.
- E. Dorey, N. Chang, Q. Y. Liu, Z. Yang and W. Zhang, Apolipoprotein E, amyloid-beta, and neuroinflammation in Alzheimer's disease, Neurosci. Bull., 2014, 30, 317–330 CAS.
- L. M. Tai, S. Mehra, V. Shete, S. Estus, G. W. Rebeck, G. Bu and M. J. LaDu, Soluble apoE/Abeta complex: mechanism and therapeutic target for APOE4-induced AD risk, Mol. Neurodegener., 2014, 9, 2 CrossRef PubMed.
- G. S. Zaman and F. Zaman, Relationship between postprandial endotoxemia in nonobese postmenopausal women and diabetic nonobese postmenopausal women, J. Nat. Sci., Biol. Med., 2015, 6, 89–93 CrossRef CAS PubMed.
- J. L. Ding and B. Ho, Endotoxin detection–from limulus amebocyte lysate to recombinant factor C, Subcell. Biochem., 2010, 53, 187–208 CAS.
- M. Lan, J. Wu, W. Liu, W. Zhang, J. Ge, H. Zhang, J. Sun, W. Zhao and P. Wang, Copolythiophene-derived colorimetric and fluorometric sensor for visually supersensitive determination of lipopolysaccharide, J. Am. Chem. Soc., 2012, 134, 6685–6694 CrossRef CAS PubMed.
- W. Su, S. E. Kim, M. Cho, J. D. Nam, W. S. Choe and Y. Lee, Selective detection of endotoxin using an impedance aptasensor with electrochemically deposited gold nanoparticles, Innate Immun., 2013, 19, 388–397 CrossRef PubMed.
- A. P. Das, P. S. Kumar and S. Swain, Recent advances in biosensor based endotoxin detection, Biosens. Bioelectron., 2014, 51, 62–75 CrossRef CAS PubMed.
- R. L. Kitchens, G. Wolfbauer, J. J. Albers and R. S. Munford, Plasma lipoproteins promote the release of bacterial lipopolysaccharide from the monocyte cell surface, J. Biol. Chem., 1999, 274, 34116–34122 CrossRef CAS PubMed.
- W. Khovidhunkit, M. S. Kim, R. A. Memon, J. K. Shigenaga, A. H. Moser, K. R. Feingold and C. Grunfeld, Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host, J. Lipid Res., 2004, 45, 1169–1196 CrossRef CAS PubMed.
- J. H. M. Levels, P. R. Abraham, E. P. van Barreveld, J. C. M. Meijers and S. J. G. van Deventer, Distribution and kinetics of lipoprotein-bound lipoteichoic acid, Infect. Immun., 2003, 71, 3280–3284 CrossRef CAS.
- P. Fraunberger, S. Schaefer, K. Werdan, A. K. Walli and D. Seidel, Reduction of circulating cholesterol and apolipoprotein levels during sepsis, Clin. Chem. Lab. Med., 1999, 37, 357–362 CrossRef CAS PubMed.
- J. H. M. Levels, P. Geurts, H. Karlsson, R. Marée, S. Ljunggren, L. Fornander, L. Wehenkel, M. Lindahl, E. S. G. Stroes, J. A. Kuivenhoven and J. C. M. Meijers, High-density lipoprotein proteome dynamics in human endotoxemia, Proteome Sci., 2011, 9, 34 CrossRef CAS PubMed.
- R. F. Wilson, J. F. Barletta and J. G. Tyburski, Hypocholesterolemia in sepsis and critically ill or injured patients, Crit. Care, 2003, 7, 413–414 CrossRef PubMed.
- J. Y. Chien, J. S. Jerng, C. J. Yu and P. C. Yang, Low serum level of high-density lipoprotein cholesterol is a poor prognostic factor for severe sepsis, Crit. Care Med., 2005, 33, 1688–1693 CrossRef CAS.
- A. L. Catapano, A. Pirillo, F. Bonacina and G. D. Norata, HDL in innate and adaptive immunity, Cardiovasc. Res., 2014, 103, 372–383 CrossRef CAS PubMed.
- L. Guo, J. Ai, Z. Zheng, D. A. Howatt, A. Daugherty, B. Huang and X. A. Li, High density lipoprotein protects against polymicrobe-induced sepsis in mice, J. Biol. Chem., 2013, 288, 17947–17953 CrossRef CAS PubMed.
- A. Pirillo, A. L. Catapano and G. D. Norata, HDL in infectious diseases and sepsis, Handb. Exp. Pharmacol., 2015, 224, 483–508 Search PubMed.
- G. D. Hitchens and D. B. Kell, Uncouplers can shuttle rapidly between localised energy coupling sites during photophosphorylation by chromatophores of Rhodopseudomonas capsulata N22, Biochem. J., 1983, 212, 25–30 CrossRef CAS.
- G. D. Hitchens and D. B. Kell, On the functional unit of energy coupling in photophosphorylation by bacterial chromatophores, Biochim. Biophys. Acta, 1983, 723, 308–316 CrossRef CAS.
- D. B. Kell, H. M. Ryder, A. S. Kaprelyants and H. V. Westerhoff, Quantifying heterogeneity: flow cytometry of bacterial cultures, Antonie van Leeuwenhoek, 1991, 60, 145–158 CrossRef CAS.
- H. M. Davey and D. B. Kell, Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analysis, Microbiol. Rev., 1996, 60, 641–696 CAS.
- S. Zielen, J. Trischler and R. Schubert, Lipopolysaccharide challenge: immunological effects and safety in humans, Expert Rev. Clin. Immunol., 2015, 11, 409–418 CrossRef CAS PubMed.
- C. M. Harris and D. B. Kell, The estimation of microbial biomass, Biosensors, 1985, 1, 17–84 CrossRef CAS.
- S. W. Watson, T. J. Novitsky, H. L. Quinby and F. W. Valois, Determination of bacterial number and biomass in the marine environment, Appl. Environ. Microbiol., 1977, 33, 940–946 CAS.
- F. C. Pearson, M. E. Weary, H. E. Sargent, T. J. Novitsky, H. Lin, G. Lindsay, R. N. Berzofsky, A. L. Lane, J. D. Wilson, J. F. Cooper, E. J. Helme, C. W. Twohy, H. I. Basch, M. A. Rech, J. W. Slade and M. P. Winegar, Comparison of several control standard endotoxins to the National Reference Standard Endotoxin–an HIMA collaborative study, Appl. Environ. Microbiol., 1985, 50, 91–93 CAS.
- J. Andrä, P. Garidel, A. Majerle, R. Jerala, R. Ridge, E. Paus, T. Novitsky, M. H. Koch and K. Brandenburg, Biophysical characterization of the interaction of Limulus polyphemus endotoxin neutralizing protein with lipopolysaccharide, Eur. J. Biochem., 2004, 271, 2037–2046 CrossRef PubMed.
- P. A. Ketchum and T. J. Novitsky, Assay of endotoxin by Limulus amebocyte lysate, Methods Mol. Med., 2000, 36, 3–12 CAS.
- I. Mattsby-Baltzer, K. Lindgren, B. Lindholm and L. Edebo, Endotoxin shedding by enterobacteria: free and cell-bound endotoxin differ in Limulus activity, Infect. Immun., 1991, 59, 689–695 CAS.
- T. J. Novitsky, Limitations of the Limulus amebocyte lysate test in demonstrating circulating lipopolysaccharides, Ann. N. Y. Acad. Sci., 1998, 851, 416–421 CrossRef CAS PubMed.
- T. J. Novitsky, Biomedical Applications of Limulus Amebocyte Lysate, Biology and Conservation of Horseshoe Crabs, 2009, pp. 315–329 Search PubMed.
- A. Albillos, A. de la Hera, M. González, J. L. Moya, J. L. Calleja, J. Monserrat, L. Ruiz-del-Arbol and M. Alvarez-Mon, Increased lipopolysaccharide binding protein in cirrhotic patients with marked immune and hemodynamic derangement, Hepatology, 2003, 37, 208–217 CrossRef CAS PubMed.
- C. Elsing, S. Ernst, N. Kayali, W. Stremmel and S. Harenberg, Lipopolysaccharide binding protein, interleukin-6 and C-reactive protein in acute gastrointestinal infections: value as biomarkers to reduce unnecessary antibiotic therapy, Infection, 2011, 39, 327–331 CrossRef CAS PubMed.
- N. T. Funderburg, S. R. Stubblefield Park, H. C. Sung, G. Hardy, B. Clagett, J. Ignatz-Hoover, C. V. Harding, P. Fu, J. A. Katz, M. M. Lederman and A. D. Levine, Circulating CD4+ and CD8+ T cells are activated in inflammatory bowel disease and are associated with plasma markers of inflammation, Immunology, 2013, 140, 87–97 CrossRef CAS PubMed.
- H. Zhou, J. Hu, Q. Zhu, S. Yang, Y. Zhang, R. Gao, L. Liu, Y. Wang, Q. Zhen, Q. Lv and Q. Li, Lipopolysaccharide-binding protein cannot independently predict type 2 diabetes mellitus: a nested case-control study, J. Diabetes, 2015 DOI:10.1111/1753-0407.12281.
- T. Vollmer, C. Piper, K. Kleesiek and J. Dreier, Lipopolysaccharide-binding protein: a new biomarker for infectious endocarditis?, Clin. Chem., 2009, 55, 295–304 CAS.
- L. Sun, Z. Yu, X. Ye, S. Zou, H. Li, D. Yu, H. Wu, Y. Chen, J. Dore, K. Clement, F. B. Hu and X. Lin, A marker of endotoxemia is associated with obesity and related metabolic disorders in apparently healthy Chinese, Diabetes Care, 2010, 33, 1925–1932 CrossRef CAS PubMed.
- J. Zweigner, H. J. Gramm, O. C. Singer, K. Wegscheider and R. R. Schumann, High concentrations of lipopolysaccharide-binding protein in serum of patients with severe sepsis or septic shock inhibit the lipopolysaccharide response in human monocytes, Blood, 2001, 98, 3800–3808 CrossRef CAS PubMed.
- T. D. Sandahl, H. Gronbaek, H. J. Moller, S. Stoy, K. L. Thomsen, A. K. Dige, J. Agnholt, S. Hamilton-Dutoit, S. Thiel and H. Vilstrup, Hepatic macrophage activation and the LPS pathway in patients with alcoholic hepatitis: a prospective cohort study, Am. J. Gastroenterol., 2014, 109, 1749–1756 CrossRef CAS PubMed.
- E. A. Tsalkidou, E. Roilides, S. Gardikis, G. Trypsianis, A. Kortsaris, A. Chatzimichael and I. Tentes, Lipopolysaccharide-binding protein: a potential marker of febrile urinary tract infection in childhood, Pediatr. Nephrol., 2013, 28, 1091–1097 CrossRef PubMed.
- R. Berner, B. Fürll, F. Stelter, J. Dröse, H. P. Müller and C. Schütt, Elevated levels of lipopolysaccharide-binding protein and soluble CD14 in plasma in neonatal early-onset sepsis, Clin. Diagn. Lab. Immunol., 2002, 9, 440–445 CAS.
- J. L. Leante-Castellanos, L. G. de Guadiana-Romualdo, C. Fuentes-Gutiérrez, A. Hernando-Holgado, A. Garcia-González and E. Jiménez-Santos, The value of lipopolysaccharide binding protein for diagnosis of late-onset neonatal sepsis in very low birth weight infants, J. Perinat. Med., 2015, 43, 253–257 CAS.
- A. H. M. Froon, M. A. Dentener, J. W. M. Greve, G. Ramsay and W. A. Buurman, Lipopolysaccharide toxicity-regulating proteins in bacteremia, J. Infect. Dis., 1995, 171, 1250–1257 CrossRef CAS PubMed.
- S. M. Opal, P. J. Scannon, J. L. Vincent, M. White, S. F. Carroll, J. E. Palardy, N. A. Parejo, J. P. Pribble and J. H. Lemke, Relationship between plasma levels of lipopolysaccharide (LPS) and LPS-binding protein in patients with severe sepsis and septic shock, J. Infect. Dis., 1999, 180, 1584–1589 CrossRef CAS PubMed.
- N. Lamping, R. Dettmer, N. W. Schröder, D. Pfeil, W. Hallatschek, R. Burger and R. R. Schumann, LPS-binding protein protects mice from septic shock caused by LPS or Gram-negative bacteria, J. Clin. Invest., 1998, 101, 2065–2071 CrossRef CAS PubMed.
- J. Villar, L. Pérez-Méndez, E. Espinosa, C. Flores, J. Blanco, A. Muriel, S. Basaldúa, M. Muros, L. Blanch, A. Artigas and R. M. Kacmarek, Grecia and Gen-Sep Groups, Serum lipopolysaccharide binding protein levels predict severity of lung injury and mortality in patients with severe sepsis, PLoS One, 2009, 4, e6818 Search PubMed.
- M. Mierzchala, M. Krzystek-Korpacka, A. Gamian and G. Durek, Quantitative indices of dynamics in concentrations of lipopolysaccharide-binding protein (LBP) as prognostic factors in severe sepsis/septic shock patients – Comparison with CRP and procalcitonin, Clin. Biochem., 2011, 44, 357–363 CrossRef CAS PubMed.
- H. Ghanim, S. Abuaysheh, C. L. Sia, K. Korzeniewski, A. Chaudhuri, J. M. Fernandez-Real and P. Dandona, Increase in plasma endotoxin concentrations and the expression of Toll-like receptors and suppressor of cytokine signaling-3 in mononuclear cells after a high-fat, high-carbohydrate meal: implications for insulin resistance, Diabetes Care, 2009, 32, 2281–2287 CrossRef CAS PubMed.
- P. W. Hunt, J. Brenchley, E. Sinclair, J. M. McCune, M. Roland, K. Page-Shafer, P. Hsue, B. Emu, M. Krone, H. Lampiris, D. Douek, J. N. Martin and S. G. Deeks, Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy, J. Infect. Dis., 2008, 197, 126–133 CrossRef PubMed.
- A. L. Harte, N. F. da Silva, S. J. Creely, K. C. McGee, T. Billyard, E. M. Youssef-Elabd, G. Tripathi, E. Ashour, M. S. Abdalla, H. M. Sharada, A. I. Amin, A. D. Burt, S. Kumar, C. P. Day and P. G. McTernan, Elevated endotoxin levels in non-alcoholic fatty liver disease, J. Inflammation, 2010, 7, 15 CrossRef PubMed.
- J. C. Marshall, P. M. Walker, D. M. Foster, D. Harris, M. Ribeiro, J. Paice, A. D. Romaschin and A. N. Derzko, Measurement of endotoxin activity in critically ill patients using whole blood neutrophil dependent chemiluminescence, Crit. Care, 2002, 6, 342–348 CrossRef.
- M. Nymark, P. J. Pussinen, A. M. Tuomainen, C. Forsblom, P. H. Groop, M. Lehto and FinnDiane Study Group, Serum lipopolysaccharide activity is associated with the progression of kidney disease in finnish patients with type 1 diabetes, Diabetes Care, 2009, 32, 1689–1693 CrossRef CAS PubMed.
- O. S. Al-Attas, N. M. Al-Daghri, K. Al-Rubeaan, N. F. da Silva, S. L. Sabico, S. Kumar, P. G. McTernan and A. L. Harte, Changes in endotoxin levels in T2DM subjects on anti-diabetic therapies, Cardiovasc. Diabetol., 2009, 8, 20 CrossRef PubMed.
- C. J. Wiedermann, S. Kiechl, S. Dunzendorfer, P. Schratzberger, G. Egger, F. Oberhollenzer and J. Willeit, Association of endotoxemia with carotid atherosclerosis and cardiovascular disease: prospective results from the Bruneck study, J. Am. Coll. Cardiol., 1999, 34, 1975–1981 CrossRef CAS.
- A. A. Nash, R. G. Dalziel and J. R. Fitzgerald, Mims' pathogenesis of infectious disease, Academic Press, New York, 6th edn, 2015 Search PubMed.
- L. Margulis and M. J. Chapman, Endosymbioses: cyclical and permanent in evolution, Trends Microbiol., 1998, 6, 342–345 CrossRef CAS ; discussion 345–346.
- E. C. Cocking, P. J. Stone and M. R. Davey, Symbiosome-like intracellular colonization of cereals and other crop plants by nitrogen-fixing bacteria for reduced inputs of synthetic nitrogen fertilizers, Sci. China, Ser. C: Life Sci., 2005, 48(suppl 2), 888–896 CAS.
- J. L. Knoth, S. H. Kim, G. J. Ettl and S. L. Doty, Biological nitrogen fixation and biomass accumulation within poplar clones as a result of inoculations with diazotrophic endophyte consortia, New Phytol., 2014, 201, 599–609 CrossRef CAS PubMed.
- B. Foxman, The epidemiology of urinary tract infection, Nat. Rev. Urol., 2010, 7, 653–660 CrossRef PubMed.
- S. Heytens, A. De Sutter, D. De Backer, G. Verschraegen and T. Christiaens, Cystitis: symptomatology in women with suspected uncomplicated urinary tract infection, J. Womens Health (Larchmt), 2011, 20, 1117–1121 CrossRef PubMed.
- J. Marschall, L. Zhang, B. Foxman, D. K. Warren, J. P. Henderson and CDC Prevention Epicenters Program, Both host and pathogen factors predispose to Escherichia coli urinary-source bacteremia in hospitalized patients, Clin. Infect. Dis., 2012, 54, 1692–1698 CrossRef PubMed.
- G. J. Domingue, G. M. Ghoniem, K. L. Bost, C. Fermin and L. G. Human, Dormant microbes in interstitial cystitis, J. Urol., 1995, 153, 1321–1326 CrossRef CAS.
- B. Foxman, Recurring urinary tract infection: incidence and risk factors, Am. J. Public Health, 1990, 80, 331–333 CrossRef CAS PubMed.
- B. Foxman, Epidemiology of urinary tract infections: incidence, morbidity, and economic costs, Am. J. Med., 2002, 113(suppl 1A), 5S–13S CrossRef.
- C. F. Marrs, L. Zhang and B. Foxman, Escherichia coli mediated urinary tract infections: are there distinct uropathogenic E. coli (UPEC) pathotypes?, FEMS Microbiol. Lett., 2005, 252, 183–190 CrossRef CAS PubMed.
- T. J. Hannan, I. U. Mysorekar, C. S. Hung, M. L. Isaacson-Schmid and S. J. Hultgren, Early severe inflammatory responses to uropathogenic E. coli predispose to chronic and recurrent urinary tract infection, PLoS Pathog., 2010, 6, e1001042 Search PubMed.
- D. A. Hunstad and S. S. Justice, Intracellular lifestyles and immune evasion strategies of uropathogenic Escherichia coli, Annu. Rev. Microbiol., 2010, 64, 203–221 CrossRef CAS PubMed.
- K. Ejrnæs, Bacterial characteristics of importance for recurrent urinary tract infections caused by Escherichia coli, Dan. Med. Bull., 2011, 58, B4187 Search PubMed.
- T. J. Hannan, M. Totsika, K. J. Mansfield, K. H. Moore, M. A. Schembri and S. J. Hultgren, Host–pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection, FEMS Microbiol. Rev., 2012, 36, 616–648 CrossRef CAS PubMed.
