Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Effects of estrone and organic carbon exposure on the transformation of estrone†
David T.
Tan
,
William A.
Arnold
and
Paige J.
Novak
*
Department of Civil, Environmental, Geo-Engineering, University of Minnesota, 500 Pillsbury Drive SE, Minneapolis, Minnesota 55455, USA. E-mail: novak010@umn.edu; Fax: +612 626 7750; Tel: +612 626 9846
First published on 23rd April 2015
Abstract
Exposure of biomass to estrone (E1) and alternate organic substrates was studied to determine whether cometabolism or multiple substrate utilization is an operating mechanism for the transformation of E1 and if feeding intervals affect the selection of E1 degrading bacteria. Biomass generated in membrane bioreactors (MBRs) was capable of degrading E1 regardless of E1 exposure. Nevertheless, pre-exposed biomass had higher E1 transformation rates (P = 0.05) and un-exposed biomass showed a clear lag phase (6 h) prior to E1 tranformation. These results are consistent with and strongly suggest metabolic transformation of E1 via multiple substrate utilization. In the feeding interval study, longer intervals between feeding periods selected for E1 degraders at high organic carbon loads (100 mg COD L−1 d−1; P = 0.018), but had no effect at low organic carbon loads (30 mg COD L−1 d−1; P = 0.32). A lag phase was observed in E1 transformation during famine periods but was absent during feast periods. This result indicates that the presence of other organic carbon substrates speeds the transformation of E1. This research is the first to demonstrate evidence for the role of multiple substrate utilization in the transformation of E1 and suggests operating conditions to improve selection for and activity of E1 degrading bacteria.
Water impact statementEstrone (E1) is a critical micropollutant due to its ubiquity and effects on aquatic life. The effects of microbial exposure to E1 and alternate organic substrates were studied to determine if E1-degradation by a mixed microbial community proceeds via multiple substrate utilization or cometabolism. Additional work was done examining the effects of feeding cycles in conjunction with organic carbon concentrations. Removal patterns show that E1 transformation proceeds metabolically (multiple substrate utilization), while feeding cycle duration effects were dependent on organic carbon concentration. These results suggest potential approaches for improving E1 removal in wastewater treatment systems. |
1. Introduction
The ubiquitous nature1 and adverse ecological effects2–4 of estrogens in surface water are well known. Municipal wastewater treatment plants (WWTPs) are one of the primary sources of estrogens and other micropollutants to the environment.5 Improving and optimizing micropollutant removal in municpal WWTPs is therefore critical. Estrone (E1) is of particular concern because it is a major contributor to estrogenic activity in treated wastewater6,7 and observed E1 removal rates across WWTPs vary widely.8–11 Nevertheless, simple transformations of steroidal estrogens result in a dramatic drop in estrogenicity,12 precluding the need to stimulate complete mineralization. A more fundamental understanding of micropollutant transformation, and in particular E1 transformation, is needed to provide the scientific underpinning for improved WWTP operation.Because micropollutants are, by definition, present at low concentrations that are unlikely to sustain growth, it is thought that microbial transformation of these compounds takes place either fortuitiously, not benefiting the microbe and induced by another substrate (cometabolism),13 or that these micropollutants are metabolized in conjunction with other substrates that cumulatively enable growth (multiple substrate utilization).14 As a micropollutant, E1 is unlikely to sustain microorganisms specialized in E1 degradation15 and so E1 transformation in WWTPs has been attributed to both cometabolism and multiple substrate utilization.16–18 While cometabolic transformation of E1 by the ammonia monooxygenase enzyme has been refuted,19 it is not clear whether the bacteria responsible for E1 transformation in wastewater treatment are capable of utilizing this compound as a carbon and/or energy source, or if transformation is cometabolic. Furthermore, identifying key microbes to study the mechanism for the initial step of E1 transformation in wastewater treatment is difficult because of a wide variety of possible microbial candidates,20–22 strain-level E1 transformation ability,21,23 and possible initial transformation products that do not require carbon uptake.24