- B. Foxman, Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden, Infectious disease clinics of North America, 2014, 28, 1–13 CrossRef PubMed.
- A. Brauner, S. H. Jacobson and I. Kuhn, Urinary Escherichia coli causing recurrent infections – a prospective follow-up of biochemical phenotypes, Clin. Nephrol., 1992, 38, 318–323 CAS.
- T. A. Russo, A. Stapleton, S. Wenderoth, T. M. Hooton and W. E. Stamm, Chromosomal restriction fragment length polymorphism analysis of Escherichia coli strains causing recurrent urinary tract infections in young women, J. Infect. Dis., 1995, 172, 440–445 CrossRef CAS PubMed.
- R. Ikäheimo, A. Siitonen, T. Heiskanen, U. Kärkkäinen, P. Kuosmanen, P. Lipponen and P. H. Mäkelä, Recurrence of urinary tract infection in a primary care setting: analysis of a 1-year follow-up of 179 women, Clin. Infect. Dis., 1996, 22, 91–99 CrossRef.
- D. A. Rosen, J. S. Pinkner, J. M. Jones, J. N. Walker, S. Clegg and S. J. Hultgren, Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression, Infect. Immun., 2008, 76, 3337–3345 CrossRef CAS PubMed.
- Y. Luo, Y. Ma, Q. Zhao, L. Wang, L. Guo, L. Ye, Y. Zhang and J. Yang, Similarity and divergence of phylogenies, antimicrobial susceptibilities, and virulence factor profiles of Escherichia coli isolates causing recurrent urinary tract infections that persist or result from reinfection, J. Clin. Microbiol., 2012, 50, 4002–4007 CrossRef CAS PubMed.
- J. M. Bower, D. S. Eto and M. A. Mulvey, Covert operations of uropathogenic Escherichia coli within the urinary tract, Traffic, 2005, 6, 18–31 CrossRef CAS PubMed.
- S. S. Justice, C. Hung, J. A. Theriot, D. A. Fletcher, G. G. Anderson, M. J. Footer and S. J. Hultgren, Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 1333–1338 CrossRef CAS PubMed.
- I. U. Mysorekar and S. J. Hultgren, Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 14170–14175 CrossRef CAS PubMed.
- D. A. Rosen, T. M. Hooton, W. E. Stamm, P. A. Humphrey and S. J. Hultgren, Detection of intracellular bacterial communities in human urinary tract infection, PLoS Med., 2007, 4, e329 CrossRef PubMed.
- B. K. Dhakal, R. R. Kulesus and M. A. Mulvey, Mechanisms and consequences of bladder cell invasion by uropathogenic Escherichia coli, Eur. J. Clin. Invest., 2008, 38(suppl 2), 2–11 CrossRef CAS PubMed.
- D. J. Schwartz, S. L. Chen, S. J. Hultgren and P. C. Seed, Population dynamics and niche distribution of uropathogenic Escherichia coli during acute and chronic urinary tract infection, Infect. Immun., 2011, 79, 4250–4259 CrossRef CAS PubMed.
- X. R. Wu, T. T. Sun and J. J. Medina, In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 9630–9635 CrossRef CAS.
- D. B. Kell, P. D. Dobson and S. G. Oliver, Pharmaceutical drug transport: the issues and the implications that it is essentially carrier-mediated only, Drug Discovery Today, 2011, 16, 704–714 CrossRef CAS PubMed.
- D. B. Kell, P. D. Dobson, E. Bilsland and S. G. Oliver, The promiscuous binding of pharmaceutical drugs and their transporter-mediated uptake into cells: what we (need to) know and how we can do so, Drug Discovery Today, 2013, 18, 218–239 CrossRef CAS PubMed.
- D. B. Kell and S. G. Oliver, How drugs get into cells: tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion, Front. Pharmacol., 2014, 5, 231 Search PubMed.
- D. B. Kell, What would be the observable consequences if phospholipid bilayer diffusion of drugs into cells is negligible?, Trends Pharmacol. Sci., 2015, 36, 15–21 CrossRef CAS PubMed.
- E. J. Arking, D. M. Appelt, J. T. Abrams, S. Kolbe, A. P. Hudson and B. J. Balin, Ultrastructural Analysis of Chlamydia pneumoniae in the Alzheimer's Brain, Pathogenesis, 1999, 1, 201–211 Search PubMed.
- B. J. Balin, H. C. Gerard, E. J. Arking, D. M. Appelt, P. J. Branigan, J. T. Abrams, J. A. Whittum-Hudson and A. P. Hudson, Identification and localization of Chlamydia pneumoniae in the Alzheimer's brain, Med. Microbiol. Immunol., 1998, 187, 23–42 CrossRef CAS.
- B. J. Balin and D. M. Appelt, Role of infection in Alzheimer's disease, J. Am. Osteopath. Assoc., 2001, 101, S1–S6 CAS.
- B. J. Balin, C. S. Little, C. J. Hammond, D. M. Appelt, J. A. Whittum-Hudson, H. C. Gerard and A. P. Hudson, Chlamydophila pneumoniae and the etiology of late-onset Alzheimer's disease, J. Alzheimer's Dis., 2008, 13, 371–380 CAS.
- H. C. Gérard, U. Dreses-Werringloer, K. S. Wildt, S. Deka, C. Oszust, B. J. Balin, W. H. Frey 2nd, E. Z. Bordayo, J. A. Whittum-Hudson and A. P. Hudson, Chlamydophila (Chlamydia) pneumoniae in the Alzheimer's brain, FEMS Immunol. Med. Microbiol., 2006, 48, 355–366 CrossRef PubMed.
- C. J. Hammond, L. R. Hallock, R. J. Howanski, D. M. Appelt, C. S. Little and B. J. Balin, Immunohistological detection of Chlamydia pneumoniae in the Alzheimer's disease brain, BMC Neurosci., 2010, 11, 121 CrossRef PubMed.
- R. F. Itzhaki, M. A. Wozniak, D. M. Appelt and B. J. Balin, Infiltration of the brain by pathogens causes Alzheimer's disease, Neurobiol. Aging, 2004, 25, 619–627 CrossRef CAS PubMed.
- C. Lim, C. J. Hammond, S. T. Hingley and B. J. Balin, Chlamydia pneumoniae infection of monocytes in vitro stimulates innate and adaptive immune responses relevant to those in Alzheimer's disease, J. Neuroinflammation, 2014, 11, 5 CrossRef PubMed.
- C. S. Little, C. J. Hammond, A. MacIntyre, B. J. Balin and D. M. Appelt, Chlamydia pneumoniae induces Alzheimer-like amyloid plaques in brains of BALB/c mice, Neurobiol. Aging, 2004, 25, 419–429 CrossRef CAS.
- C. S. Little, T. A. Joyce, C. J. Hammond, H. Matta, D. Cahn, D. M. Appelt and B. J. Balin, Detection of bacterial antigens and Alzheimer's disease-like pathology in the central nervous system of BALB/c mice following intranasal infection with a laboratory isolate of Chlamydia pneumoniae, Front. Aging Neurosci., 2014, 6, 304 Search PubMed.
- C. B. Dobson, M. A. Wozniak and R. F. Itzhaki, Do infectious agents play a role in dementia?, Trends Microbiol., 2003, 11, 312–317 CrossRef CAS.
- R. F. Itzhaki and M. A. Wozniak, Alzheimer's disease, the neuroimmune axis, and viral infection, J. Neuroimmunol., 2004, 156, 1–2 CrossRef CAS PubMed.
- R. F. Itzhaki and M. A. Wozniak, Alzheimer's disease-like changes in herpes simplex virus type 1 infected cells: the case for antiviral therapy, Rejuvenation Res., 2008, 11, 319–320 CrossRef PubMed.
- R. F. Itzhaki and M. A. Wozniak, Herpes simplex virus type 1 in Alzheimer's disease: the enemy within, J. Alzheimer's Dis., 2008, 13, 393–405 CAS.
- R. F. Itzhaki, S. L. Cosby and M. A. Wozniak, Herpes simplex virus type 1 and Alzheimer's disease: the autophagy connection, J. NeuroVirol., 2008, 14, 1–4 CrossRef CAS PubMed.
- R. F. Itzhaki and M. A. Wozniak, Alzheimer's disease and infection: do infectious agents contribute to progression of Alzheimer's disease?, Alzheimer's Dementia, 2010, 6, 83–84 CrossRef PubMed ; author reply 85.
- R. F. Itzhaki and M. A. Wozniak, Could antivirals be used to treat Alzheimer's disease?, Future Microbiol., 2012, 7, 307–309 CrossRef CAS PubMed.
- M. A. Wozniak and R. F. Itzhaki, Antiviral agents in Alzheimer's disease: hope for the future?, Ther. Adv. Neurol. Disord., 2010, 3, 141–152 CrossRef CAS PubMed.
- M. A. Wozniak, A. L. Frost, C. M. Preston and R. F. Itzhaki, Antivirals reduce the formation of key Alzheimer's disease molecules in cell cultures acutely infected with herpes simplex virus type 1, PLoS One, 2011, 6, e25152 CAS.
- M. A. Wozniak, A. L. Frost and R. F. Itzhaki, The helicase-primase inhibitor BAY 57-1293 reduces the Alzheimer's disease-related molecules induced by herpes simplex virus type 1, Antiviral Res., 2013, 99, 401–404 CrossRef CAS PubMed.
- J. Miklossy, Alzheimer's disease – a spirochetosis?, NeuroReport, 1993, 4, 841–848 CrossRef CAS.
- J. Miklossy, S. Kasas, R. C. Janzer, F. Ardizzoni and H. Van der Loos, Further ultrastructural evidence that spirochaetes may play a role in the aetiology of Alzheimer's disease, NeuroReport, 1994, 5, 1201–1204 CrossRef CAS.
- J. Miklossy, K. Khalili, L. Gern, R. L. Ericson, P. Darekar, L. Bolle, J. Hurlimann and B. J. Paster, Borrelia burgdorferi persists in the brain in chronic lyme neuroborreliosis and may be associated with Alzheimer disease, J. Alzheimer's Dis., 2004, 6, 639–649 Search PubMed ; discussion 673–681.
- J. Miklossy, A. Kis, A. Radenovic, L. Miller, L. Forro, R. Martins, K. Reiss, N. Darbinian, P. Darekar, L. Mihaly and K. Khalili, Beta-amyloid deposition and Alzheimer's type changes induced by Borrelia spirochetes, Neurobiol. Aging, 2006, 27, 228–236 CrossRef CAS PubMed.
- J. Miklossy, Chronic inflammation and amyloidogenesis in Alzheimer's disease – role of Spirochetes, J. Alzheimer's Dis., 2008, 13, 381–391 CAS.
- J. Miklossy, Emerging roles of pathogens in Alzheimer disease, Expert Rev. Mol. Med., 2011, 13, e30 CrossRef PubMed.
- J. Miklossy, Alzheimer's disease – a neurospirochetosis. Analysis of the evidence following Koch's and Hill's criteria, J. Neuroinflammation, 2011, 8, 90 CrossRef PubMed.
- J. Miklossy, Historic evidence to support a causal relationship between spirochetal infections and Alzheimer's disease, Front. Aging Neurosci., 2015, 7, 46 Search PubMed.
- J. Miklossy, Biology and neuropathology of dementia in syphilis and Lyme disease, Handb. Clin. Neurol., 2008, 89, 825–844 Search PubMed.
- J. Miklossy, S. Kasas, A. D. Zurn, S. McCall, S. Yu and P. L. McGeer, Persisting atypical and cystic forms of Borrelia burgdorferi and local inflammation in Lyme neuroborreliosis, J. Neuroinflammation, 2008, 5, 40 CrossRef PubMed.
- J. Miklossy, Chronic or late lyme neuroborreliosis: analysis of evidence compared to chronic or late neurosyphilis, Open Neurol. J., 2012, 6, 146–157 CrossRef PubMed.
- O. Miman, O. Y. Kusbeci, O. C. Aktepe and Z. Cetinkaya, The probable relation between Toxoplasma gondii and Parkinson's disease, Neurosci. Lett., 2010, 475, 129–131 CrossRef CAS PubMed.
- O. Y. Kusbeci, O. Miman, M. Yaman, O. C. Aktepe and S. Yazar, Could Toxoplasma gondii have any role in Alzheimer disease?, Alzheimer Dis. Assoc. Disord., 2011, 25, 1–3 CrossRef PubMed.
- E. F. Torrey, J. J. Bartko and R. H. Yolken, Toxoplasma gondii and other risk factors for schizophrenia: an update, Schizophr. Bull., 2012, 38, 642–647 CrossRef PubMed.
- R. Diaz Heijtz, S. Wang, F. Anuar, Y. Qian, B. Björkholm, A. Samuelsson, M. L. Hibberd, H. Forssberg and S. Pettersson, Normal gut microbiota modulates brain development and behavior, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3047–3052 CrossRef PubMed.
- J. F. Cryan and T. G. Dinan, Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour, Nat. Rev. Neurosci., 2012, 13, 701–712 CrossRef CAS PubMed.
- P. Forsythe, W. A. Kunze and J. Bienenstock, On communication between gut microbes and the brain, Curr. Opin. Gastroenterol., 2012, 28, 557–562 CrossRef PubMed.
- X. Chen, R. D'Souza and S. T. Hong, The role of gut microbiota in the gut-brain axis: current challenges and perspectives, Protein Cell, 2013, 4, 403–414 CrossRef PubMed.
- J. A. Foster and K. A. McVey Neufeld, Gut-brain axis: how the microbiome influences anxiety and depression, Trends Neurosci., 2013, 36, 305–312 CrossRef CAS PubMed.
- M. Lyte, Microbial endocrinology in the microbiome-gut-brain axis: how bacterial production and utilization of neurochemicals influence behavior, PLoS Pathog., 2013, 9, e1003726 CrossRef PubMed.
- A. J. Montiel-Castro, R. M. González-Cervantes, G. Bravo-Ruiseco and G. Pacheco-López, The microbiota-gut-brain axis: neurobehavioral correlates, health and sociality, Front. Integr. Neurosci., 2013, 7, 70 Search PubMed.
- P. Bercik and S. M. Collins, The effects of inflammation, infection and antibiotics on the microbiota-gut-brain axis, Adv. Exp. Med. Biol., 2014, 817, 279–289 CrossRef CAS.
- G. De Palma, S. M. Collins and P. Bercik, The microbiota-gut-brain axis in functional gastrointestinal disorders, Gut Microbes, 2014, 5, 419–429 CrossRef PubMed.
- G. De Palma, S. M. Collins, P. Bercik and E. F. Verdu, The microbiota-gut-brain axis in gastrointestinal disorders: stressed bugs, stressed brain or both?, J. Physiol., 2014, 592, 2989–2997 CrossRef CAS PubMed.
- M. G. Gareau, Microbiota-gut-brain axis and cognitive function, Adv. Exp. Med. Biol., 2014, 817, 357–371 CrossRef CAS.
- P. Holzer and A. Farzi, Neuropeptides and the microbiota-gut-brain axis, Adv. Exp. Med. Biol., 2014, 817, 195–219 CrossRef CAS.
- M. Lyte, Microbial endocrinology and the microbiota-gut-brain axis, Adv. Exp. Med. Biol., 2014, 817, 3–24 CrossRef CAS.
- J. Bienenstock, W. Kunze and P. Forsythe, Microbiota and the gut-brain axis, Nutr. Rev., 2015, 73(suppl 1), 28–31 CrossRef PubMed.
- M. Carabotti, A. Scirocco, M. A. Maselli and C. Severi, The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems, Ann. Gastroenterol., 2015, 28, 203–209 Search PubMed.
- X. Cong, W. A. Henderson, J. Graf and J. M. McGrath, Early Life Experience and Gut Microbiome: The Brain-Gut-Microbiota Signaling System, Adv. Neonatal Care, 2015 DOI:10.1097/ANC.0000000000000191.
- R. A. Luna and J. A. Foster, Gut brain axis: diet microbiota interactions and implications for modulation of anxiety and depression, Curr. Opin. Biotechnol., 2015, 32, 35–41 CrossRef CAS PubMed.
- E. A. Mayer, K. Tillisch and A. Gupta, Gut/brain axis and the microbiota, J. Clin. Invest., 2015, 125, 926–938 CrossRef PubMed.
- V. Ridaura and Y. Belkaid, Gut microbiota: the link to your second brain, Cell, 2015, 161, 193–194 CrossRef CAS PubMed.
- M. P. Sherman, H. Zaghouani and V. Niklas, Gut microbiota, the immune system, and diet influence the neonatal gut-brain axis, Pediatr. Res., 2015, 77, 127–135 CrossRef PubMed.
- S. L. Byler, G. W. Boehm, J. D. Karp, R. A. Kohman, A. J. Tarr, T. Schallert and T. M. Barth, Systemic lipopolysaccharide plus MPTP as a model of dopamine loss and gait instability in C57Bl/6J mice, Behav. Brain Res., 2009, 198, 434–439 CrossRef CAS PubMed.
- L. Hritcu, A. Ciobica, M. Stefan, M. Mihasan, L. Palamiuc and T. Nabeshima, Spatial memory deficits and oxidative stress damage following exposure to lipopolysaccharide in a rodent model of Parkinson's disease, Neurosci. Res., 2011, 71, 35–43 CrossRef CAS PubMed.
- L. Hritcu and A. Ciobica, Intranigral lipopolysaccharide administration induced behavioral deficits and oxidative stress damage in laboratory rats: relevance for Parkinson's disease, Behav. Brain Res., 2013, 253, 25–31 CrossRef CAS PubMed.
- M. Liu and G. Bing, Lipopolysaccharide animal models for Parkinson's disease, Parkinson's Dis., 2011, 2011, 327089 Search PubMed.
- Z. Zhang, K. Zhang, X. Du and Y. Li, Neuroprotection of desferrioxamine in lipopolysaccharide-induced nigrostriatal dopamine neuron degeneration, Mol. Med. Rep., 2012, 5, 562–566 CAS.
- R. M. Santiago, J. Barbieiro, M. M. Lima, P. A. Dombrowski, R. Andreatini and M. A. B. F. Vital, Depressive-like behaviors alterations induced by intranigral MPTP, 6-OHDA, LPS and rotenone models of Parkinson's disease are predominantly associated with serotonin and dopamine, Prog. Neuropsychopharmacol. Biol. Psychiatry, 2010, 34, 1104–1114 CrossRef CAS PubMed.
- Q. He, W. Yu, J. Wu, C. Chen, Z. Lou, Q. Zhang, J. Zhao, J. Wang and B. Xiao, Intranasal LPS-mediated Parkinson's model challenges the pathogenesis of nasal cavity and environmental toxins, PLoS One, 2013, 8, e78418 CAS.
- D. B. Hoban, E. Connaughton, C. Connaughton, G. Hogan, C. Thornton, P. Mulcahy, T. C. Moloney and E. Dowd, Further characterisation of the LPS model of Parkinson's disease: a comparison of intra-nigral and intra-striatal lipopolysaccharide administration on motor function, microgliosis and nigrostriatal neurodegeneration in the rat, Brain, Behav., Immun., 2013, 27, 91–100 CrossRef CAS PubMed.
- C. B. Lawrence, D. Brough and E. M. Knight, Obese mice exhibit an altered behavioural and inflammatory response to lipopolysaccharide, Dis. Models & Mech., 2012, 5, 649–659 CAS.
- J. Cohen, The immunopathogenesis of sepsis, Nature, 2002, 420, 885–891 CrossRef CAS PubMed.
- C. Nathan, Points of control in inflammation, Nature, 2002, 420, 846–852 CrossRef CAS PubMed.
- D. Annane, P. Aegerter, M. C. Jars-Guincestre, B. Guidet and CUB-Réa Network, Current epidemiology of septic shock: the CUB-Réa Network, Am. J. Respir. Crit. Care Med., 2003, 168, 165–172 CrossRef PubMed.
- J. A. Russell, Management of sepsis, N. Engl. J. Med., 2006, 355, 1699–1713 CrossRef CAS PubMed.
- G. Suntharalingam, M. R. Perry, S. Ward, S. J. Brett, A. Castello-Cortes, M. D. Brunner and N. Panoskaltsis, Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412, N. Engl. J. Med., 2006, 355, 1018–1028 CrossRef CAS PubMed.
- S. Sriskandan and D. M. Altmann, The immunology of sepsis, J. Pathol., 2008, 214, 211–223 CrossRef CAS PubMed.
- L. Ulloa, M. Brunner, L. Ramos and E. A. Deitch, Scientific and clinical challenges in sepsis, Curr. Pharm. Des., 2009, 15, 1918–1935 CrossRef CAS.
- N. R. London, W. Zhu, F. A. Bozza, M. C. Smith, D. M. Greif, L. K. Sorensen, L. Chen, Y. Kaminoh, A. C. Chan, S. F. Passi, C. W. Day, D. L. Barnard, G. A. Zimmerman, M. A. Krasnow and D. Y. Li, Targeting Robo4-dependent slit signaling to survive the cytokine storm in sepsis and influenza, Sci. Transl. Med., 2010, 2, 23ra19 Search PubMed.
- J. Hadem, C. Hafer, A. S. Schneider, O. Wiesner, G. Beutel, T. Fuehner, T. Welte, M. M. Hoeper and J. T. Kielstein, Therapeutic plasma exchange as rescue therapy in severe sepsis and septic shock: retrospective observational single-centre study of 23 patients, BMC Anesthesiol., 2014, 14, 24 CrossRef PubMed.
- M. M. Levy, M. P. Fink, J. C. Marshall, E. Abraham, D. Angus, D. Cook, J. Cohen, S. M. Opal, J. L. Vincent, G. Ramsay and Sccm/Esicm/Accp/Ats/Sis, 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference, Crit. Care Med., 2003, 31, 1250–1256 CrossRef PubMed.
- W. M. Dunne Jr., Laboratory diagnosis of sepsis? No SIRS, not just yet, J. Clin. Microbiol., 2015, 53, 2404–2409 CrossRef PubMed.
- D. H. Lewis, D. L. Chan, D. Pinheiro, E. Armitage-Chan and O. A. Garden, The immunopathology of sepsis: pathogen recognition, systemic inflammation, the compensatory anti-inflammatory response, and regulatory T cells, J. Vet. Intern. Med., 2012, 26, 457–482 CrossRef CAS PubMed.
- F. Ratzinger, M. Schuardt, K. Eichbichler, I. Tsirkinidou, M. Bauer, H. Haslacher, D. Mitteregger, M. Binder and H. Burgmann, Utility of sepsis biomarkers and the infection probability score to discriminate sepsis and systemic inflammatory response syndrome in standard care patients, PLoS One, 2013, 8, e82946 Search PubMed.
- R. P. Dellinger, M. M. Levy, A. Rhodes, D. Annane, H. Gerlach, S. M. Opal, J. E. Sevransky, C. L. Sprung, I. S. Douglas, R. Jaeschke, T. M. Osborn, M. E. Nunnally, S. R. Townsend, K. Reinhart, R. M. Kleinpell, D. C. Angus, C. S. Deutschman, F. R. Machado, G. D. Rubenfeld, S. A. Webb, R. J. Beale, J. L. Vincent, R. Moreno and Surviving Sepsis Campaign Guidelines Committee including the Pediatric Subgroup, Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012, Crit. Care Med., 2013, 41, 580–637 CrossRef PubMed.
- American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference, Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis, Crit. Care Med., 1992, 20, 864–874 CrossRef.
- R. C. Bone, R. A. Balk, F. B. Cerra, R. P. Dellinger, A. M. Fein, W. A. Knaus, R. M. Schein and W. J. Sibbald, Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine, Chest, 1992, 101, 1644–1655 CrossRef CAS.
- G. S. Martin, D. M. Mannino and M. Moss, The effect of age on the development and outcome of adult sepsis, Crit. Care Med., 2006, 34, 15–21 CrossRef.
- R. H. K. Eng, S. M. Smith, P. Fan-Havard and T. Ogbara, Effect of antibiotics on endotoxin release from Gram-negative bacteria, Diagn. Microbiol. Infect. Dis., 1993, 16, 185–189 CrossRef CAS.
- J. M. Prins, S. J. H. van Deventer, E. J. Kuijper and P. Speelman, Clinical relevance of antibiotic-induced endotoxin release, Antimicrob. Agents Chemother., 1994, 38, 1211–1218 CrossRef CAS.
- J. J. Jackson and H. Kropp, Differences in mode of action of beta-lactam antibiotics influence morphology, LPS release and in vivo antibiotic efficacy, J. Endotoxin Res., 1996, 3, 201–218 CAS.
- D. C. Morrison, S. E. Bucklin, M. C. Leeson and M. Norimatsu, Contribution of soluble endotoxin released from Gram-negative bacteria by antibiotics to the pathogenesis of experimental sepsis in mice, J. Endotoxin Res., 1996, 3, 237–243 CAS.
- T. Kirikae, M. Nakano and D. C. Morrison, Antibiotic-induced endotoxin release from bacteria and its clinical significance, Microbiol. Immunol., 1997, 41, 285–294 CrossRef CAS PubMed.
- R. G. Holzheimer, Antibiotic induced endotoxin release and clinical sepsis: a review, J. Chemother., 2001, 13(spec no. 1), 159–172 CrossRef CAS PubMed.
- P. M. Lepper, T. K. Held, E. M. Schneider, E. Bölke, H. Gerlach and M. Trautmann, Clinical implications of antibiotic-induced endotoxin release in septic shock, Intensive Care Med., 2002, 28, 824–833 CrossRef CAS PubMed.
- M. Trautmann, C. Scheibe, N. Wellinghausen, O. Holst and P. M. Lepper, Low endotoxin release from Escherichia coli and Bacteroides fragilis during exposure to moxifloxacin, Chemotherapy, 2010, 56, 364–370 CrossRef CAS PubMed.
- S. M. Walters, V. S. Dubey, N. R. Jeffrey and D. R. Dixon, Antibiotic-induced Porphyromonas gingivalis LPS release and inhibition of LPS-stimulated cytokines by antimicrobial peptides, Peptides, 2010, 31, 1649–1653 CrossRef CAS PubMed.
- S. Tanabe, M. Yoshioka, D. Hinode and D. Grenier, Subinhibitory concentrations of tetracyclines induce lipopolysaccharide shedding by Porphyromonas gingivalis and modulate the host inflammatory response, J. Periodontal Res., 2014, 49, 603–608 CrossRef CAS PubMed.
- G. Guerrier and E. D'Ortenzio, The Jarisch–Herxheimer reaction in leptospirosis: a systematic review, PLoS One, 2013, 8, e59266 CAS.
- P. Kadam, N. A. Gregory, B. Zelger and J. A. Carlson, Delayed Onset of the Jarisch-Herxheimer Reaction in Doxycycline-Treated Disease: a Case Report and Review of its Histopathology and Implications for Pathogenesis, Am. J. Dermatopathol., 2015, 37, e68–e74 CrossRef PubMed.
- D. Fekade, K. Knox, K. Hussein, A. Melka, D. G. Lalloo, R. E. Coxon and D. A. Warrell, Prevention of Jarisch–Herxheimer reactions by treatment with antibodies against tumor necrosis factor alpha, N. Engl. J. Med., 1996, 335, 311–315 CrossRef CAS PubMed.
- P. W. Taylor, Bactericidal and bacteriolytic activity of serum against Gram-negative bacteria, Microbiol. Rev., 1983, 47, 46–83 CAS.
- T. J. Iwashyna, E. W. Ely, D. M. Smith and K. M. Langa, Long-term cognitive impairment and functional disability among survivors of severe sepsis, JAMA, J. Am. Med. Assoc., 2010, 304, 1787–1794 CrossRef CAS PubMed.
- S. Yende, W. Linde-Zwirble, F. Mayr, L. A. Weissfeld, S. Reis and D. C. Angus, Risk of cardiovascular events in survivors of severe sepsis, Am. J. Respir. Crit. Care Med., 2014, 189, 1065–1074 CrossRef PubMed.
- T. J. Iwashyna, C. R. Cooke, H. Wunsch and J. M. Kahn, Population burden of long-term survivorship after severe sepsis in older Americans, J. Am. Geriatr. Soc., 2012, 60, 1070–1077 CrossRef PubMed.
- S. Yende, T. J. Iwashyna and D. C. Angus, Interplay between sepsis and chronic health, Trends Mol. Med., 2014, 20, 234–238 CrossRef PubMed.
- M. R. Boocock and J. R. Coggins, Kinetics of 5-enolpyruvylshikimate-3-phosphate synthase inhibition by glyphosate, FEBS Lett., 1983, 154, 127–133 CrossRef CAS.
- J. L. Rubin, C. G. Gaines and R. A. Jensen, Glyphosate inhibition of 5-enolpyruvylshikimate 3-phosphate synthase from suspension-cultured cells of Nicotiana silvestris, Plant Physiol., 1984, 75, 839–845 CrossRef CAS PubMed.
- H. C. Steinrucken and N. Amrhein, 5-Enolpyruvylshikimate-3-phosphate synthase of Klebsiella pneumoniae 2. Inhibition by glyphosate [N-(phosphonomethyl)glycine], Eur. J. Biochem., 1984, 143, 351–357 CrossRef CAS PubMed.
- G. M. Kishore and D. M. Shah, Amino acid biosynthesis inhibitors as herbicides, Annu. Rev. Biochem., 1988, 57, 627–663 CrossRef CAS PubMed.
- A. Cornish-Bowden, Why is uncompetitive inhibition so rare? A possible explanation, with implications for the design of drugs and pesticides, FEBS Lett., 1986, 203, 3–6 CrossRef CAS.
- D. B. Kell, Metabolomics, modelling and machine learning in systems biology: towards an understanding of the languages of cells. The 2005 Theodor Bücher lecture, FEBS J., 2006, 273, 873–894 CrossRef CAS PubMed.
- A. Currin, N. Swainston, P. J. Day and D. B. Kell, Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently, Chem. Soc. Rev., 2015, 44, 1172–1239 RSC.
- A. Goldbeter and D. E. Koshland Jr., An amplified sensitivity arising from covalent modification in biological systems, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 6840–6844 CrossRef CAS.
- A. Goldbeter and D. E. Koshland Jr., Ultrasensitivity in biochemical systems controlled by covalent modification. Interplay between zero-order and multistep effects, J. Biol. Chem., 1984, 259, 14441–14447 CAS.
- M. L. Cárdenas and A. Cornish-Bowden, Characteristics necessary for an interconvertible enzyme cascade to generate a highly sensitive response to an effector, Biochem. J., 1989, 257, 339–345 CrossRef.
- A. Cornish-Bowden, Fundamentals of enzyme kinetics, Portland Press, London, 2nd edn, 1995 Search PubMed.
- F. Ortega, L. Acerenza, H. V. Westerhoff, F. Mas and M. Cascante, Product dependence and bifunctionality compromise the ultrasensitivity of signal transduction cascades, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 1170–1175 CrossRef CAS PubMed.
- Y. Li and A. Goldbeter, Pulsatile signaling in intercellular communication. Periodic stimuli are more efficient than random or chaotic signals in a model based on receptor desensitization, Biophys. J., 1992, 61, 161–171 CrossRef CAS.
- L. Ashall, C. A. Horton, D. E. Nelson, P. Paszek, S. Ryan, K. Sillitoe, C. V. Harper, D. G. Spiller, J. F. Unitt, D. S. Broomhead, D. B. Kell, D. Rand, V. Sée and M. R. H. White, Pulsatile stimulation determines timing and specificity of NFkappa-B-dependent transcription, Science, 2009, 324, 242–246 CrossRef CAS PubMed.
- J. J. Tyson, K. C. Chen and B. Novak, Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell, Curr. Opin. Cell. Biol., 2003, 15, 221–231 CrossRef CAS.
- D. B. Kell and J. D. Knowles, The role of modeling in systems biology, in System modeling in cellular biology: from concepts to nuts and bolts, ed. Z. Szallasi, J. Stelling and V. Periwal, MIT Press, Cambridge, 2006, pp. 3–18 Search PubMed.
- H. Y. Hsu and M. H. Wen, Lipopolysaccharide-mediated reactive oxygen species and signal transduction in the regulation of interleukin-1 gene expression, J. Biol. Chem., 2002, 277, 22131–22139 CrossRef CAS PubMed.
- C. A. Dinarello, Interleukin-1 in the pathogenesis and treatment of inflammatory diseases, Blood, 2011, 117, 3720–3732 CrossRef CAS PubMed.
- C. A. Dinarello, A clinical perspective of IL-1beta as the gatekeeper of inflammation, Eur. J. Immunol., 2011, 41, 1203–1217 CrossRef CAS PubMed.
- S. U. Seo, N. Kamada, R. Munoz-Planillo, Y. G. Kim, D. Kim, Y. Koizumi, M. Hasegawa, S. D. Himpsl, H. P. Browne, T. D. Lawley, H. L. Mobley, N. Inohara and G. Nunez, Distinct Commensals Induce Interleukin-1beta via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury, Immunity, 2015, 42, 744–755 CrossRef CAS PubMed.
- J. R. Bethea, G. Y. Gillespie and E. N. Benveniste, Interleukin-1 beta induction of TNF-alpha gene expression: involvement of protein kinase C, J. Cell. Physiol., 1992, 152, 264–273 CrossRef CAS PubMed.
- A. L. Hopkins, J. S. Mason and J. P. Overington, Can we rationally design promiscuous drugs?, Curr. Opin. Struct. Biol., 2006, 16, 127–136 CrossRef CAS PubMed.
- A. L. Hopkins, Network pharmacology: the next paradigm in drug discovery, Nat. Chem. Biol., 2008, 4, 682–690 CrossRef CAS PubMed.
- D. B. Kell, Finding novel pharmaceuticals in the systems biology era using multiple effective drug targets, phenotypic screening, and knowledge of transporters: where drug discovery went wrong and how to fix it, FEBS J., 2013, 280, 5957–5980 CrossRef CAS PubMed.
- D. B. Kell and H. V. Westerhoff, Metabolic control theory: its role in microbiology and biotechnology, FEMS Microbiol. Rev., 1986, 39, 305–320 CrossRef CAS PubMed.
- A. Cornish-Bowden, J.-H. S. Hofmeyr and M. L. Cárdenas, Strategies for manipulating metabolic fluxes in biotechnology, Bioorg. Chem., 1995, 23, 439–449 CrossRef CAS.
- D. A. Fell, Understanding the control of metabolism, Portland Press, London, 1996 Search PubMed.
- D. A. Fell, Increasing the flux in metabolic pathways: a metabolic control analysis perspective, Biotechnol. Bioeng., 1998, 58, 121–124 CrossRef CAS.
- C. R. H. Raetz and C. Whitfield, Lipopolysaccharide endotoxins, Annu. Rev. Biochem., 2002, 71, 635–700 CrossRef CAS PubMed.
- M. Caroff and D. Karibian, Structure of bacterial lipopolysaccharides, Carbohydr. Res., 2003, 338, 2431–2447 CrossRef CAS PubMed.
- S. Müller-Loennies, L. Brade, C. R. MacKenzie, F. E. Di Padova and H. Brade, Identification of a cross-reactive epitope widely present in lipopolysaccharide from enterobacteria and recognized by the cross-protective monoclonal antibody WN1 222-5, J. Biol. Chem., 2003, 278, 25618–25627 CrossRef PubMed.
- O. Holst, The structures of core regions from enterobacterial lipopolysaccharides – an update, FEMS Microbiol. Lett., 2007, 271, 3–11 CrossRef CAS PubMed.
- P. Swain, S. K. Nayak, P. K. Nanda and S. Dash, Biological effects of bacterial lipopolysaccharide (endotoxin) in fish: a review, Fish Shellfish Immunol., 2008, 25, 191–201 CrossRef CAS PubMed.
- C. De Castro, M. Parrilli, O. Holst and A. Molinaro, Microbe-associated molecular patterns in innate immunity: extraction and chemical analysis of Gram-negative bacterial lipopolysaccharides, Methods Enzymol., 2010, 480, 89–115 CAS.
- C. Whitfield, Biosynthesis and assembly of capsular polysaccharides in Escherichia coli, Annu. Rev. Biochem., 2006, 75, 39–68 CrossRef CAS PubMed.
- X. Wang and P. J. Quinn, Lipopolysaccharide: biosynthetic pathway and structure modification, Prog. Lipid Res., 2010, 49, 97–107 CrossRef CAS PubMed.
- E. Fahy, S. Subramaniam, H. A. Brown, C. K. Glass, A. H. Merrill Jr., R. C. Murphy, C. R. Raetz, D. W. Russell, Y. Seyama, W. Shaw, T. Shimizu, F. Spener, G. van Meer, M. S. VanNieuwenhze, S. H. White, J. L. Witztum and E. A. Dennis, A comprehensive classification system for lipids, J. Lipid Res., 2005, 46, 839–861 CrossRef CAS PubMed.
- C. R. H. Raetz, T. A. Garrett, C. M. Reynolds, W. A. Shaw, J. D. Moore, D. C. Smith Jr., A. A. Ribeiro, R. C. Murphy, R. J. Ulevitch, C. Fearns, D. Reichart, C. K. Glass, C. Benner, S. Subramaniam, R. Harkewicz, R. C. Bowers-Gentry, M. W. Buczynski, J. A. Cooper, R. A. Deems and E. A. Dennis, Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4, J. Lipid Res., 2006, 47, 1097–1111 CrossRef CAS PubMed.
- N. M. Kelly, L. Young and A. S. Cross, Differential induction of tumor necrosis factor by bacteria expressing rough and smooth lipopolysaccharide phenotypes, Infect. Immun., 1991, 59, 4491–4496 CAS.
- E. Klipp, R. Herwig, A. Kowald, C. Wierling and H. Lehrach, Systems biology in practice: concepts, implementation and clinical application, Wiley/VCH, Berlin, 2005 Search PubMed.
- S. Hoops, S. Sahle, R. Gauges, C. Lee, J. Pahle, N. Simus, M. Singhal, L. Xu, P. Mendes and U. Kummer, COPASI: a COmplex PAthway SImulator, Bioinformatics, 2006, 22, 3067–3074 CrossRef CAS PubMed.
- B. Ø. Palsson, Systems biology: constraint-based reconstruction and analysis, Cambridge University Press, Cambridge, 2015 Search PubMed.
- A. Nowotny, Relationship of structure and biological activity of bacterial endotoxins, Naturwissenschaften, 1971, 58, 397–409 CrossRef CAS.