Distinguishing between the mechanisms of cometabolism and multiple substrate utilization is not merely a theoretical exercise, because either would have important and different implications for process optimization. Biomass exposure to E1 and/or other organic carbon substrates will result in different E1 removal patterns depending on the transformation mechanism. If cometabolism is the key E1 degradation mechanism in wastewater, prior exposure to estrogens will not affect removal rates, but the presence of cometabolites will be crucial. Alternatively, if multiple substrate utilization is the key mechanism, exposure to E1 will provide a theoretical competitive advantage, as has been suggested by others,25 though it may not be large enough to detect. Additionally, organic carbon exposure would affect E1 removal by multiple substrate utilizers, theorized to be slower growers,26 resulting in beneficial or harmful community selection. In fact, a previous study showed supporting evidence for this with organic carbon having positive effects via biomass growth but eventual selection against E1 degraders after repeated doses of high organic carbon concentrations.27
The purpose of this research was to determine which mechanism, cometabolism or multiple substrate utilization, controls the transformation of E1 and to explore the effects of biomass exposure to E1 or other organic carbon substrates on E1 transformation. Due to the aforementioned difficulties of identifying key E1 degrading bacteria, changes in removal rates and patterns in mixed-microbial communities were used to identify the key mechanism. To accomplish this, loss of E1 was measured to obtain the desired information regarding the initial step of E1 transformation. Two sets of studies were performed. First, biomass generated in a membrane bioreactor (MBR) with and without exposure to E1 was used in kinetic studies to determine whether cometabolism or multiple substrate utilization was the key mechanism involved in E1 transformation. Next, experiments were performed to determine if the adverse selection effects observed in a previous study conducted in our laboratory27 could be mitigated by reducing organic carbon concentrations or varying the period between feeding cycles. Additionally, transformation rates during feast and famine periods during these cycles were compared. Together, these studies provide a much clearer picture of E1 removal in wastewater treatment systems.
2. Materials and methods
2.1 Chemicals and synthetic wastewater
E1 and deuterated or 13C-labeled E1 were obtained from Sigma and Cambridge Isotopes, respectively. The recipe for synthetic wastewater was adapted from Boeije et al.28 and contained (per L): 75 mg urea, 11 mg ammonium chloride, 12 mg sodium uric acid, 25 mg magnesium phosphate dibasic trihydrate, and 20 mg potassium phosphate tribasic. The synthetic wastewater also contained a carbon source made up of the following (per L, for 100 mg chemical oxygen demand (COD) per L nominal concentration): 6 mg bacteriological peptone, 51 mg sodium acetate, 6 mg dry meat extract, 17 mg glycerine, 21 mg potato starch, and 25 mg low fat milk powder. The carbon source was diluted or concentrated for carbon feeds of various strengths.2.2 Sludge seed
Biomass used to start each experiment (batch and MBR) was taken from the Metropolitan Wastewater Treatment Plant in St. Paul, Minnesota, which operates with both seasonal nitrification and biological phosphorus removal. A single sample of activated sludge was triple-washed with phosphate-buffered saline, divided into 3.5 mL aliquots, and cryopreserved in 15% glycerol (v/v) at −80 °C until use. A single sludge aliquot was used to seed each reactor.2.3 Batch systems
Batch systems were used for E1 exposure and feast-famine experiments. Excess aeration via air sparging was provided so that reactors were saturated with dissolved oxygen. pH values over the course of the experiments ranged from 7.2 to 7.6. Reactors were continuously mixed with stir bars.2.4 E1 exposure experiment