- E. T. Rietschel, T. Kirikae, F. U. Schade, U. Mamat, G. Schmidt, H. Loppnow, A. J. Ulmer, U. Zahringer, U. Seydel and F. Di Padova, et al., Bacterial endotoxin: molecular relationships of structure to activity and function, FASEB J., 1994, 8, 217–225 CAS.
- S. I. Miller, R. K. Ernst and M. W. Bader, LPS, TLR4 and infectious disease diversity, Nat. Rev. Microbiol., 2005, 3, 36–46 CrossRef CAS PubMed.
- M. S. Trent, C. M. Stead, A. X. Tran and J. V. Hankins, Diversity of endotoxin and its impact on pathogenesis, J. Endotoxin Res., 2006, 12, 205–223 CrossRef CAS PubMed.
- Y. Tsutsumi-Ishii, K. Shimada, H. Daida, R. Toman and I. Nagaoka, Low potency of Chlamydophila LPS to activate human mononuclear cells due to its reduced affinities for CD14 and LPS-binding protein, Int. Immunol., 2008, 20, 199–208 CrossRef CAS PubMed.
- J. C. Hurley, Diagnosis of endotoxemia with Gram-negative bacteremia is bacterial species dependent: a meta-analysis of clinical studies, J. Clin. Microbiol., 2009, 47, 3826–3831 CrossRef PubMed.
- X. Wang and P. J. Quinn, Endotoxins: lipopolysaccharides of Gram-negative bacteria, Subcell. Biochem., 2010, 53, 3–25 CAS.
- J. C. Hurley and S. M. Opal, Prognostic value of endotoxemia in patients with Gram-negative bacteremia is bacterial species dependent, J. Innate Immun., 2013, 5, 555–564 CrossRef CAS PubMed.
- C. Slocum, S. R. Coats, N. Hua, C. Kramer, G. Papadopoulos, E. O. Weinberg, C. V. Gudino, J. A. Hamilton, R. P. Darveau and C. A. Genco, Distinct lipid A moieties contribute to pathogen-induced site-specific vascular inflammation, PLoS Pathog., 2014, 10, e1004215 Search PubMed.
- M. P. Fink, Animal models of sepsis and its complications, Kidney Int., 2008, 74, 991–993 CrossRef PubMed.
- M. P. Fink, Animal models of sepsis, Virulence, 2014, 5, 143–153 CrossRef PubMed.
- D. R. Webb, Animal models of human disease: inflammation, Biochem. Pharmacol., 2014, 87, 121–130 CrossRef CAS PubMed.
- D. E. Nelson, V. See, G. Nelson and M. R. White, Oscillations in transcription factor dynamics: a new way to control gene expression, Biochem. Soc. Trans., 2004, 32, 1090–1092 CrossRef CAS PubMed.
- A. E. C. Ihekwaba, D. S. Broomhead, R. Grimley, N. Benson, M. R. H. White and D. B. Kell, Synergistic control of oscillations in the NF-κB signalling pathway, IEE Proc.: Syst. Biol., 2005, 152, 153–160 CrossRef CAS.
- D. A. Turner, P. Paszek, D. J. Woodcock, D. E. Nelson, C. A. Horton, Y. Wang, D. G. Spiller, D. A. Rand, M. R. H. White and C. V. Harper, Physiological levels of TNFalpha stimulation induce stochastic dynamics of NF-kappaB responses in single living cells, J. Cell Sci., 2010, 123, 2834–2843 CrossRef CAS PubMed.
- G. Lahav, N. Rosenfeld, A. Sigal, N. Geva-Zatorsky, A. J. Levine, M. B. Elowitz and U. Alon, Dynamics of the p53-Mdm2 feedback loop in individual cells, Nat. Genet., 2004, 36, 147–150 CrossRef CAS PubMed.
- G. Lahav, Oscillations by the p53-Mdm2 feedback loop, Adv. Exp. Med. Biol., 2008, 641, 28–38 CrossRef CAS.
- E. Batchelor, A. Loewer, C. Mock and G. Lahav, Stimulus-dependent dynamics of p53 in single cells, Mol. Syst. Biol., 2011, 7, 488 CrossRef PubMed.
- J. E. Purvis, K. W. Karhohs, C. Mock, E. Batchelor, A. Loewer and G. Lahav, p53 dynamics control cell fate, Science, 2012, 336, 1440–1444 CrossRef CAS PubMed.
- D. Brough, R. A. Le Feuvre, Y. Iwakura and N. J. Rothwell, Purinergic (P2X7) receptor activation of microglia induces cell death via an interleukin-1-independent mechanism, Mol. Cell. Neurosci., 2002, 19, 272–280 CrossRef CAS PubMed.
- R. A. Le Feuvre, D. Brough, Y. Iwakura, K. Takeda and N. J. Rothwell, Priming of macrophages with lipopolysaccharide potentiates P2X7-mediated cell death via a caspase-1-dependent mechanism, independently of cytokine production, J. Biol. Chem., 2002, 277, 3210–3218 CrossRef CAS PubMed.
- M. E. Edye, G. Lopez-Castejon, S. M. Allan and D. Brough, Acidosis drives damage-associated molecular pattern (DAMP)-induced interleukin-1 secretion via a caspase-1-independent pathway, J. Biol. Chem., 2013, 288, 30485–30494 CrossRef CAS PubMed.
- A. J. Lee, K. J. Cho and J. H. Kim, MyD88-BLT2-dependent cascade contributes to LPS-induced interleukin-6 production in mouse macrophage, Exp. Mol. Med., 2015, 47, e156 CrossRef CAS PubMed.
- H. Mizutani, Y. Ishihara, A. Izawa, Y. Fujihara, S. Kobayashi, H. Gotou, E. Okabe, H. Takeda, Y. Ozawa, Y. Kamiya, H. Kamei, T. Kikuchi, G. Yamamoto, A. Mitani, T. Nishihara and T. Noguchi, Lipopolysaccharide of Aggregatibacter actinomycetemcomitans up-regulates inflammatory cytokines, prostaglandin E2 synthesis and osteoclast formation in interleukin-1 receptor antagonist-deficient mice, J. Periodontal Res., 2013, 48, 748–756 CAS.
- A. Meng, X. Zhang and Y. Shi, Role of p38 MAPK and STAT3 in lipopolysaccharide-stimulated mouse alveolar macrophages, Exp. Ther. Med., 2014, 8, 1772–1776 CAS.
- D. Brough, P. Pelegrin and N. J. Rothwell, Pannexin-1-dependent caspase-1 activation and secretion of IL-1 beta is regulated by zinc, Eur. J. Immunol., 2009, 39, 352–358 CrossRef CAS PubMed.
- M. Lee, S. H. Kim, H. K. Lee, Y. Cho, J. Kang and S. H. Sung, ent-kaurane and ent-pimarane diterpenes from Siegesbeckia pubescens inhibit lipopolysaccharide-induced nitric oxide production in BV2 microglia, Biol. Pharm. Bull., 2014, 37, 152–157 CAS.
- B. G. Small, B. W. McColl, R. Allmendinger, R. Pahle, G. Lopez-Castejon, N. J. Rothwell, J. Knowles, P. Mendes, D. Brough and D. B. Kell, Efficient discovery of anti-inflammatory small molecule combinations using evolutionary computing, Nat. Chem. Biol., 2011, 7, 902–908 CrossRef CAS PubMed.
- B. G. Taksande, C. T. Chopde, M. J. Umekar and N. R. Kotagale, Agmatine attenuates lipopolysaccharide induced anorexia and sickness behavior in rats, Pharmacol., Biochem. Behav., 2015, 132, 108–114 CrossRef CAS PubMed.
- F. Gao, Z. Liu, W. Ren and W. Jiang, Acute lipopolysaccharide exposure facilitates epileptiform activity via enhanced excitatory synaptic transmission and neuronal excitability in vitro, Neuropsychiatr. Dis. Treat., 2014, 10, 1489–1495 CrossRef CAS PubMed.
- K. Chatzivasileiou, C. A. Lux, G. Steinhoff and H. Lang, Dental follicle progenitor cells responses to Porphyromonas gingivalis LPS, J. Cell. Mol. Med., 2013, 17, 766–773 CrossRef CAS PubMed.
- S. Braesch-Andersen, S. Paulie, C. Smedman, S. Mia and M. Kumagai-Braesch, ApoE production in human monocytes and its regulation by inflammatory cytokines, PLoS One, 2013, 8, e79908 Search PubMed.
- J. D. Rempel, J. Packiasamy, H. J. Dean, J. McGavock, A. Janke, M. Collister, B. Wicklow and E. A. Sellers, Preliminary analysis of immune activation in early onset type 2 diabetes, Int. J. Circumpolar Health, 2013, 72, 21190 CrossRef PubMed.
- L. Fernández-Bertolín, J. Mullol, M. Fuentes-Prado, J. Roca-Ferrer, I. Alobid, C. Picado and L. Pujols, Effect of lipopolysaccharide on glucocorticoid receptor function in control nasal mucosa fibroblasts and in fibroblasts from patients with chronic rhinosinusitis with nasal polyps and asthma, PLoS One, 2015, 10, e0125443 Search PubMed.
- C. Sandersen, D. Bienzle, S. Cerri, T. Franck, S. Derochette, P. Neven, A. Mouytis-Mickalad and D. Serteyn, Effect of inhaled hydrosoluble curcumin on inflammatory markers in broncho-alveolar lavage fluid of horses with LPS-induced lung neutrophilia, Multidiscip. Respir. Med., 2015, 10, 16 CrossRef PubMed.
- D. Lamarque, A. P. Moran, Z. Szepes, J. C. Delchier and B. J. Whittle, Cytotoxicity associated with induction of nitric oxide synthase in rat duodenal epithelial cells in vivo by lipopolysaccharide of Helicobacter pylori: inhibition by superoxide dismutase, Br. J. Pharmacol., 2000, 130, 1531–1538 CrossRef CAS PubMed.
- K. L. Comstock, K. A. Krown, M. T. Page, D. Martin, P. Ho, M. Pedraza, E. N. Castro, N. Nakajima, C. C. Glembotski, P. J. E. Quintana and R. A. Sabbadini, LPS-Induced TNF-alpha release from and apoptosis in rat cardiomyocytes: Obligatory role for CD14 in mediating the LPS response, J. Mol. Cell. Cardiol., 1998, 30, 2761–2775 CrossRef CAS PubMed.
- Q. Xiong, Q. Ru, L. Chen, K. Yue, X. Tian, B. Ma, L. Liu, R. Wu, C. Xu, M. Pi and C. Li, Combined Effects of Fine Particulate Matter and Lipopolysaccharide on Apoptotic Responses in NR8383 Macrophages, J. Toxicol. Environ. Health, Part A, 2015, 78, 443–452 CrossRef CAS PubMed.
- P. C. Li, Y. C. Tien, C. H. Day, P. Pai, W. W. Kuo, T. S. Chen, C. H. Kuo, C. H. Tsai, D. T. Ju and C. Y. Huang, Impact of LPS-induced cardiomyoblast cell apoptosis inhibited by earthworm extracts, Cardiovasc. Toxicol., 2015, 15, 172–179 CrossRef PubMed.
- L. Zhong, X. L. Zhou, Y. S. Liu, Y. M. Wang, F. Ma, B. L. Guo, Z. Q. Yan and Q. Y. Zhang, Estrogen receptor alpha mediates the effects of notoginsenoside R1 on endotoxin-induced inflammatory and apoptotic responses in H9c2 cardiomyocytes, Mol. Med. Rep., 2015, 12, 119–126 CAS.
- C. Guo, L. Yuan, J. G. Wang, F. Wang, X. K. Yang, F. H. Zhang, J. L. Song, X. Y. Ma, Q. Cheng and G. H. Song, Lipopolysaccharide (LPS) induces the apoptosis and inhibits osteoblast differentiation through JNK pathway in MC3T3-E1 cells, Inflammation, 2014, 37, 621–631 CrossRef CAS PubMed.
- M. Takahashi, A. Ota, S. Karnan, E. Hossain, Y. Konishi, L. Damdindorj, H. Konishi, T. Yokochi, M. Nitta and Y. Hosokawa, Arsenic trioxide prevents nitric oxide production in lipopolysaccharide – stimulated RAW 264.7 by inhibiting a TRIF-dependent pathway, Cancer Sci., 2013, 104, 165–170 CrossRef CAS PubMed.
- J. Xaus, M. Comalada, A. F. Valledor, J. Lloberas, F. López-Soriano, J. M. Argilés, C. Bogdan and A. Celada, LPS induces apoptosis in macrophages mostly through the autocrine production of TNF-alpha, Blood, 2000, 95, 3823–3831 CAS.
- J. Zhao, X. Li, M. Zou, J. He, Y. Han, D. Wu, H. Yang and J. Wu, miR-135a inhibition protects A549 cells from LPS-induced apoptosis by targeting Bcl-2, Biochem. Biophys. Res. Commun., 2014, 452, 951–957 CrossRef CAS PubMed.
- A. L. Blomkalns, L. L. Stoll, W. Shaheen, S. A. Romig-Martin, E. W. Dickson, N. L. Weintraub and G. M. Denning, Low level bacterial endotoxin activates two distinct signaling pathways in human peripheral blood mononuclear cells, J. Inflammation, 2011, 8, 4 CrossRef CAS PubMed.
- X. J. Dai, N. Li, L. Yu, Z. Y. Chen, R. Hua, X. Qin and Y. M. Zhang, Activation of BV2 microglia by lipopolysaccharide triggers an inflammatory reaction in PC12 cell apoptosis through a toll-like receptor 4-dependent pathway, Cell Stress Chaperones, 2015, 20, 321–331 CrossRef CAS PubMed.
- J. Li, J. He and C. Yu, Chitosan oligosaccharide inhibits LPS-induced apoptosis of vascular endothelial cells through the BKCa channel and the p38 signaling pathway, Int. J. Mol. Med., 2012, 30, 157–164 CAS.
- I. S. Kim, H. M. Ko, S. Koppula, B. W. Kim and D. K. Choi, Protective effect of Chrysanthemum indicum Linne against 1-methyl-4-phenylpridinium ion and lipopolysaccharide-induced cytotoxicity in cellular model of Parkinson's disease, Food Chem. Toxicol., 2011, 49, 963–973 CrossRef CAS PubMed.
- P. Matzinger, The danger model: a renewed sense of self, Science, 2002, 296, 301–305 CrossRef CAS PubMed.
- T. Pradeu and E. L. Cooper, The danger theory: 20 years later, Front. Immunol., 2012, 3, 287 Search PubMed.
- C. A. Janeway Jr., Approaching the asymptote? Evolution and revolution in immunology, Cold Spring Harbor Symp. Quant. Biol., 1989, 54(pt 1), 1–13 CrossRef.
- D. L. Tang, R. Kang, C. B. Coyne, H. J. Zeh and M. T. Lotze, PAMPs and DAMPs: signal 0s that spur autophagy and immunity, Immunol. Rev., 2012, 249, 158–175 CrossRef CAS PubMed.
- A. Poltorak, X. L. He, I. Smirnova, M. Y. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton and B. Beutler, Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene, Science, 1998, 282, 2085–2088 CrossRef CAS.
- A. Poltorak, P. Ricciardi-Castagnoli, S. Citterio and B. Beutler, Physical contact between lipopolysaccharide and Toll-like receptor 4 revealed by genetic complementation, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 2163–2167 CrossRef CAS PubMed.
- B. Beutler, X. Du and A. Poltorak, Identification of Toll-like receptor 4 (Tlr4) as the sole conduit for LPS signal transduction: genetic and evolutionary studies, J. Endotoxin Res., 2001, 7, 277–280 CrossRef CAS PubMed.
- Y. C. Lu, W. C. Yeh and P. S. Ohashi, LPS/TLR4 signal transduction pathway, Cytokine, 2008, 42, 145–151 CrossRef CAS PubMed.
- B. S. Park, D. H. Song, H. M. Kim, B. S. Choi, H. Lee and J. O. Lee, The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex, Nature, 2009, 458, 1191–1195 CrossRef CAS PubMed.
- L. A. J. O'Neill, D. Golenbock and A. G. Bowie, The history of Toll-like receptors – redefining innate immunity, Nat. Rev. Immunol., 2013, 13, 453–460 CrossRef PubMed.
- K. Fassbender, S. Walter, S. Kuhl, R. Landmann, K. Ishii, T. Bertsch, A. K. Stalder, F. Muehlhauser, Y. Liu, A. J. Ulmer, S. Rivest, A. Lentschat, E. Gulbins, M. Jucker, M. Staufenbiel, K. Brechtel, J. Walter, G. Multhaup, B. Penke, Y. Adachi, T. Hartmann and K. Beyreuther, The LPS receptor (CD14) links innate immunity with Alzheimer's disease, FASEB J., 2004, 18, 203–205 CAS.
- Y. Liu, S. Walter, M. Stagi, D. Cherny, M. Letiembre, W. Schulz-Schaeffer, H. Heine, B. Penke, H. Neumann and K. Fassbender, LPS receptor (CD14): a receptor for phagocytosis of Alzheimer's amyloid peptide, Brain, 2005, 128, 1778–1789 CrossRef PubMed.
- E. G. Reed-Geaghan, J. C. Savage, A. G. Hise and G. E. Landreth, CD14 and toll-like receptors 2 and 4 are required for fibrillar A{beta}-stimulated microglial activation, J. Neurosci., 2009, 29, 11982–11992 CrossRef CAS PubMed.
- F. L. Heppner, R. M. Ransohoff and B. Becher, Immune attack: the role of inflammation in Alzheimer disease, Nat. Rev. Neurosci., 2015, 16, 358–372 CrossRef CAS PubMed.
- L. Verstrepen, T. Bekaert, T. L. Chau, J. Tavernier, A. Chariot and R. Beyaert, TLR-4, IL-1R and TNF-R signaling to NF-kappa B: variations on a common theme, Cell. Mol. Life Sci., 2008, 65, 2964–2978 CrossRef CAS PubMed.
- L. A. J. O'Neill, C. E. Bryant and S. L. Doyle, Therapeutic targeting of toll-like receptors for infectious and inflammatory diseases and cancer, Pharmacol. Rev., 2009, 61, 177–197 CrossRef PubMed.
- O. Takeuchi and S. Akira, Pattern Recognition Receptors and Inflammation, Cell, 2010, 140, 805–820 CrossRef CAS PubMed.
- T. Kawai and S. Akira, Toll-like receptors and their crosstalk with other innate receptors in infection and immunity, Immunity, 2011, 34, 637–650 CrossRef CAS PubMed.
- P. P. Tak and G. S. Firestein, NF-kappaB: a key role in inflammatory diseases, J. Clin. Invest., 2001, 107, 7–11 CrossRef CAS PubMed.
- J. Ding, D. Song, X. Ye and S. F. Liu, A pivotal role of endothelial-specific NF-kappaB signaling in the pathogenesis of septic shock and septic vascular dysfunction, J. Immunol., 2009, 183, 4031–4038 CrossRef CAS PubMed.
- S. Vallabhapurapu and M. Karin, Regulation and function of NF-kappaB transcription factors in the immune system, Annu. Rev. Immunol., 2009, 27, 693–733 CrossRef CAS PubMed.
- A. R. Noort, P. P. Tak and S. W. Tas, Non-canonical NF-kappaB signaling in rheumatoid arthritis: Dr Jekyll and Mr Hyde?, Arthritis Res. Ther., 2015, 17, 15 Search PubMed.
- J. A. Kellum, L. Kong, M. P. Fink, L. A. Weissfeld, D. M. Yealy, M. R. Pinsky, J. Fine, A. Krichevsky, R. L. Delude, D. C. Angus and I. M. S. I. Gen, Understanding the inflammatory cytokine response in pneumonia and sepsis: results of the Genetic and Inflammatory Markers of Sepsis (GenIMS) Study, Arch. Intern. Med., 2007, 167, 1655–1663 CrossRef CAS PubMed.
- J. A. Hagar, D. A. Powell, Y. Aachoui, R. K. Ernst and E. A. Miao, Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock, Science, 2013, 341, 1250–1253 CrossRef CAS PubMed.
- N. Kayagaki, M. T. Wong, I. B. Stowe, S. R. Ramani, L. C. Gonzalez, S. Akashi-Takamura, K. Miyake, J. Zhang, W. P. Lee, A. Muszynski, L. S. Forsberg, R. W. Carlson and V. M. Dixit, Noncanonical Inflammasome Activation by Intracellular LPS Independent of TLR4, Science, 2013, 341, 1246–1249 CrossRef CAS PubMed.
- J. Shi, Y. Zhao, Y. Wang, W. Gao, J. Ding, P. Li, L. Hu and F. Shao, Inflammatory caspases are innate immune receptors for intracellular LPS, Nature, 2014, 514, 187–192 CrossRef CAS PubMed.
- A. Denes, G. Coutts, N. Lénárt, S. M. Cruickshank, P. Pelegrin, J. Skinner, N. Rothwell, S. M. Allan and D. Brough, AIM2 and NLRC4 inflammasomes contribute with ASC to acute brain injury independently of NLRP3, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4050–4055 CrossRef CAS PubMed.
- E. Latz, T. S. Xiao and A. Stutz, Activation and regulation of the inflammasomes, Nat. Rev. Immunol., 2013, 13, 397–411 CrossRef CAS PubMed.
- M. Chamaillard, M. Hashimoto, Y. Horie, J. Masumoto, S. Qiu, L. Saab, Y. Ogura, A. Kawasaki, K. Fukase, S. Kusumoto, M. A. Valvano, S. J. Foster, T. W. Mak, G. Nuñez and N. Inohara, An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid, Nat. Immunol., 2003, 4, 702–707 CrossRef CAS PubMed.
- M. Chamaillard, N. Inohara and G. Nuñez, Battling enteroinvasive bacteria: Nod1 comes to the rescue, Trends Microbiol., 2004, 12, 529–532 CrossRef CAS PubMed.
- S. E. Girardin, I. G. Boneca, J. Viala, M. Chamaillard, A. Labigne, G. Thomas, D. J. Philpott and P. J. Sansonetti, Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection, J. Biol. Chem., 2003, 278, 8869–8872 CrossRef CAS PubMed.
- S. E. Girardin, I. G. Boneca, L. A. Carneiro, A. Antignac, M. Jéhanno, J. Viala, K. Tedin, M. K. Taha, A. Labigne, U. Zähringer, A. J. Coyle, P. S. DiStefano, J. Bertin, P. J. Sansonetti and D. J. Philpott, Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan, Science, 2003, 300, 1584–1587 CrossRef CAS PubMed.
- S. Traub, N. Kubasch, S. Morath, M. Kresse, T. Hartung, R. R. Schmidt and C. Hermann, Structural requirements of synthetic muropeptides to synergize with lipopolysaccharide in cytokine induction, J. Biol. Chem., 2004, 279, 8694–8700 CrossRef CAS PubMed.
- I. G. Boneca, The role of peptidoglycan in pathogenesis, Curr. Opin. Microbiol., 2005, 8, 46–53 CrossRef CAS PubMed.
- I. Tattoli, L. H. Travassos, L. A. Carneiro, J. G. Magalhaes and S. E. Girardin, The Nodosome: Nod1 and Nod2 control bacterial infections and inflammation, Semin. Immunopathol., 2007, 29, 289–301 CrossRef CAS PubMed.
- L. Le Bourhis, S. Benko and S. E. Girardin, Nod1 and Nod2 in innate immunity and human inflammatory disorders, Biochem. Soc. Trans., 2007, 35, 1479–1484 CrossRef CAS PubMed.
- M. A. Boudreau, J. F. Fisher and S. Mobashery, Messenger functions of the bacterial cell wall-derived muropeptides, Biochemistry, 2012, 51, 2974–2990 CrossRef CAS PubMed.
- W. Deng and J. Xie, NOD2 signaling and role in pathogenic mycobacterium recognition, infection and immunity, Cell. Physiol. Biochem., 2012, 30, 953–963 CrossRef CAS PubMed.
- J. W. Johnson, J. F. Fisher and S. Mobashery, Bacterial cell-wall recycling, Ann. N. Y. Acad. Sci., 2013, 1277, 54–75 CrossRef CAS PubMed.
- J. Dworkin, The medium is the message: interspecies and interkingdom signaling by peptidoglycan and related bacterial glycans, Annu. Rev. Microbiol., 2014, 68, 137–154 CrossRef CAS PubMed.
- M. S. Siegrist, B. M. Swarts, D. M. Fox, S. A. Lim and C. R. Bertozzi, Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface, FEMS Microbiol. Rev., 2015, 39, 184–202 CrossRef PubMed.
- W. Strober, P. J. Murray, A. Kitani and T. Watanabe, Signalling pathways and molecular interactions of NOD1 and NOD2, Nat. Rev. Immunol., 2006, 6, 9–20 CrossRef CAS PubMed.
- K. Kobayashi, N. Inohara, L. D. Hernandez, J. E. Galan, G. Nunez, C. A. Janeway, R. Medzhitov and R. A. Flavell, RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems, Nature, 2002, 416, 194–199 CrossRef CAS PubMed.
- J. G. Magalhaes, J. Lee, K. Geddes, S. Rubino, D. J. Philpott and S. E. Girardin, Essential role of Rip2 in the modulation of innate and adaptive immunity triggered by Nod1 and Nod2 ligands, Eur. J. Immunol., 2011, 41, 1445–1455 CrossRef CAS PubMed.
- G. V. Mukamolova, A. G. Murzin, E. G. Salina, G. R. Demina, D. B. Kell, A. S. Kaprelyants and M. Young, Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation, Mol. Microbiol., 2006, 59, 84–98 CrossRef CAS PubMed.
- N. H. Keep, J. M. Ward, M. Cohen-Gonsaud and B. Henderson, Wake up! Peptidoglycan lysis and bacterial non-growth states, Trends Microbiol., 2006, 14, 271–276 CrossRef CAS PubMed.
- D. E. Smith, B. Clémençon and M. A. Hediger, Proton-coupled oligopeptide transporter family SLC15: physiological, pharmacological and pathological implications, Mol. Aspects Med., 2013, 34, 323–336 CrossRef CAS PubMed.
- R. S. Hotchkiss and I. E. Karl, The pathophysiology and treatment of sepsis, N. Engl. J. Med., 2003, 348, 138–150 CrossRef CAS PubMed.
- L. Hoareau, K. Bencharif, P. Rondeau, R. Murumalla, P. Ravanan, F. Tallet, P. Delarue, M. Cesari, R. Roche and F. Festy, Signaling pathways involved in LPS induced TNFalpha production in human adipocytes, J. Inflammation, 2010, 7, 1 CrossRef PubMed.
- Y. Kobayashi, A. Iwata, K. Suzuki, A. Suto, S. Kawashima, Y. Saito, T. Owada, M. Kobayashi, N. Watanabe and H. Nakajima, B and T lymphocyte attenuator inhibits LPS-induced endotoxic shock by suppressing Toll-like receptor 4 signaling in innate immune cells, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5121–5126 CrossRef CAS PubMed.
- L. Koch, D. Frommhold, K. Buschmann, N. Kuss, J. Poeschl and P. Ruef, LPS- and LTA-induced expression of IL-6 and TNF-alpha in neonatal and adult blood: role of MAPKs and NF-kappaB, Mediators Inflammation, 2014, 2014, 283126 Search PubMed.
- T. Wada and J. M. Penninger, Mitogen-activated protein kinases in apoptosis regulation, Oncogene, 2004, 23, 2838–2849 CrossRef CAS PubMed.
- N. D. Perkins, NF-kappaB: tumor promoter or suppressor?, Trends Cell Biol., 2004, 14, 64–69 CrossRef CAS PubMed.
- N. D. Perkins, Integrating cell-signalling pathways with NF-kappaB and IKK function, Nat. Rev. Mol. Cell Biol., 2007, 8, 49–62 CrossRef CAS PubMed.
- Y. Ben-Neriah and M. Karin, Inflammation meets cancer, with NF-kappaB as the matchmaker, Nat. Immunol., 2011, 12, 715–723 CrossRef CAS PubMed.
- M. Guma, D. Stepniak, H. Shaked, M. E. Spehlmann, S. Shenouda, H. Cheroutre, I. Vicente-Suarez, L. Eckmann, M. F. Kagnoff and M. Karin, Constitutive intestinal NF-kappaB does not trigger destructive inflammation unless accompanied by MAPK activation, J. Exp. Med., 2011, 208, 1889–1900 CrossRef CAS PubMed.
- J. J. Haddad and N. E. Abdel-Karim, NF-kappaB cellular and molecular regulatory mechanisms and pathways: therapeutic pattern or pseudoregulation?, Cell. Immunol., 2011, 271, 5–14 CrossRef CAS PubMed.
- Y. Wang, P. Paszek, C. A. Horton, D. B. Kell, M. R. H. White, D. S. Broomhead and M. R. Muldoon, Interactions among oscillatory pathways in NF- kappa B signalling, BMC Syst. Biol., 2011, 5, 23 CrossRef CAS PubMed.
- J. A. DiDonato, F. Mercurio and M. Karin, NF-kappaB and the link between inflammation and cancer, Immunol. Rev., 2012, 246, 379–400 CrossRef PubMed.
- N. D. Perkins, The diverse and complex roles of NF-kappaB subunits in cancer, Nat. Rev. Cancer, 2012, 12, 121–132 CAS.
- Y. Wang, P. Paszek, C. A. Horton, H. Yue, M. R. H. White, D. B. Kell, M. R. Muldoon and D. S. Broomhead, A systematic survey of the response of a model NF-kappaB signalling pathway to TNFalpha stimulation, J. Theor. Biol., 2012, 297, 137–147 CrossRef CAS PubMed.
- F. Pinheiro da Silva and V. Nizet, Cell death during sepsis: integration of disintegration in the inflammatory response to overwhelming infection, Apoptosis, 2009, 14, 509–521 CrossRef PubMed.
- D. Moquin and F. K. Chan, The molecular regulation of programmed necrotic cell injury, Trends Biochem. Sci., 2010, 35, 434–441 CrossRef CAS PubMed.
- F. K. Chan, Fueling the flames: mammalian programmed necrosis in inflammatory diseases, Cold Spring Harbor Perspect. Biol., 2012, 4, a008805 Search PubMed.
- S. Ardestani, D. L. Deskins and P. P. Young, Membrane TNF-alpha-activated programmed necrosis is mediated by Ceramide-induced reactive oxygen species, J. Mol. Signaling, 2013, 8, 12 CrossRef PubMed.
- G. W. Dorn, 2nd, Molecular mechanisms that differentiate apoptosis from programmed necrosis, Toxicol. Pathol., 2013, 41, 227–234 CrossRef PubMed.
- M. Dagenais, T. Douglas and M. Saleh, Role of programmed necrosis and cell death in intestinal inflammation, Curr. Opin. Gastroenterol., 2014, 30, 566–575 CrossRef CAS PubMed.
- A. Degterev, W. Zhou, J. L. Maki and J. Yuan, Assays for necroptosis and activity of RIP kinases, Methods Enzymol., 2014, 545, 1–33 CAS.
- H. Sridharan and J. W. Upton, Programmed necrosis in microbial pathogenesis, Trends Microbiol., 2014, 22, 199–207 CrossRef CAS PubMed.
- L. Sun and X. Wang, A new kind of cell suicide: mechanisms and functions of programmed necrosis, Trends Biochem. Sci., 2014, 39, 587–593 CrossRef CAS PubMed.
- F. K. Chan, N. F. Luz and K. Moriwaki, Programmed necrosis in the cross talk of cell death and inflammation, Annu. Rev. Immunol., 2015, 33, 79–106 CrossRef CAS PubMed.
- M. Feoktistova and M. Leverkus, Programmed necrosis and necroptosis signalling, FEBS J., 2015, 282, 19–31 CrossRef CAS PubMed.
- S. Jouan-Lanhouet, F. Riquet, L. Duprez, T. Vanden Berghe, N. Takahashi and P. Vandenabeele, Necroptosis, in vivo detection in experimental disease models, Semin. Cell Dev. Biol., 2014, 35, 2–13 CrossRef CAS PubMed.
- M. Pasparakis and P. Vandenabeele, Necroptosis and its role in inflammation, Nature, 2015, 517, 311–320 CrossRef CAS PubMed.
- S. L. Fink and B. T. Cookson, Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells, Infect. Immun., 2005, 73, 1907–1916 CrossRef CAS PubMed.
- S. L. Fink and B. T. Cookson, Pyroptosis and host cell death responses during Salmonella infection, Cell. Microbiol., 2007, 9, 2562–2570 CrossRef CAS PubMed.
- H. Ashida, H. Mimuro, M. Ogawa, T. Kobayashi, T. Sanada, M. Kim and C. Sasakawa, Cell death and infection: a double-edged sword for host and pathogen survival, J. Cell Biol., 2011, 195, 931–942 CrossRef CAS PubMed.
- K. Labbé and M. Saleh, Pyroptosis: A Caspase-1-Dependent Programmed Cell Death and a Barrier to Infection, Prog. Inflammation Res., 2011, 17–36 Search PubMed.
- T. Bergsbaken, S. L. Fink and B. T. Cookson, Pyroptosis: host cell death and inflammation, Nat. Rev. Microbiol., 2009, 7, 99–109 CrossRef CAS PubMed.
- K. R. Bortoluci and R. Medzhitov, Control of infection by pyroptosis and autophagy: role of TLR and NLR, Cell. Mol. Life Sci., 2010, 67, 1643–1651 CrossRef CAS PubMed.
- O. Kepp, L. Galluzzi, L. Zitvogel and G. Kroemer, Pyroptosis – a cell death modality of its kind?, Eur. J. Immunol., 2010, 40, 627–630 CrossRef CAS PubMed.
- P. Vandenabeele, L. Galluzzi, T. Vanden Berghe and G. Kroemer, Molecular mechanisms of necroptosis: an ordered cellular explosion, Nat. Rev. Mol. Cell Biol., 2010, 11, 700–714 CrossRef CAS PubMed.
- C. R. Lupfer and T. D. Kanneganti, The role of inflammasome modulation in virulence, Virulence, 2012, 3, 262–270 CrossRef PubMed.
- Y. Aachoui, V. Sagulenko, E. A. Miao and K. J. Stacey, Inflammasome-mediated pyroptotic and apoptotic cell death, and defense against infection, Curr. Opin. Microbiol., 2013, 16, 319–326 CrossRef CAS PubMed.
- L. D. Cunha and D. S. Zamboni, Subversion of inflammasome activation and pyroptosis by pathogenic bacteria, Front. Cell. Infect. Microbiol., 2013, 3, 76 Search PubMed.
- G. Doitsh, N. L. Galloway, X. Geng, Z. Yang, K. M. Monroe, O. Zepeda, P. W. Hunt, H. Hatano, S. Sowinski, I. Muñoz-Arias and W. C. Greene, Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection, Nature, 2013, 505, 509–514 CrossRef PubMed.
- C. N. LaRock and B. T. Cookson, Burning down the house: cellular actions during pyroptosis, PLoS Pathog., 2013, 9, e1003793 Search PubMed.
- S. J. Dixon, K. M. Lemberg, M. R. Lamprecht, R. Skouta, E. M. Zaitsev, C. E. Gleason, D. N. Patel, A. J. Bauer, A. M. Cantley, W. S. Yang, B. Morrison 3rd and B. R. Stockwell, Ferroptosis: an iron-dependent form of nonapoptotic cell death, Cell, 2012, 149, 1060–1072 CrossRef CAS PubMed.
- P. Guan, Z. H. Shi, Y. Q. Li and N. Wang, Ferroptosis: a new mechanism of cell death, Prog. Biochem. Biophys., 2013, 40, 137–140 Search PubMed.
- S. Toyokuni, Role of iron in carcinogenesis: cancer as a ferrotoxic disease, Cancer Sci., 2009, 100, 9–16 CrossRef CAS PubMed.
- L. R. Zacharski, Ferrotoxic disease: the next great public health challenge, Clin. Chem., 2014, 60, 1362–1364 CAS.
- E. Pretorius, N. Vermeulen, J. Bester, B. Lipinski and D. B. Kell, A novel method for assessing the role of iron and its functional chelation in fibrin fibril formation: the use of scanning electron microscopy, Toxicol. Mech. Methods, 2013, 23, 352–359 CrossRef CAS PubMed.
- E. Pretorius, J. Bester, N. Vermeulen, B. Lipinski, G. S. Gericke and D. B. Kell, Profound morphological changes in the erythrocytes and fibrin networks of patients with hemochromatosis or with hyperferritinemia, and their normalization by iron chelators and other agents, PLoS One, 2014, 9, e85271 Search PubMed.
- E. Pretorius, A. C. Swanepoel, A. V. Buys, N. Vermeulen, W. Duim and D. B. Kell, Eryptosis as a marker of Parkinson's disease, Aging, 2014, 6, 788–819 CAS.
- E. Pretorius, J. Bester, N. Vermeulen, S. Alummoottil, P. Soma, A. V. Buys and D. B. Kell, Poorly controlled type 2 diabetes is accompanied by significant morphological and ultrastructural changes in both erythrocytes and in thrombin-generated fibrin: implications for diagnostics, Cardiovasc. Diabetol., 2015, 13, 30 CrossRef PubMed.
- L. Koch, S. Hofer, M. A. Weigand, D. Frommhold, J. Poeschl and P. Ruef, Inhibition of LPS-Induced Activation of Coagulation by p38 MAPK Inhibitor, ISRN Hematol., 2012, 2012, 762614 CrossRef PubMed.
- L. C. Wu, X. Lin and H. Sun, Tanshinone IIA protects rabbits against LPS-induced disseminated intravascular coagulation (DIC), Acta Pharmacol. Sin., 2012, 33, 1254–1259 CrossRef CAS PubMed.
- P. X. Yu, Q. J. Zhou, W. W. Zhu, Y. H. Wu, L. C. Wu, X. Lin, M. H. Chen and B. T. Qiu, Effects of quercetin on LPS-induced disseminated intravascular coagulation (DIC) in rabbits, Thromb. Res., 2013, 131, e270–e273 CrossRef CAS PubMed.
- Z. Wu, J. N. Li, Z. Q. Bai and X. Lin, Antagonism by salvianolic acid B of lipopolysaccharide-induced disseminated intravascular coagulation in rabbits, Clin. Exp. Pharmacol. Physiol., 2014, 41, 502–508 CrossRef CAS PubMed.