Continuous flow MBRs were operated as described in a previous paper27 to culture biomass with and without exposure to E1. Briefly, MBRs (150 mL) were operated with an HRT of 8 h and an SRT of 10 d and fed a synthetic wastewater with a COD of 100 mg L−1 for a 30 d period (3 SRTs). MBRs were either exposed or not exposed to E1 over this 30 d period (10 μg L−1 E1 in the influent or no E1 in the influent, respectively). Each paired treatment was run in triplicate, with each pair sharing the same sludge seed. Following the 30 d period, the reactors were sacrificed and the biomass recovered by centrifuging. The biomass from each MBR was resuspended in a 1 L batch reactor, and biomass concentration in each reactor was determined via a volatile suspended solids (VSS) test.29 Additional biomass was collected for Illumina analysis. Previously collected effluent from the MBRs was used as reactor liquor. This effluent had pH of 7.4, ammonia concentration of 18.2 mg L−1, and COD of 12 mg L−1. E1 was added to each batch reactor at a concentration of 10 μg L−1, and E1 removal was monitored over a 22 h period.2.5 Feast-famine experiment
The impact of the interval between COD addition on E1 removal was examined using batch reactors. Two experiments were performed. In each experiment a 10 L batch reactor was seeded with cryopreserved activated sludge, synthetic wastewater with 100 mg L−1 COD, and 10 μg L−1 E1. After a 5 d period, this 10 L reactor was split into 3 sets of triplicate 1.0 L reactors. The reactors received 10 μg L−1 of E1 on day 0 and were operated for 12 d following one of the two feeding regimes shown in Table 1, resulting in exposure to a total of either 180 or 600 mg L−1 COD over a 6 d experimental cycle.| Low organic carbon | High organic carbon | |||
|---|---|---|---|---|
| Feeding schedule (in mg per L COD) | Total organic carbon added over 6 d cycle (mg COD per L) | Feeding schedule (in mg L−1 COD) | Total organic carbon added over 6 d cycle (mg COD per L) | |
| 1d feed | 30 each day | 180 | 100 each day | 600 |
| 3d feed | 90 on days 0, 3, 6, 9, 12 | 180 | 300 on days 0, 3, 6, 9, 12 | 600 |
| 6d feed | 180 on days 0, 6, 12 | 180 | 600 on days 0, 6, 12 | 600 |
| Kinetic studies | Days 6, 11, 12 | Days 6, 12 | ||
E1 (10 μg L−1) was added periodically to each reactor (Table 1) and the E1 removal rate at that time point was determined over a period of 12 h. Biomass was also collected at the time of each E1 kinetic study and quantified by VSS. The experiments were identical with the following exceptions: (1) different total COD concentrations were fed over a 6 d experimental cycle (Table 1) and (2) an E1 transformation study was performed on day 11 during the low organic carbon experiment, but was not performed during the high organic carbon experiment.
2.6 Sample processing and analysis
Detailed procedures for E1 and DNA sampling and analysis are provided in the ESI.† Briefly, E1 samples were processed via solid phase extraction and silica gel cleanup based on the method by Ternes et al.30 Processed samples were analyzed via LC-MS as described in Tan et al.27 or by LC-MS-MS with methods adapted from Di Carro et al.31DNA samples were extracted using the FastDNA spin kit (MP Biomedicals, Solon, OH), the 16S region amplified using primers described by Muyzer et al.,32 and prepared for Illumina sequencing as described by Bartem et al.33 Illumina sequencing was carried out on an Illumina MiSeq platform at the University of Minnesota Genomics Center (Saint Paul, MN, USA). Illumina sequence reads were processed using Quantitative Insights Into Microbial Ecology (QIIME)34 and clustered using uclust.35 Reference sequences for each OTU were compared to the Greengenes reference database.36 UNIFRAC was used for principle coordinate analysis of communities.37
2.7 Data analysis
E1 removal rates and confidence intervals were obtained from the linear regression function in Microsoft Excel, while the student t-test (paired t-tests of E1 degradation rates for each pair of exposed/non-exposed reactors seeded with the same sludge seed in the E1 exposure experiments, paired t-tests to compare reactor performance in feeding cycles over time, pooled t-tests to compare different reactor conditions) were performed using the relevant function in the same program.Lag phases for E1 degradation in the first study were visually distinct, and were identified by plotting degradation curves on a log scale to determine when first-order degradation began. Lag phases for E1 degradation in the second study were determined using residual analysis of linear regressions (log scale), omitting data points until residuals no longer showed a bias.