- J. Simmons and J. F. Pittet, The coagulopathy of acute sepsis, Curr. Opin. Anaesthesiol., 2015, 28, 227–236 CrossRef CAS PubMed.
- S. Bellary, W. W. Arden, R. W. Schwartz and K. W. Anderson, Effect of lipopolysaccharide, leukocytes, and monoclonal anti-lipid A antibodies on erythrocyte membrane elastance, Shock, 1995, 3, 132–136 CrossRef CAS.
- J. M. B. Pöschl, C. Leray, P. Ruef, J. P. Cazenave and O. Linderkamp, Endotoxin binding to erythrocyte membrane and erythrocyte deformability in human sepsis and in vitro, Crit. Care Med., 2003, 31, 924–928 CrossRef PubMed.
- J. Mestres, E. Gregori-Puigjané, S. Valverde and R. V. Solé, The topology of drug-target interaction networks: implicit dependence on drug properties and target families, Mol. BioSyst., 2009, 5, 1051–1057 RSC.
- J. U. Peters, P. Schnider, P. Mattei and M. Kansy, Pharmacological promiscuity: dependence on compound properties and target specificity in a set of recent Roche compounds, ChemMedChem, 2009, 4, 680–686 CrossRef CAS PubMed.
- D. A. Price, J. Blagg, L. Jones, N. Greene and T. Wager, Physicochemical drug properties associated with in vivo toxicological outcomes: a review, Expert Opin. Drug Metab. Toxicol., 2009, 5, 921–931 CrossRef CAS PubMed.
- P. D. Leeson and J. R. Empfield, Reducing the risk of drug attrition associated with physicochemical properties, Annu. Rep. Med. Chem., 2010, 45, 393–407 CAS.
- F. Saleem, T. C. Bjorndahl, C. L. Ladner, R. Perez-Pineiro, B. N. Ametaj and D. S. Wishart, Lipopolysaccharide induced conversion of recombinant prion protein, Prion, 2014, 8, 221–232 CrossRef CAS.
- A. V. Gyulkhandanyan, A. Mutlu, J. Freedman and V. Leytin, Mitochondrial permeability transition pore (MPTP)-dependent and -independent pathways of mitochondrial membrane depolarization, cell shrinkage and microparticle formation during platelet apoptosis, Br. J. Haematol., 2015, 169, 142–145 CrossRef CAS PubMed.
- M. J. Mooberry and N. S. Key, Microparticle analysis in disorders of hemostasis and thrombosis, Cytometry, Part A, 2015 DOI:10.1002/cyto.a.22647.
- R. M. Thushara, M. Hemshekhar, Basappa, K. Kemparaju, K. S. Rangappa and K. S. Girish, Biologicals, platelet apoptosis and human diseases: an outlook, Crit. Rev. Oncol. Hematol., 2015, 93, 149–158 CrossRef CAS PubMed.
- C. Yang, W. Xiong, Q. Qiu, H. Tahiri, C. Gagnon, G. Liu and P. Hardy, Generation of lymphocytic microparticles and detection of their proapoptotic effect on airway epithelial cells, J. Visualized Exp., 2015, e52651 Search PubMed.
- E. Niccolai, G. Emmi, D. Squatrito, E. Silvestri, L. Emmi, A. Amedei and D. Prisco, Microparticles: Bridging the Gap between Autoimmunity and Thrombosis, Semin. Thromb. Hemostasis, 2015, 41, 413–422 CrossRef CAS PubMed.
- W. Kolowos, U. S. Gaipl, A. Sheriff, R. E. Voll, P. Heyder, P. Kern, J. R. Kalden and M. Herrmann, Microparticles shed from different antigen-presenting cells display an individual pattern of surface molecules and a distinct potential of allogeneic T-cell activation, Scand. J. Immunol., 2005, 61, 226–233 CrossRef CAS PubMed.
- R. Nieuwland and A. Sturk, Why do cells release vesicles?, Thromb. Res., 2010, 125(suppl 1), S49–51 CrossRef CAS PubMed.
- C. Beyer and D. S. Pisetsky, The role of microparticles in the pathogenesis of rheumatic diseases, Nat. Rev. Rheumatol., 2010, 6, 21–29 CrossRef CAS PubMed.
- S. M. Camus, J. A. De Moraes, P. Bonnin, P. Abbyad, S. Le Jeune, F. Lionnet, L. Loufrani, L. Grimaud, J. C. Lambry, D. Charue, L. Kiger, J. M. Renard, C. Larroque, H. Le Clésiau, A. Tedgui, P. Bruneval, C. Barja-Fidalgo, A. Alexandrou, P. L. Tharaux, C. M. Boulanger and O. P. Blanc-Brude, Circulating cell membrane microparticles transfer heme to endothelial cells and trigger vaso-occlusions in sickle cell disease, Blood, 2015, 125, 3805–3814 CrossRef CAS PubMed.
- F. Ciesielski, B. Davis, M. Rittig, B. B. Bonev and P. O'Shea, Receptor-independent interaction of bacterial lipopolysaccharide with lipid and lymphocyte membranes; the role of cholesterol, PLoS One, 2012, 7, e38677 CAS.
- M. Lamkanfi and V. M. Dixit, Inflammasomes and their roles in health and disease, Annu. Rev. Cell Dev. Biol., 2012, 28, 137–161 CrossRef CAS PubMed.
- M. Lamkanfi and V. M. Dixit, Mechanisms and functions of inflammasomes, Cell, 2014, 157, 1013–1022 CrossRef CAS PubMed.
- P. H. V. Saavedra, D. Demon, H. Van Gorp and M. Lamkanfi, Protective and detrimental roles of inflammasomes in disease, Semin. Immunopathol., 2015, 37, 313–322 CrossRef CAS PubMed.
- Z. Badran, X. Struillou, C. Verner, T. Clee, M. Rakic, M. C. Martinez and A. Soueidan, Periodontitis as a risk factor for systemic disease: are microparticles the missing link?, Med. Hypotheses, 2015, 84, 555–556 CrossRef PubMed.
- A. Berezin, A. Zulli, S. Kerrigan, D. Petrovic and P. Kruzliak, Predictive role of circulating endothelial-derived microparticles in cardiovascular diseases, Clin. Biochem., 2015, 42, 562–568 CrossRef PubMed.
- J. Rodríguez-Carrio, M. Alperi-López, P. Lopez, S. Alonso-Castro, S. R. Carro-Esteban, F. J. Ballina-Garcíá and A. Suarez, Altered profile of circulating microparticles in rheumatoid arthritis patients, Clin. Sci., 2015, 128, 437–448 CrossRef PubMed.
- M. A. E. K. Salem, A. A. M. Adly, E. A. R. Ismail, Y. W. Darwish and H. A. Kamel, Platelets microparticles as a link between micro- and macro-angiopathy in young patients with type 1 diabetes, Platelets, 2015, 1–7 CAS.
- J. M. Alam and M. Yamazaki, Spontaneous insertion of lipopolysaccharide into lipid membranes from aqueous solution, Chem. Phys. Lipids, 2011, 164, 166–174 CrossRef CAS PubMed.
- F. Ciesielski, D. C. Griffin, M. Rittig, I. Moriyon and B. B. Bonev, Interactions of lipopolysaccharide with lipid membranes, raft models – a solid state NMR study, Biochim. Biophys. Acta, 2013, 1828, 1731–1742 CrossRef CAS PubMed.
- K. Nomura, M. Maeda, K. Sugase and S. Kusumoto, Lipopolysaccharide induces raft domain expansion in membrane composed of a phospholipid-cholesterol-sphingomyelin ternary system, Innate Immun., 2011, 17, 256–268 CrossRef CAS PubMed.
- R. J. Berckmans, R. Nieuwland, P. P. Tak, A. N. Böing, F. P. H. T. M. Romijn, M. C. Kraan, F. C. Breedveld, C. E. Hack and A. Sturk, Cell-derived microparticles in synovial fluid from inflamed arthritic joints support coag-ulation exclusively via a factor VII-dependent mechanism, Arthritis Rheum., 2002, 46, 2857–2866 CrossRef CAS PubMed.
- E. F. Midura, P. L. Jernigan, J. W. Kuethe, L. A. Friend, R. Veile, A. T. Makley, C. C. Caldwell and M. D. Goodman, Microparticles impact coagulation after traumatic brain injury, J. Surg. Res., 2015, 197, 25–31 CrossRef PubMed.
- K. Labbé and M. Saleh, Cell death in the host response to infection, Cell Death Differ., 2008, 15, 1339–1349 CrossRef PubMed.
- G. Yeretssian, K. Labbé and M. Saleh, Molecular regulation of inflammation and cell death, Cytokine, 2008, 43, 380–390 CrossRef CAS PubMed.
- S. D. Kobayashi, K. M. Rigby and F. R. DeLeo, Bacteria-induced Host Cell Death, Bact. Pathog., 2012, 317–362 CAS.
- E. Clark, C. Hoare, J. Tanianis-Hughes, G. L. Carlson and G. Warhurst, Interferon gamma induces translocation of commensal Escherichia coli across gut epithelial cells via a lipid raft-mediated process, Gastroenterology, 2005, 128, 1258–1267 CrossRef CAS PubMed.
- A. T. Blikslager, A. J. Moeser, J. L. Gookin, S. L. Jones and J. Odle, Restoration of barrier function in injured intestinal mucosa, Physiol. Rev., 2007, 87, 545–564 CrossRef CAS PubMed.
- A. Fasano, Zonulin and its regulation of intestinal barrier function: the biological door to inflammation, autoimmunity, and cancer, Physiol. Rev., 2011, 91, 151–175 CrossRef CAS PubMed.
- T. H. Frazier, J. K. DiBaise and C. J. McClain, Gut microbiota, intestinal permeability, obesity-induced inflammation, and liver injury, JPEN, J. Parenter. Enteral Nutr., 2011, 35, 14S–20S CrossRef CAS PubMed.
- L. J. John, M. Fromm and J. D. Schulzke, Epithelial barriers in intestinal inflammation, Antioxid. Redox Signaling, 2011, 15, 1255–1270 CrossRef CAS PubMed.
- Y. Ilan, Leaky gut and the liver: a role for bacterial translocation in nonalcoholic steatohepatitis, World J. Gastroenterol., 2012, 18, 2609–2618 CrossRef PubMed.
- T. F. S. Teixeira, M. C. Collado, C. L. L. F. Ferreira, J. Bressan and C. G. di Peluzio Mdo, Potential mechanisms for the emerging link between obesity and increased intestinal permeability, Nutr. Res., 2012, 32, 637–647 CrossRef CAS PubMed.
- T. F. S. Teixeira, N. C. S. Souza, P. G. Chiarello, S. C. C. Franceschini, J. Bressan, C. L. L. F. Ferreira and C. G. di Peluzio Mdo, Intestinal permeability parameters in obese patients are correlated with metabolic syndrome risk factors, Clin. Nutr., 2012, 31, 735–740 CrossRef CAS PubMed.
- N. Ijssennagger, C. Belzer, G. J. Hooiveld, J. Dekker, S. W. van Mil, M. Muller, M. Kleerebezem and R. van der Meer, Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 10038–10043 CrossRef CAS PubMed.
- X. Li and M. A. Atkinson, The role for gut permeability in the pathogenesis of type 1 diabetes – a solid or leaky concept?, Pediatr. Diabetes, 2015 DOI:10.1111/pedi.12305.
- P. Yu and C. M. Martin, Increased gut permeability and bacterial translocation in Pseudomonas pneumonia-induced sepsis, Crit. Care Med., 2000, 28, 2573–2577 CrossRef CAS.
- J. M. Brenchley, D. A. Price and D. C. Douek, HIV disease: fallout from a mucosal catastrophe?, Nat. Immunol., 2006, 7, 235–239 CrossRef CAS PubMed.
- P. Vassallo and R. G. Trohman, Prescribing amiodarone: an evidence-based review of clinical indications, JAMA, J. Am. Med. Assoc., 2007, 298, 1312–1322 CrossRef CAS PubMed.
- W. Jiang, M. M. Lederman, P. Hunt, S. F. Sieg, K. Haley, B. Rodriguez, A. Landay, J. Martin, E. Sinclair, A. I. Asher, S. G. Deeks, D. C. Douek and J. M. Brenchley, Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection, J. Infect. Dis., 2009, 199, 1177–1185 CrossRef CAS PubMed.
- D. Kevans, W. Turpin, K. Madsen, J. Meddings, K. Shestopaloff, W. Xu, G. Moreno-Hagelsieb, A. Griffiths, M. S. Silverberg, A. Paterson, K. Croitoru and G. E. M. Project, Determinants of intestinal permeability in healthy first-degree relatives of individuals with Crohn's disease, Inflammatory Bowel Dis., 2015, 21, 879–887 CrossRef PubMed.
- X. Han, M. P. Fink, R. Yang and R. L. Delude, Increased iNOS activity is essential for intestinal epithelial tight junction dysfunction in endotoxemic mice, Shock, 2004, 21, 261–270 CrossRef CAS PubMed.
- M. Schietroma, F. Carlei, S. Cappelli and G. Amicucci, Intestinal permeability and systemic endotoxemia after laparotomic or laparoscopic cholecystectomy, Ann. Surg., 2006, 243, 359–363 CrossRef PubMed.
- J. Zhaowei, M. J. D'Souza and C. W. Oettinger, Reversal of LPS induced endothelial cell TNF synthesis and increased permeability with microencapsulated antisense oligomers to NF-kappaB, J. Microencapsulation, 2007, 24, 596–607 CrossRef CAS PubMed.
- C. T. Capaldo and A. Nusrat, Cytokine regulation of tight junctions, Biochim. Biophys. Acta, 2009, 1788, 864–871 CrossRef CAS PubMed.
- M. Schietroma, B. Pessia, F. Carlei, E. M. Cecilia and G. Amicucci, Intestinal permeability, systemic endotoxemia, and bacterial translocation after open or laparoscopic resection for colon cancer: a prospective randomized study, Int. J. Colorectal Dis., 2013, 28, 1651–1660 CrossRef PubMed.
- Z. Ruan, S. Liu, Y. Zhou, S. Mi, G. Liu, X. Wu, K. Yao, H. Assaad, Z. Deng, Y. Hou, G. Wu and Y. Yin, Chlorogenic acid decreases intestinal permeability and increases expression of intestinal tight junction proteins in weaned rats challenged with LPS, PLoS One, 2014, 9, e97815 Search PubMed.
- C. Funke, S. A. Schneider, D. Berg and D. B. Kell, Genetics and iron in the systems biology of Parkinson's disease and some related disorders, Neurochem. Int., 2013, 62, 637–652 CrossRef CAS PubMed.
- N. Singh, The role of iron in prion disease and other neurodegenerative diseases, PLoS Pathog., 2014, 10, e1004335 Search PubMed.
- G. G. Tedeschi, D. Amici and M. Paparelli, Incorporation of nucleosides and amino-acids in human erythrocyte suspensions: possible relation with a diffuse infection of mycoplasms or bacteria in the L form, Nature, 1969, 222, 1285–1286 CrossRef CAS PubMed.
- G. G. Tedeschi, A. Bondi, M. Paparelli and G. Sprovieri, Electron microscopical evidence of the evolution of corynebacteria-like microorganisms within human erythrocytes, Experientia, 1978, 34, 458–460 CrossRef CAS.
- R. W. McLaughlin, H. Vali, P. C. Lau, R. G. E. Palfree, A. De Ciccio, M. Sirois, D. Ahmad, R. Villemur, M. Desrosiers and E. C. S. Chan, Are there naturally occurring pleomorphic bacteria in the blood of healthy humans?, J. Clin. Microbiol., 2002, 40, 4771–4775 CrossRef.
- O. H. Munz, S. Sela, B. S. Baker, C. E. M. Griffiths, A. V. Powles and L. Fry, Evidence for the presence of bacteria in the blood of psoriasis patients, Arch. Dermatol. Res., 2010, 302, 495–498 CrossRef PubMed.
- P. A. Leppäluoto, Bacterial vaginosis: what is physiological in vaginal bacteriology? An update and opinion, Acta Obstet. Gynecol. Scand., 2011, 90, 1302–1306 CrossRef PubMed.
- R. W. Hyman, M. Fukushima, H. Jiang, E. Fung, L. Rand, B. Johnson, K. C. Vo, A. B. Caughey, J. F. Hilton, R. W. Davis and L. C. Giudice, Diversity of the vaginal microbiome correlates with preterm birth, Reprod. Sci., 2014, 21, 32–40 CrossRef PubMed.
- D. B. DiGiulio, B. J. Callahan, P. J. McMurdie, E. K. Costello, D. J. Lyell, A. Robaczewska, C. L. Sun, D. S. Goltsman, R. J. Wong, G. Shaw, D. K. Stevenson, S. P. Holmes and D. A. Relman, Temporal and spatial variation of the human microbiota during pregnancy, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11060–11065 CrossRef CAS PubMed.
- K. Aagaard, J. Ma, K. M. Antony, R. Ganu, J. Petrosino and J. Versalovic, The placenta harbors a unique microbiome, Sci. Transl. Med., 2014, 6, 237ra265 Search PubMed.
- R. Amarasekara, R. W. Jayasekara, H. Senanayake and V. H. Dissanayake, Microbiome of the placenta in pre-eclampsia supports the role of bacteria in the multifactorial cause of pre-eclampsia, J. Obstet. Gynaecol. Res., 2015, 41, 662–669 CrossRef CAS PubMed.
- B. Cao, M. J. Stout, I. Lee and I. U. Mysorekar, Placental Microbiome and Its Role in Preterm Birth, Neoreviews, 2014, 15, e537–e545 CrossRef PubMed.
- V. Blanc, F. O Valle, E. Pozo, A. Puertas, R. Leon and F. Mesa, Oral bacteria in placental tissues: increased molecular detection in pregnant periodontitis patients, Oral Dis., 2015 DOI:10.1111/odi.12364.
- J. Zheng, X. Xiao, Q. Zhang, L. Mao, M. Yu and J. Xu, The Placental Microbiome Varies in Association with Low Birth Weight in Full-Term Neonates, Nutrients, 2015, 7, 6924–6937 CrossRef PubMed.
- C. Gardella, D. E. Riley, J. Hitti, K. Agnew, J. N. Krieger and D. Eschenbach, Identification and sequencing of bacterial rDNAs in culture-negative amniotic fluid from women in premature labor, Am. J. Perinatol., 2004, 21, 319–323 CrossRef PubMed.
- D. B. DiGiulio, R. Romero, H. P. Amogan, J. P. Kusanovic, E. M. Bik, F. Gotsch, C. J. Kim, O. Erez, S. Edwin and D. A. Relman, Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation, PLoS One, 2008, 3, e3056 Search PubMed.
- D. B. DiGiulio, M. T. Gervasi, R. Romero, E. Vaisbuch, S. Mazaki-Tovi, J. P. Kusanovic, K. S. Seok, R. Gomez, P. Mittal, F. Gotsch, T. Chaiworapongsa, E. Oyarzun, C. J. Kim and D. A. Relman, Microbial invasion of the amniotic cavity in pregnancies with small-for-gestational-age fetuses, J. Perinat. Med., 2010, 38, 495–502 Search PubMed.
- D. B. DiGiulio, M. Gervasi, R. Romero, S. Mazaki-Tovi, E. Vaisbuch, J. P. Kusanovic, K. S. Seok, R. Gomez, P. Mittal, F. Gotsch, T. Chaiworapongsa, E. Oyarzun, C. J. Kim and D. A. Relman, Microbial invasion of the amniotic cavity in preeclampsia as assessed by cultivation and sequence-based methods, J. Perinat. Med., 2010, 38, 503–513 CAS.
- D. B. DiGiulio, R. Romero, J. P. Kusanovic, R. Gomez, C. J. Kim, K. S. Seok, F. Gotsch, S. Mazaki-Tovi, E. Vaisbuch, K. Sanders, E. M. Bik, T. Chaiworapongsa, E. Oyarzun and D. A. Relman, Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes, Am. J. Reprod. Immunol., 2010, 64, 38–57 Search PubMed.
- D. B. DiGiulio, Diversity of microbes in amniotic fluid, Semin. Fetal Neonatal. Med., 2012, 17, 2–11 CrossRef PubMed.
- R. Romero, J. Miranda, T. Chaiworapongsa, P. Chaemsaithong, F. Gotsch, Z. Dong, A. I. Ahmed, B. H. Yoon, S. S. Hassan, C. J. Kim, S. J. Korzeniewski and L. Yeo, A novel molecular microbiologic technique for the rapid diagnosis of microbial invasion of the amniotic cavity and intra-amniotic infection in preterm labor with intact membranes, Am. J. Reprod. Immunol., 2014, 71, 330–358 CrossRef CAS PubMed.
- L. F. Stinson, D. J. Ireland, M. W. Kemp, M. S. Payne, S. J. Stock, J. P. Newnham and J. A. Keelan, Effects of cytokine-suppressive anti-inflammatory drugs on inflammatory activation in ex vivo human and ovine fetal membranes, Reproduction, 2014, 147, 313–320 CrossRef CAS PubMed.
- P. Y. Ng, D. J. Ireland and J. A. Keelan, Drugs to block cytokine signaling for the prevention and treatment of inflammation-induced preterm birth, Front. Immunol., 2015, 6, 166 Search PubMed.
- R. Romero, J. Miranda, J. P. Kusanovic, T. Chaiworapongsa, P. Chaemsaithong, A. Martinez, F. Gotsch, Z. Dong, A. I. Ahmed, M. Shaman, K. Lannaman, B. H. Yoon, S. S. Hassan, C. J. Kim, S. J. Korzeniewski, L. Yeo and Y. M. Kim, Clinical chorioamnionitis at term I: microbiology of the amniotic cavity using cultivation and molecular techniques, J. Perinat. Med., 2015, 43, 19–36 CAS.
- T. Nakatsuji, H. I. Chiang, S. B. Jiang, H. Nagarajan, K. Zengler and R. L. Gallo, The microbiome extends to subepidermal compartments of normal skin, Nat. Commun., 2013, 4, 1431 CrossRef PubMed.
- T. Trotta, C. Porro, R. Calvello and M. A. Panaro, Biological role of Toll-like receptor-4 in the brain, J. Neuroimmunol., 2014, 268, 1–12 CrossRef CAS PubMed.
- A. Ebringer, T. Rashid and C. Wilson, Rheumatoid arthritis, Proteus, anti-CCP antibodies and Karl Popper, Autoimmun. Rev., 2010, 9, 216–223 CrossRef CAS PubMed.
- A. Ebringer, Rheumatoid Arthritis and Proteus, Springer, London, 2012 Search PubMed.
- A. Ebringer and T. Rashid, Rheumatoid arthritis is caused by a Proteus urinary tract infection, APMIS, 2014, 122, 363–368 CrossRef CAS PubMed.
- X. Zhang, D. Zhang, H. Jia, Q. Feng, D. Wang, D. Liang, X. Wu, J. Li, L. Tang, Y. Li, Z. Lan, B. Chen, Y. Li, H. Zhong, H. Xie, Z. Jie, W. Chen, S. Tang, X. Xu, X. Wang, X. Cai, S. Liu, Y. Xia, J. Li, X. Qiao, J. Y. Al-Aama, H. Chen, L. Wang, Q. J. Wu, F. Zhang, W. Zheng, Y. Li, M. Zhang, G. Luo, W. Xue, L. Xiao, J. Li, W. Chen, X. Xu, Y. Yin, H. Yang, J. Wang, K. Kristiansen, L. Liu, T. Li, Q. Huang, Y. Li and J. Wang, The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment, Nat. Med., 2015, 21, 895–905 CrossRef CAS PubMed.
- D. W. Dresser and A. M. Popham, Induction of an IgM anti-(bovine)-IgG response in mice by bacterial lipopolysaccharide, Nature, 1976, 264, 552–554 CrossRef CAS PubMed.
- S. Izui, R. A. Eisenberg and F. J. Dixon, IgM rheumatoid factors in mice injected with bacterial lipopolysaccharides, J. Immunol., 1979, 122, 2096–2102 CAS.
- P. J. Meredith, J. A. Kristie and R. L. Walford, Aging increases expression of LPS-induced autoantibody-secreting B cells, J. Immunol., 1979, 123, 87–91 CAS.
- M. Fujiwara, A. Kariyone and M. Kimura, Tolerance inducibility and the elicitation of autoantibodies by LPS in aged NZB mice, J. Clin. Lab. Immunol., 1980, 3, 185–188 CAS.
- R. Dziarski, Preferential induction of autoantibody secretion in polyclonal activation by peptidoglycan and lipopolysaccharide. I. In vitro studies, J. Immunol., 1982, 128, 1018–1025 CAS.
- R. Dziarski, Preferential induction of autoantibody secretion in polyclonal activation by peptidoglycan and lipopolysaccharide. II. In vivo studies, J. Immunol., 1982, 128, 1026–1030 CAS.
- R. Dziarski, Comparison of in vitro and in vivo mitogenic and polyclonal antibody and autoantibody responses to peptidoglycan, LPS, protein A, PWM, PHA and Con A in normal and autoimmune mice, J. Clin. Lab. Immunol., 1985, 16, 93–109 CAS.
- L. M. Hang, M. T. Aguado, F. J. Dixon and A. N. Theofilopoulos, Induction of severe autoimmune disease in normal mice by simultaneous action of multiple immunostimulators, J. Exp. Med., 1985, 161, 423–428 CrossRef CAS.
- D. N. Posnett and J. Edinger, When do microbes stimulate rheumatoid factor?, J. Exp. Med., 1997, 185, 1721–1723 CrossRef CAS.
- M. Satoh, V. M. Shaheen, M. Shaw, H. B. Richards and W. H. Reeves, Role of bacterial lipopolysaccharide (LPS) in the induction of Lupus autoantibodies by pristane, Arthritis Rheum., 1998, 41, S178–S178 Search PubMed.
- R. D. Yammani, M. A. Leyva, R. N. Jennings and K. M. Haas, C4 Deficiency is a predisposing factor for Streptococcus pneumoniae-induced autoantibody production, J. Immunol., 2014, 193, 5434–5443 CrossRef CAS PubMed.
- V. Petrušić, N. Todorović, I. Živković, R. Dimitrijević, L. Muhandes, I. Rajnpreht and L. Dimitrijević, Autoantibody response and pregnancy-related pathology induced by combined LPS and tetanus toxoid hyperimmunization in BALB/c and C57BL/6 mice, Autoimmunity, 2015, 48, 87–99 CrossRef PubMed.
- S. M. Yentis, N. Soni and P. G. Riches, In vitro effects of HA-1A (Centoxin) on cytokine production in whole blood from intensive care unit patients, Br. J. Anaesth., 1994, 73, 805–811 CrossRef CAS PubMed.
- L. Marks, The birth pangs of monoclonal antibody therapeutics: the failure and legacy of Centoxin, MAbs, 2012, 4, 403–412 CrossRef PubMed.
- P. J. Albert, A. D. Proal and T. G. Marshall, Vitamin D: the alternative hypothesis, Autoimmun. Rev., 2009, 8, 639–644 CrossRef CAS PubMed.
- G. P. Blaney, P. J. Albert and A. D. Proal, Vitamin D metabolites as clinical markers in autoimmune and chronic disease, Ann. N. Y. Acad. Sci., 2009, 1173, 384–390 CrossRef CAS PubMed.
- A. D. Proal, P. J. Albert and T. Marshall, Autoimmune disease in the era of the metagenome, Autoimmun. Rev., 2009, 8, 677–681 CrossRef CAS PubMed.
- A. D. Proal, P. J. Albert, G. P. Blaney, I. A. Lindseth, C. Benediktsson and T. G. Marshall, Immunostimulation in the era of the metagenome, Cell. Mol. Immunol., 2011, 8, 213–225 CrossRef CAS PubMed.
- A. D. Proal, P. J. Albert, T. G. Marshall, G. P. Blaney and I. A. Lindseth, Immunostimulation in the treatment for chronic fatigue syndrome/myalgic encephalomyelitis, Immunol. Res., 2013, 56, 398–412 CrossRef CAS PubMed.
- M. Maes, I. Mihaylova and J. C. Leunis, Increased serum IgA and IgM against LPS of enterobacteria in chronic fatigue syndrome (CFS): indication for the involvement of Gram-negative enterobacteria in the etiology of CFS and for the presence of an increased gut-intestinal permeability, J. Affective Disord., 2007, 99, 237–240 CrossRef CAS PubMed.
- M. Maes, M. Kubera and J. C. Leunis, The gut-brain barrier in major depression: intestinal mucosal dysfunction with an increased translocation of LPS from gram negative enterobacteria (leaky gut) plays a role in the inflammatory pathophysiology of depression, Neuroendocrinol. Lett., 2008, 29, 117–124 Search PubMed.
- M. Maes, Leaky gut in chronic fatigue syndrome: a review, Activitas Nervosa Superior Rediviva, 2009, 51, 21–28 Search PubMed.
- M. Maes, M. Kubera, J. C. Leunis and M. Berk, Increased IgA and IgM responses against gut commensals in chronic depression: further evidence for increased bacterial translocation or leaky gut, J. Affective Disord., 2012, 141, 55–62 CrossRef CAS PubMed.
- M. Maes, F. N. M. Twisk, M. Kubera, K. Ringel, J. C. Leunis and M. Geffard, Increased IgA responses to the LPS of commensal bacteria is associated with inflammation and activation of cell-mediated immunity in chronic fatigue syndrome, J. Affective Disord., 2012, 136, 909–917 CrossRef CAS PubMed.
- M. Lyte, The role of catecholamines in Gram-negative sepsis, Med. Hypotheses, 1992, 37, 255–258 CrossRef CAS.
- M. Lyte and S. Ernst, Catecholamine-induced growth of Gram-negative bacteria, Life Sci., 1992, 50, 203–212 CrossRef CAS.
- M. Lyte, The role of microbial endocrinology in infectious disease, J. Endocrinol., 1993, 137, 343–345 CrossRef CAS.
- P. P. E. Freestone and M. Lyte, Microbial endocrinology: experimental design issues in the study of interkingdom signalling in infectious disease, Adv. Appl. Microbiol., 2008, 64, 75–105 CAS.
- M. Lyte, Microbial endocrinology and infectious disease in the 21st century, Trends Microbiol., 2004, 12, 14–20 CrossRef CAS PubMed.
- P. P. E. Freestone, S. M. Sandrini, R. D. Haigh and M. Lyte, Microbial endocrinology: how stress influences susceptibility to infection, Trends Microbiol., 2008, 16, 55–64 CrossRef CAS PubMed.
- M. Lyte, Microbial endocrinology: host-microbiota neuroendocrine interactions influencing brain and behavior, Gut Microbes, 2014, 5, 381–389 CrossRef PubMed.
- J. D. Galley, M. C. Nelson, Z. Yu, S. E. Dowd, J. Walter, P. S. Kumar, M. Lyte and M. T. Bailey, Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota, BMC Microbiol., 2014, 14, 189 CrossRef PubMed.
- M. Lyte, The effect of stress on microbial growth, Anim. Health Res. Rev., 2014, 15, 172–174 CrossRef PubMed.
- S. W. Brown, R. T. Meyers, K. M. Brennan, J. M. Rumble, N. Narasimhachari, E. F. Perozzi, J. J. Ryan, J. K. Stewart and K. Fischer-Stenger, Catecholamines in a macrophage cell line, J. Neuroimmunol., 2003, 135, 47–55 CrossRef CAS.
- K. L. Engler, M. L. Rudd, J. J. Ryan, J. K. Stewart and K. Fischer-Stenger, Autocrine actions of macrophage-derived catecholamines on interleukin-1 beta, J. Neuroimmunol., 2005, 160, 87–91 CrossRef CAS PubMed.
- M. A. Flierl, D. Rittirsch, B. A. Nadeau, A. J. Chen, J. V. Sarma, F. S. Zetoune, S. R. McGuire, R. P. List, D. E. Day, L. M. Hoesel, H. Gao, N. Van Rooijen, M. S. Huber-Lang, R. R. Neubig and P. A. Ward, Phagocyte-derived catecholamines enhance acute inflammatory injury, Nature, 2007, 449, 721–725 CrossRef CAS PubMed.
- M. A. Flierl, D. Rittirsch, B. A. Nadeau, J. V. Sarma, D. E. Day, A. B. Lentsch, M. S. Huber-Lang and P. A. Ward, Upregulation of phagocyte-derived catecholamines augments the acute inflammatory response, PLoS One, 2009, 4, e4414 Search PubMed.
- P. P. Freestone, M. Lyte, C. P. Neal, A. F. Maggs, R. D. Haigh and P. H. Williams, The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin, J. Bacteriol., 2000, 182, 6091–6098 CrossRef CAS.
- P. P. E. Freestone, R. D. Haigh, P. H. Williams and M. Lyte, Involvement of enterobactin in norepinephrine-mediated iron supply from transferrin to enterohaemorrhagic Escherichia coli, FEMS Microbiol. Lett., 2003, 222, 39–43 CrossRef CAS.
- S. M. Sandrini, R. Shergill, J. Woodward, R. Muralikuttan, R. D. Haigh, M. Lyte and P. P. Freestone, Elucidation of the mechanism by which catecholamine stress hormones liberate iron from the innate immune defense proteins transferrin and lactoferrin, J. Bacteriol., 2010, 192, 587–594 CrossRef CAS PubMed.
- R. J. Abergel, M. C. Clifton, J. C. Pizarro, J. A. Warner, D. K. Shuh, R. K. Strong and K. N. Raymond, The siderocalin/enterobactin interaction: a link between mammalian immunity and bacterial iron transport, J. Am. Chem. Soc., 2008, 130, 11524–11534 CrossRef CAS PubMed.
- D. W. Reid, G. J. Anderson and I. L. Lamont, Role of lung iron in determining the bacterial and host struggle in cystic fibrosis, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2009, 297, L795–802 CrossRef CAS PubMed.
- M. Nairz, A. Schroll, T. Sonnweber and G. Weiss, The struggle for iron – a metal at the host–pathogen interface, Cell. Microbiol., 2010, 12, 1691–1702 CrossRef CAS PubMed.
- A. K. Sia, B. E. Allred and K. N. Raymond, Siderocalins: Siderophore binding proteins evolved for primary pathogen host defense, Curr. Opin. Chem. Biol., 2013, 17, 150–157 CrossRef CAS PubMed.
- N. Leon-Sicairos, R. Reyes-Cortes, A. M. Guadrón-Llanos, J. Madueña-Molina, C. Leon-Sicairos and A. Canizalez-Román, Strategies of Intracellular Pathogens for Obtaining Iron from the Environment, BioMed Res. Int., 2015, 2015, 476534 Search PubMed.
- F. Meziani, X. Delabranche, P. Asfar and F. Toti, Bench-to-bedside review: circulating microparticles – a new player in sepsis?, Crit. Care, 2010, 14, 236 CrossRef PubMed.
- V. L. Reid and N. R. Webster, Role of microparticles in sepsis, Br. J. Anaesth., 2012, 109, 503–513 CrossRef CAS PubMed.
- D. Annane, E. Bellissant and J. M. Cavaillon, Septic shock, Lancet, 2005, 365, 63–78 CrossRef CAS.
- R. S. Parker and G. Clermont, Systems engineering medicine: engineering the inflammation response to infectious and traumatic challenges, J. R. Soc., Interface, 2010, 7, 989–1013 CrossRef CAS PubMed.
- M. P. Fink and H. S. Warren, Strategies to improve drug development for sepsis, Nat. Rev. Drug Discovery, 2014, 13, 741–758 CrossRef CAS PubMed.
- J. C. Marshall, Why have clinical trials in sepsis failed?, Trends Mol. Med., 2014, 20, 195–203 CrossRef PubMed.
- J. B. German, L. A. Gillies, J. T. Smilowitz, A. M. Zivkovic and S. M. Watkins, Lipidomics and lipid profiling in metabolomics, Curr. Opin. Lipidol., 2007, 18, 66–71 CAS.
- M. J. O. Wakelam, T. R. Pettitt and A. D. Postle, Lipidomic analysis of signaling pathways, Methods Enzymol., 2007, 432, 233–246 CAS.
- M. Orešič, V. A. Hanninen and A. Vidal-Puig, Lipidomics: a new window to biomedical frontiers, Trends Biotechnol., 2008, 26, 647–652 CrossRef PubMed.
- M. Sud, E. Fahy, D. Cotter, E. A. Dennis and S. Subramaniam, LIPID MAPS-Nature Lipidomics Gateway: An Online Resource for Students and Educators Interested in Lipids, J. Chem. Educ., 2012, 89, 291–292 CrossRef CAS PubMed.
- J. M. Foster, P. Moreno, A. Fabregat, H. Hermjakob, C. Steinbeck, R. Apweiler, M. J. O. Wakelam and J. A. Vizcaino, LipidHome: a database of theoretical lipids optimized for high throughput mass spectrometry lipidomics, PLoS One, 2013, 8, e61951 CAS.
- M. S. Köberlin, B. Snijder, L. X. Heinz, C. L. Baumann, A. Fauster, G. I. Vladimer, A. C. Gavin and G. Superti-Furga, A Conserved Circular Network of Coregulated Lipids Modulates Innate Immune Responses, Cell, 2015, 162, 170–183 CrossRef PubMed.
- E. J. Ziegler, C. J. Fisher Jr., C. L. Sprung, R. C. Straube, J. C. Sadoff, G. E. Foulke, C. H. Wortel, M. P. Fink, R. P. Dellinger and N. N. Teng, et al., Treatment of Gram-negative bacteremia and septic shock with HA-1A human monoclonal antibody against endotoxin. A randomized, double-blind, placebo-controlled trial. The HA-1A Sepsis Study Group, N. Engl. J. Med., 1991, 324, 429–436 CrossRef CAS PubMed.
- J. P. A. Ioannidis, Why most published research findings are false, PLoS Med., 2005, 2, e124 CrossRef PubMed.
- D. Broadhurst and D. B. Kell, Statistical strategies for avoiding false discoveries in metabolomics and related experiments, Metabolomics, 2006, 2, 171–196 CrossRef CAS.