3. Results and discussion
3.1 Effect of E1 exposure on E1 transformation: evidence for multiple substrate utilizers
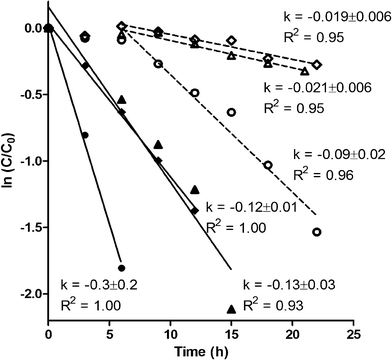
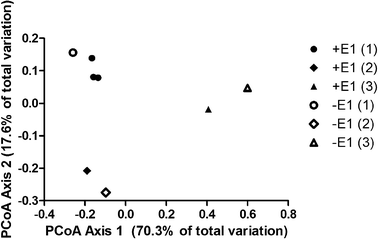
E1 transformation took place in both reactors with and without prior exposure to E1 (Fig. 1). Prior E1 exposure, however, correlated with higher removal rates and the lack of a lag phase. The transformation of E1 in all reactors clearly shows that E1 degraders do not require the presence of E1 to grow. A lag phase of 6 h was observed in cultures without prior E1 exposure, suggesting induction of E1 transformation enzymes. Indeed, growth of minor communities is unlikely to explain the end of the lag phase due to the low amount of E1 present, and in particular, the short time frame over which the lag period ended. This stands in contrast to typical cometabolic patterns where lag phases are not observed apart from inhibition or enzyme induction by cometabolites.38–40 Neither would explain the presence or absence of lag phase in our study as reactor liquor was identical across reactors. The higher E1 removal rates, shown through paired t-tests, in biomass with prior exposure to E1 (P = 0.05) suggests that either (1) prolonged exposure to E1 resulted in higher up-regulation of E1 transformation enzymes than the short-term exposure achieved, and/or (2) the ability to degrade E1 provided a competitive advantage within some niche, resulting in higher growth than would be expected based on the energy available in E1 alone. Both explanations point toward metabolic transformation of E1.Principal coordinate analysis of Illumina sequences from the mature MBRs, shown in Fig. 2, showed no broad community effects from E1 exposure. Instead, the initial sludge seed appeared to drive variation in communities (Fig. S1†). Analysis of operational taxonomic units at the genus level and at the species level for abundant genus (data not shown), did not show enrichment resulting from E1 exposure, suggesting that (1) exposure to E1 resulted in up-regulation of transformation enzymes rather than a larger population of E1 degraders, (2) E1 degraders are a very small fraction of the population, and/or (3) multiple genus/species may degrade E1. The inability to detect enrichment of rare OTUs is unsurprising, due to the large number of reactor replicates that would have been required.41
The E1 degradation patterns observed for communities with and without prior E1 exposure demonstrate that multiple substrate utilization is involved in the transformation of E1. This hypothesis has been suggested by others.25,26,42 Prior evidence for the role of multiple substrate utilization in E1 transformation includes: higher degradation rates associated with municipal wastewater biomass compared to industrial wastewater biomass;43 improved E1 transformation with extended exposure to E1;25 possible metabolic transformation of E1;44 and higher rates of estrogen degradation in the presence of diverse food sources, which may favor multiple substrate utilization.42 The results shown herein, however, are the first that we are aware of that directly compare the effect of E1 exposure on similarly cultured biomass, providing strong evidence for the role of multiple substrate utilization in the biodegradation of E1.
3.2 Effect of carbon exposure on E1 transformation
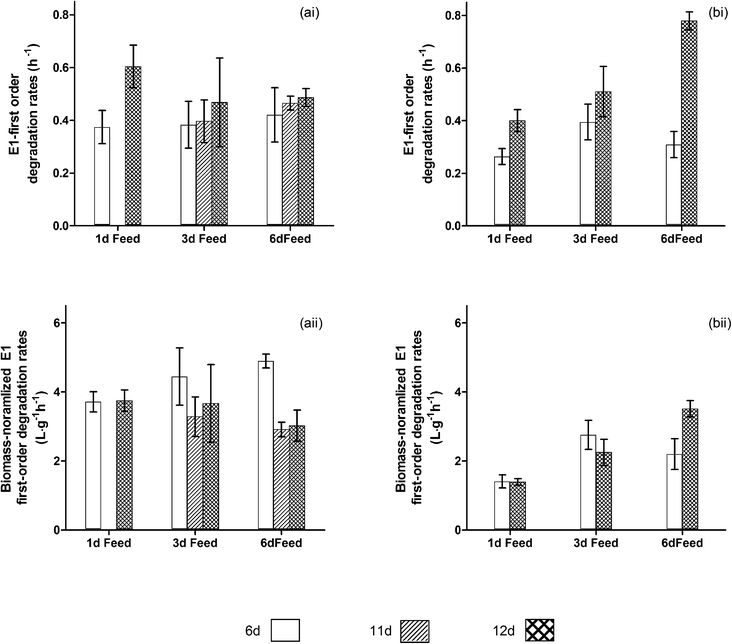
Fig. 3 shows that longer intervals between feeding cycles were beneficial for reactors receiving higher COD loads, suggesting that feeding cycles can be important for selection of E1 degraders. Among the reactors receiving higher COD loads, those fed every 6 d were the only reactors to have a statistically significant increase in overall E1 removal rates between days 6 and 12 (P = 0.018). Biomass normalized rates for this set of reactors also appeared to increase (P = 0.073). This suggests that the change in performance is not a result of only biomass growth, but also was a result of community development. At day 12, these reactors had significantly higher removal rates than those fed every day (P = 0.002) and rates also appeared higher than those fed every 3 d (P = 0.057).Conversely, among the reactors receiving lower COD loads, the longer interval between feeding did not confer any advantage, suggesting that the importance of feeding cycles for selection of E1 degraders depends on the abundance of food. Overall and biomass-normalized removal rates were similar across these reactors on day 12 (P = 0.32 and 0.75 respectively). For this group of reactors, only those fed daily showed an increase in overall E1 removal rates (P = 0.017). This change appears to be attributable to the increase in biomass, because biomass-normalized removal rates did not change (P = 0.85). Additionally, biomass-normalized removal rates dropped between days 6 and 12 for the 6 d feed reactors (P = 0.020). It is not clear why this occurred, but overall removal rates did not change.
Famine conditions appeared to induce a lag phase prior to E1 removal, suggesting that exposure to some minimal level of organic carbon was beneficial to E1 transformation. A clear 4 h lag phase was observed in E1 transformation experiments for the 6 d feed reactors on day 11; 2–4 h lag phases seemed to occur for the 3 d feed reactors as well (Fig. S4†). No comparable lag phases were observed for any reactors during feast periods (Fig. S2–S3†). In contrast to lag phase effects, the presence of other carbon sources did not appear to affect the rate of E1 transformation, because the removal rate for a given reactor was similar on days 11 (famine) and 12 (feast). While the presence of a certain low level of organic carbon is beneficial for inducing the transformation of E1, there were no additional benefits, particularly in terms of removal rates, provided by higher organic carbon concentrations.
The difference in E1 transformation behavior in the presence and absence of exposure to other carbon sources is further evidence of multiple substrate utilization of E1 and gives further insight into this mechanism. As in the previous experiment, if cometabolism were the mechanism of transformation, no lag phase would be expected during the famine period. Additionally, during this period, slower removal rates or the absence of E1 transformation might be expected as a result of a lack of cometabolites. As such, the similarities in removal rates between day 11 (famine) and 12 (feast) are consistent with multiple substrate utilization. We had not predicted, however, a lag phase during famine conditions based on this mechanism. At least one other study has observed a similar reduced lag phase for micropollutant transformation not attributable to microbial growth and following exposure to supplementary organic carbon.45 It is known that oligotrophic stress can cause de-repression of the catabolome,46 facilitating multiple substrate utilization. The prevailing view of this behavior is that of multiple substrate utilizing bacteria producing a large variety of enzymes at all times to scavenge food.15,46 We hypothesize that in addition to de-repression and constant production of a variety of enzymes, exposure to carbon sources may up-regulate the production of this suite of enzymes. Further work needs to be performed to explore and confirm this idea, which would lead to a better understanding of the metabolic activity of bacteria under low organic carbon conditions.