- B. Derkx, J. Wittes and R. McCloskey, Randomized, placebo-controlled trial of HA-1A, a human monoclonal antibody to endotoxin, in children with meningococcal septic shock. European Pediatric Meningococcal Septic Shock Trial Study Group, Clin. Infect. Dis., 1999, 28, 770–777 CAS.
- J. D. Baumgartner and M. P. Glauser, Immunotherapy of endotoxemia and septicemia, Immunobiology, 1993, 187, 464–477 CrossRef CAS.
- E. J. Helmerhorst, J. J. Maaskant and B. J. Appelmelk, Anti-lipid A monoclonal antibody centoxin (HA-1A) binds to a wide variety of hydrophobic ligands, Infect. Immun., 1998, 66, 870–873 CAS.
- S. Ekins, Predicting undesirable drug interactions with promiscuous proteins in silico, Drug Discovery Today, 2004, 9, 276–285 CrossRef CAS.
- P. D. Leeson and B. Springthorpe, The influence of drug-like concepts on decision-making in medicinal chemistry, Nat. Rev. Drug Discovery, 2007, 6, 881–890 CrossRef CAS PubMed.
- P. D. Leeson and S. A. St-Gallay, The influence of the ‘organizational factor’ on compound quality in drug discovery, Nat. Rev. Drug Discovery, 2011, 10, 749–765 CrossRef CAS PubMed.
- N. S. Tan, M. L. Ng, Y. H. Yau, P. K. W. Chong, B. Ho and J. L. Ding, Definition of endotoxin binding sites in horseshoe crab factor C recombinant sushi proteins and neutralization of endotoxin by sushi peptides, FASEB J., 2000, 14, 1801–1813 CrossRef CAS.
- N. S. Tan, B. Ho and J. L. Ding, High-affinity LPS binding domain(s) in recombinant factor C of a horseshoe crab neutralizes LPS-induced lethality, FASEB J., 2000, 14, 859–870 CrossRef CAS.
- J. L. Ding, P. Li and B. Ho, The Sushi peptides: structural characterization and mode of action against Gram-negative bacteria, Cell. Mol. Life Sci., 2008, 65, 1202–1219 CrossRef CAS PubMed.
- Y. H. Yau, B. Ho, N. S. Tan, M. L. Ng and J. L. Ding, High therapeutic index of factor C Sushi peptides: potent antimicrobials against Pseudomonas aeruginosa, Antimicrob. Agents Chemother., 2001, 45, 2820–2825 CrossRef CAS PubMed.
- S. Leptihn, J. Y. Har, J. Chen, B. Ho, T. Wohland and J. L. Ding, Single molecule resolution of the antimicrobial action of quantum dot-labeled sushi peptide on live bacteria, BMC Biol., 2009, 7, 22 CrossRef PubMed.
- S. Leptihn, L. Guo, V. Frecer, B. Ho, J. L. Ding and T. Wohland, One step at a time: action mechanism of Sushi 1 antimicrobial peptide and derived molecules, Virulence, 2010, 1, 42–44 CrossRef PubMed.
- P. Li, M. Sun, T. Wohland, B. Ho and J. L. Ding, The molecular mechanism of interaction between sushi peptide and Pseudomonas endotoxin, Cell. Mol. Immunol., 2006, 3, 21–28 CAS.
- P. Li, M. Sun, T. Wohland, D. Yang, B. Ho and J. L. Ding, Molecular mechanisms that govern the specificity of Sushi peptides for Gram-negative bacterial membrane lipids, Biochemistry, 2006, 45, 10554–10562 CrossRef CAS PubMed.
- P. Li, M. Sun, B. Ho and J. L. Ding, The specificity of Sushi peptides for endotoxin and anionic phospholipids: potential application of POPG as an adjuvant for anti-LPS strategies, Biochem. Soc. Trans., 2006, 34, 270–272 CrossRef CAS.
- V. Frecer, B. Ho and J. L. Ding, De novo design of potent antimicrobial peptides, Antimicrob. Agents Chemother., 2004, 48, 3349–3357 CrossRef CAS PubMed.
- S. Bhattacharjya, De novo designed lipopolysaccharide binding peptides: structure based development of antiendotoxic and antimicrobial drugs, Curr. Med. Chem., 2010, 17, 3080–3093 CrossRef CAS.
- A. Skerra, Anticalins as alternative binding proteins for therapeutic use, Curr. Opin. Mol. Ther., 2007, 9, 336–344 CAS.
- M. Gebauer and A. Skerra, Anticalins small engineered binding proteins based on the lipocalin scaffold, Methods Enzymol., 2012, 503, 157–188 CAS.
- E. Eggenstein, A. Eichinger, H. J. Kim and A. Skerra, Structure-guided engineering of Anticalins with improved binding behavior and biochemical characteristics for application in radio-immuno imaging and/or therapy, J. Struct. Biol., 2014, 185, 203–214 CrossRef CAS PubMed.
- A. Richter, E. Eggenstein and A. Skerra, Anticalins: exploiting a non-Ig scaffold with hypervariable loops for the engineering of binding proteins, FEBS Lett., 2014, 588, 213–218 CrossRef CAS PubMed.
- A. Schiefner and A. Skerra, The menagerie of human lipocalins: a natural protein scaffold for molecular recognition of physiological compounds, Acc. Chem. Res., 2015, 48, 976–985 CrossRef CAS PubMed.
- E. Hailman, H. S. Lichenstein, M. M. Wurfel, D. S. Miller, D. A. Johnson, M. Kelley, L. A. Busse, M. M. Zukowski and S. D. Wright, Lipopolysaccharide (LPS)-binding protein accelerates the binding of LPS to CD14, J. Exp. Med., 1994, 179, 269–277 CrossRef CAS.
- S. Lengacher, C. V. Jongeneel, D. Le Roy, J. D. Lee, V. Kravchenko, R. J. Ulevitch, M. P. Glauser and D. Heumann, Reactivity of murine and human recombinant LPS-binding protein (LBP) within LPS and gram negative bacteria, J. Inflammation, 1995, 47, 165–172 CAS.
- P. Li, B. Ho and J. L. Ding, Recombinant factor C competes against LBP to bind lipopolysaccharide and neutralizes the endotoxicity, J. Endotoxin Res., 2007, 13, 150–157 CrossRef CAS PubMed.
- E. D. Weinberg and J. Miklossy, Iron withholding: a defense against disease, J. Alzheimer's Dis., 2008, 13, 451–463 CAS.
- G. Srinivasan, J. D. Aitken, B. Zhang, F. A. Carvalho, B. Chassaing, R. Shashidharamurthy, N. Borregaard, D. P. Jones, A. T. Gewirtz and M. Vijay-Kumar, Lipocalin 2 deficiency dysregulates iron homeostasis and exacerbates endotoxin-induced sepsis, J. Immunol., 2012, 189, 1911–1919 CrossRef CAS PubMed.
- C. Lehmann, N. Sharawi, N. Al-Banna, N. Corbett, J. W. Kuethe and C. C. Caldwell, Novel approaches to the development of anti-sepsis drugs, Expert Opin. Drug Discovery, 2014, 9, 523–531 CrossRef CAS PubMed.
- G. Luo, B. Spellberg, T. Gebremariam, H. Lee, Y. Q. Xiong, S. W. French, A. Bayer and A. S. Ibrahim, Combination therapy with iron chelation and vancomycin in treating murine staphylococcemia, Eur. J. Clin. Microbiol. Infect. Dis., 2014, 33, 845–851 CrossRef CAS PubMed.
- C. Zeng, Q. Chen, K. Zhang, Q. Chen, S. Song and X. Fang, Hepatic Hepcidin Protects against Polymicrobial Sepsis in Mice by Regulating Host Iron Status, Anesthesiology, 2015, 122, 374–386 CrossRef CAS PubMed.
- M. B. Zimmermann, C. Chassard, F. Rohner, E. K. N'Goran, C. Nindjin, A. Dostal, J. Utzinger, H. Ghattas, C. Lacroix and R. F. Hurrell, The effects of iron fortification on the gut microbiota in African children: a randomized controlled trial in Cote d'Ivoire, Am. J. Clin. Nutr., 2010, 92, 1406–1415 CrossRef CAS PubMed.
- T. Jaeggi, G. A. Kortman, D. Moretti, C. Chassard, P. Holding, A. Dostal, J. Boekhorst, H. M. Timmerman, D. W. Swinkels, H. Tjalsma, J. Njenga, A. Mwangi, J. Kvalsvig, C. Lacroix and M. B. Zimmermann, Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants, Gut, 2015, 64, 731–742 CrossRef CAS PubMed.
- N. R. Perron and J. L. Brumaghim, A review of the antioxidant mechanisms of polyphenol compounds related to iron binding, Cell Biochem. Biophys., 2009, 53, 75–100 CrossRef CAS PubMed.
- V. Chobot and F. Hadacek, Iron and its complexation by phenolic cellular metabolites: from oxidative stress to chemical weapons, Plant Signaling Behav., 2010, 5, 4–8 CrossRef CAS.
- V. Chobot, F. Hadacek and L. Kubicova, Effects of selected dietary secondary metabolites on reactive oxygen species production caused by iron(II) autoxidation, Molecules, 2014, 19, 20023–20033 CrossRef PubMed.
- J. R. O'Dell, G. Paulsen, C. E. Haire, K. Blakely, W. Palmer, S. Wees, P. J. Eckhoff, L. W. Klassen, M. Churchill, D. Doud, A. Weaver and G. F. Moore, Treatment of early seropositive rheumatoid arthritis with minocycline: four-year followup of a double-blind, placebo-controlled trial, Arthritis Rheum., 1999, 42, 1691–1695 CrossRef.
- R. M. Bonelli, A. K. Hodl, P. Hofmann and H. P. Kapfhammer, Neuroprotection in Huntington's disease: a 2-year study on minocycline, Int. Clin. Psychopharmacol., 2004, 19, 337–342 CrossRef PubMed.
- T. Miyaoka, R. Yasukawa, H. Yasuda, M. Hayashida, T. Inagaki and J. Horiguchi, Minocycline as adjunctive therapy for schizophrenia: an open-label study, Clin. Neuropharmacol., 2008, 31, 287–292 CrossRef CAS PubMed.
- F. Zhao, Y. Hua, Y. He, R. F. Keep and G. Xi, Minocycline-induced attenuation of iron overload and brain injury after experimental intracerebral hemorrhage, Stroke, 2011, 42, 3587–3593 CrossRef PubMed.
- T. Miyaoka, R. Wake, M. Furuya, K. Liaury, M. Ieda, K. Kawakami, K. Tsuchie, M. Taki, K. Ishihara, T. Araki and J. Horiguchi, Minocycline as adjunctive therapy for patients with unipolar psychotic depression: an open-label study, Prog. Neuropsychopharmacol. Biol. Psychiatry, 2012, 37, 222–226 CrossRef CAS PubMed.
- C. E. Schmitt, S. G. Fabi, T. Kukreja and J. S. Feinberg, Hypopigmented cutaneous sarcoidosis responsive to minocycline, J. Drugs Dermatol., 2012, 11, 385–389 CAS.
- J. K. Soczynska, R. B. Mansur, E. Brietzke, W. Swardfager, S. H. Kennedy, H. O. Woldeyohannes, A. M. Powell, M. S. Manierka and R. S. McIntyre, Novel therapeutic targets in depression: minocycline as a candidate treatment, Behav. Brain Res., 2012, 235, 302–317 CrossRef CAS PubMed.
- S. F. Abcouwer, C. M. Lin, S. Shanmugam, A. Muthusamy, A. J. Barber and D. A. Antonetti, Minocycline prevents retinal inflammation and vascular permeability following ischemia-reperfusion injury, J. Neuroinflammation, 2013, 10, 149 CrossRef PubMed.
- Y. Li, T. Li, H. Qi and F. Yuan, Minocycline protects against hepatic ischemia/reperfusion injury in a rat model, Biomed. Rep., 2015, 3, 19–24 Search PubMed.
- N. Garrido-Mesa, A. Zarzuelo and J. Gálvez, Minocycline: far beyond an antibiotic, Br. J. Pharmacol., 2013, 169, 337–352 CrossRef CAS PubMed.
- N. Garrido-Mesa, A. Zarzuelo and J. Gálvez, What is behind the non-antibiotic properties of minocycline?, Pharmacol. Res., 2013, 67, 18–30 CrossRef CAS PubMed.
- T. G. Marshall and F. E. Marshall, Sarcoidosis succumbs to antibiotics – implications for autoimmune disease, Autoimmun. Rev., 2004, 3, 295–300 CrossRef CAS PubMed.
- L. Xie, L. Xie and P. E. Bourne, Structure-based systems biology for analyzing off-target binding, Curr. Opin. Struct. Biol., 2011, 21, 189–199 CrossRef CAS PubMed.
- J. Ochoa-Repáraz, D. W. Mielcarz, L. E. Ditrio, A. R. Burroughs, D. M. Foureau, S. Haque-Begum and L. H. Kasper, Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis, J. Immunol., 2009, 183, 6041–6050 CrossRef PubMed.
- H. Yokote, S. Miyake, J. L. Croxford, S. Oki, H. Mizusawa and T. Yamamura, NKT cell-dependent amelioration of a mouse model of multiple sclerosis by altering gut flora, Am. J. Pathol., 2008, 173, 1714–1723 CrossRef CAS PubMed.
- J. Ochoa-Repáraz, D. W. Mielcarz, S. Begum-Haque and L. H. Kasper, Gut, bugs, and brain: role of commensal bacteria in the control of central nervous system disease, Ann. Neurol., 2011, 69, 240–247 CrossRef PubMed.
- K. Berer, M. Mues, M. Koutrolos, Z. A. Rasbi, M. Boziki, C. Johner, H. Wekerle and G. Krishnamoorthy, Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination, Nature, 2011, 479, 538–541 CrossRef CAS PubMed.
- K. Berer and G. Krishnamoorthy, Commensal gut flora and brain autoimmunity: a love or hate affair?, Acta Neuropathol., 2012, 123, 639–651 CrossRef CAS PubMed.
- Y. Wang and L. H. Kasper, The role of microbiome in central nervous system disorders, Brain, Behav., Immun., 2014, 38, 1–12 CrossRef CAS PubMed.
- J. Ochoa-Repáraz and L. H. Kasper, Gut microbiome and the risk factors in central nervous system autoimmunity, FEBS Lett., 2014, 588, 4214–4222 CrossRef PubMed.
- D. Astrauskiene and E. Bernotiene, New insights into bacterial persistence in reactive arthritis, Clin. Exp. Rheumatol., 2007, 25, 470–479 CAS.
- M. Ogrendik and N. Karagoz, Treatment of rheumatoid arthritis with roxithromycin: a randomized trial, Postgrad. Med., 2011, 123, 220–227 CrossRef PubMed.
- B. Kwiatkowska and M. Maślińska, Macrolide therapy in chronic inflammatory diseases, Mediators Inflammation, 2012, 2012, 636157 Search PubMed.
- M. Ogrendik, Rheumatoid arthritis is an autoimmune disease caused by periodontal pathogens, Int. J. Gen. Med., 2013, 6, 383–386 CrossRef PubMed.
- V. N. Saxena and J. Dogra, Long-term use of penicillin for the treatment of chronic plaque psoriasis, Eur. J. Dermatol., 2005, 15, 359–362 CAS.
- V. N. Saxena and J. Dogra, Long-term oral azithromycin in chronic plaque psoriasis: a controlled trial, Eur. J. Dermatol., 2010, 20, 329–333 CAS.
- A. A. Alzolibani and K. Zedan, Macrolides in Chronic Inflammatory Skin Disorders, Mediators Inflammation, 2012, 159354 Search PubMed.
- A. Vila-Corcoles, O. Ochoa-Gondar, T. Rodriguez-Blanco, A. Gutierrez-Perez, A. Vila-Rovira, F. Gomez, X. Raga, C. de Diego, E. Satue, E. Salsench and EPIVAC Study Group, Clinical effectiveness of pneumococcal vaccination against acute myocardial infarction and stroke in people over 60 years: the CAPAMIS study, one-year follow-up, BMC Public Health, 2012, 12, 222 CrossRef PubMed.
- A. Vila-Corcoles, O. Ochoa-Gondar, T. Rodriguez-Blanco, C. de Diego-Cabanes, E. Satue-Gracia, A. Vila-Rovira, C. Torrente Fraga and E. R. Group, Evaluating clinical effectiveness of pneumococcal vaccination in preventing stroke: the CAPAMIS Study, 3-year follow-up, J. Stroke Cerebrovasc. Dis., 2014, 23, 1577–1584 CrossRef PubMed.
- J. van der Greef, T. Hankemeier and R. N. McBurney, Metabolomics-based systems biology and personalized medicine: moving towards n = 1 clinical trials?, Pharmacogenomics, 2006, 7, 1087–1094 CrossRef CAS PubMed.
- N. B. Gabler, N. Duan, S. Vohra and R. L. Kravitz, N-of-1 trials in the medical literature: a systematic review, Med. Care, 2011, 49, 761–768 CrossRef PubMed.
- X. Chen and P. Chen, A comparison of four methods for the analysis of N-of-1 trials, PLoS One, 2014, 9, e87752 Search PubMed.
- H. H. Kong, J. Oh, C. Deming, S. Conlan, E. A. Grice, M. A. Beatson, E. Nomicos, E. C. Polley, H. D. Komarow, N. C. S. Program, P. R. Murray, M. L. Turner and J. A. Segre, Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis, Genome Res., 2012, 22, 850–859 CrossRef CAS PubMed.
- H. R. Schumacher Jr., R. R. Evans, K. G. Saag, J. Clower, W. Jennings, S. P. Weinstein, G. D. Yancopoulos, J. Wang and R. Terkeltaub, Rilonacept (interleukin-1 trap) for prevention of gout flares during initiation of uric acid-lowering therapy: results from a phase III randomized, double-blind, placebo-controlled, confirmatory efficacy study, Arthritis Care Res., 2012, 64, 1462–1470 CrossRef PubMed.
- K. L. Molnar-Kimber and C. T. Kimber, Each type of cause that initiates rheumatoid arthritis or RA flares differentially affects the response to therapy, Med. Hypotheses, 2012, 78, 123–129 CrossRef PubMed.
- V. P. Bykerk, E. Lie, S. J. Bartlett, R. Alten, A. Boonen, R. Christensen, D. E. Furst, S. Hewlett, A. L. Leong, A. Lyddiatt, L. March, J. E. May, P. Montie, A. M. Orbai, C. Pohl, M. Scholte Voshaar, T. Woodworth, C. O. Bingham 3rd and E. H. Choy, Establishing a core domain set to measure rheumatoid arthritis flares: report of the OMERACT 11 RA flare Workshop, J. Rheumatol., 2014, 41, 799–809 CrossRef PubMed.
- R. Zhang, N. F. Lahens, H. I. Ballance, M. E. Hughes and J. B. Hogenesch, A circadian gene expression atlas in mammals: Implications for biology and medicine, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 16219–16224 CrossRef CAS PubMed.
- K. de Punder and L. Pruimboom, Stress induces endotoxemia and low-grade inflammation by increasing barrier permeability, Front. Immunol., 2015, 6, 223 Search PubMed.
Footnote |
| † Paper 3 in the series “The dormant blood microbiome in chronic, inflammatory disease”. |
| This journal is © The Royal Society of Chemistry 2015 |