Recovery time from exposure to high concentrations of organic carbon may be important for the selection of E1 degraders. In a previous paper,26 we observed that repeated exposure to high levels of organic carbon selected against E1 degraders. We had also observed that cultures typically took between 3–5 d to develop E1 degrading capabilities. Consequently, these microbes might be outcompeted by faster growing bacteria when subjected to daily feeds. The current study tested the hypothesis that these effects could be mitigated by longer intervals between feeding, and the improved performance of the reactors receiving the higher COD load at 6 d intervals is consistent with this idea. A possible alternative is that the longer interval in feeding cycles may have promoted cell death and generated microbially derived organic carbon that may also enhance the activity of E1 degraders.47 The reactors exposed to lower levels of organic carbon did not seem to be subject to the same selective pressures. Interestingly, the reactors receiving lower levels of organic carbon had higher biomass to substrate yields than their counterparts (Table S1†). This phenomenon, observed previously by others as well,48 may be associated with lower growth rates.48,49 If so, this would be consistent with the idea of E1 degraders requiring conditions that favor slower growing bacteria.
3.3 E1 and carbon exposure: strategies for optimizing E1 transformation
Optimizing E1 transformation requires generating a sufficient quantity of biomass while maintaining a selection pressure for E1 degraders. Understanding that these bacteria are multiple substrate utilizers rather than specialist degraders or cometabolizers points us in certain directions for achieving optimization. This study illustrates that carbon and E1 exposure are critical for selecting for E1 degrading bacteria. While it is not practical to control E1 exposure in wastewater treatment systems, organic carbon concentrations are a key operating parameter. In this study, we observed that reactors receiving lower COD loads tended to have higher biomass-normalized removal rates. Conversely, the highest overall E1 removal rates (not biomass normalized) were observed in the 6d feed reactors receiving higher COD loads, in which more biomass was present. This is consistent with our previous paper,27 that showed that high COD concentrations, rather than high COD loads, select against E1 degraders. Conventional continuous stirred tank reactors can achieve high COD loads concomitant with low COD concentrations. While CSTRs are more typically used for industrial wastewater treatment to mitigate against toxicity, they could also have a role in improving E1 transformation in municipal wastewater. An alternate strategy of recovery time between feeding periods was explored in this study, and also appears to be a feasible option. This approach, however, may cost more in terms of sludge storage requirements. In addition to E1 and organic carbon exposure, there are further factors that may influence selection of E1 degraders that need to be further explored, including the diversity of food sources, as theorized by Racz et al.,42 and the quality of organic carbon available,47 which could have selection effects on these slower growing, multiple substrate utilizing bacteria and could also suggest alternative design approaches such as membrane bioreactors.Acknowledgements
We thank Hanna Temme for her assistance in preparing DNA samples for Illumina and for analyzing the resulting data. D.T.T. thanks the University of Minnesota for the Moos Summer Fellowship and the MnDRIVE Graduate Fellowship.References
- D. W. Kolpin, E. T. Furlong, M. T. Meyer and E. M. Thurman, Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999–2000: a national reconnaissance, Environ. Sci. Technol., 2002, 36(6), 1202–1211 CrossRef CAS.
- C. Lavelle and P. W. Sorensen, Behavioral responses of adult male and female fathead minnows to a model estrogenic effluent and its effects of exposure regime and reproductive success, Aquat. Toxicol., 2011, 101(3–4), 521–528 CrossRef CAS PubMed.
- K. Thomas, B. Erik and B. Mark, 17 alpha-ethinylestradiol reduces the competitive reproductive fitness of the male guppy (poecilia reticulata), Biol. Reprod., 2005, 72(1), 150–156 CrossRef PubMed.
- K. A. Kidd, P. J. Blanchfield, K. H. Mills, V. P. Palace, R. E. Evans, J. M. Lazorchak and R. W. Flick, Collapse of a fish population after exposure to a synthetic estrogen, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(21), 8897–8901 CrossRef CAS PubMed.
- A. C. Johnson, R. J. Williams and P. Matthiessen, The potential steroid hormone contribution of farm animals to freshwaters, the United Kingdom as a case study, Sci. Total Environ., 2006, 362(1–3), 166–178 CrossRef CAS PubMed.
- L. Salste, P. Leskinen, M. Virta and L. Kronberg, Determination of estrogens and estrogenic activity in wastewater effluent by chemical analysis and the bioluminescent yeast assay, Sci. Total Environ., 2007, 378(3), 343–351 CrossRef CAS PubMed.
- V. Gabet-Giraud, C. Miège, J. M. Choubert, S. M. Ruel and M. Coquery, Occurrence and removal of estrogens and beta blockers by various processes in wastewater treatment plants, Sci. Total Environ., 2010, 408(19), 4257–4269 CrossRef CAS PubMed.
- S. K. Khanal, B. Xie, M. L. Thompson, S. Sung, S. K. Ong and J. H. Van Leeuwen, Fate, transport, and biodegradation of natural estrogens in the environment and engineered systems, Environ. Sci. Technol., 2006, 40(21), 6537–6546 CrossRef CAS.
- M. Cargouët, D. Perdiz, A. Mouatssim-Souali, S. Tamisier-Karolak and Y. Levi, Assessment of river contamination by estrogenic compounds in Paris area (France), Sci. Total Environ., 2004, 324(1–3), 55–66 CrossRef PubMed.
- H. R. Andersen, M. Hansen, J. Kjølholt, F. Stuer-Lauridsen, T. Ternes and B. Halling-Sørensen, Assessment of the importance of sorption for steroid estrogen removal during activated sludge treatment, Chemosphere, 2005, 61(1), 139–146 CrossRef CAS PubMed.
- C. Baronti, R. Curini, G. D'Ascenzo, A. D. Corcia, A. Gentili and R. Samperi, Monitoring natural and synthetic estrogens at activated sludge sewage treatment plants and in a receiving river water, Environ. Sci. Technol., 2000, 34(24), 5059–5066 CrossRef CAS.
- L. A. Hoogenboom, L. De Haan, D. Hooijerink, G. Bor, A. J. Murk and A. Brouwer, Estrogenic activity of estradiol and its metabolites in the ER-CALUX assay with human T47D breast cells, APMIS, 2001, 109, 101–107 CrossRef CAS PubMed.
- H. Dalton and D. I. Stirling, Co-metabolism, Philos. Trans. R. Soc., B, 1982, 297, 481–496 CrossRef CAS.
- W. Harder and L. Dijkhuizen, Strategies of mixed substrate utilization in microorganisms, Philos. Trans. R. Soc., B, 1982, 297, 459–480 CrossRef CAS.
- T. Egli, How to live at very low substrate concentration, Water Res., 2010, 44(17), 4826–4837 CrossRef CAS PubMed.
- Y. X. Ren, K. Nakano, M. Nomura, N. Chiba and O. Nishimura, Effects of bacterial activity on estrogen removal in nitrifying activated sludge, Water Res., 2007, 41(14), 3089–3096 CrossRef CAS PubMed.
- T. W. Yi, W. F. Harper, R. D. Holbrook and N. G. Love, Role of particle size and ammonium oxidation in removal of 17α-ethinylestradiol in bioreactors, J. Environ. Eng., 2006, 132(11), 1527–1529 CrossRef CAS.
- J. Shi, S. Fujisawa, S. Nakai and M. Hosomi, Biodegradation of natural and synthetic estrogens by nitrifying activated sludge and ammonia-oxidizing bacterium Nitrosomonas europaea, Water Res., 2004, 38(9), 2323–2330 CrossRef CAS PubMed.
- L. S. Gaulke, S. E. Strand, T. F. Kalhorn and H. D. Stensel, 17α-ethinylestradiol transformation via abiotic nitration in the presence of ammonia oxidizing bacteria, Environ. Sci. Technol., 2008, 42(20), 7622–7627 CrossRef CAS.
- T. Yoshimoto, F. Nagai, J. Fujimoto, K. Watanabe, H. Mizukoshi, T. Makino, K. Kimura, H. Saino, H. Sawada and H. Omura, Degradation of estrogens by Rhodococcus zopfii and Rhodococcus equi isolates from activated sludge in wastewater treatment plants, Appl. Environ. Microbiol., 2004, 70(9), 5283–5289 CrossRef CAS PubMed.
- C. P. Yu, H. Roh and K. H. Chu, 17β-estradiol-degrading bacteria isolated from activated sludge, Environ. Sci. Technol., 2007, 41(2), 486–492 CrossRef CAS.
- B. Pauwels, K. Wille, H. Noppe, H. De Brabander, T. Van de Wiele, W. Verstraete and N. Boon, 17α-cometabolism by bacteria degrading estrone, 17β-estradiol, and estriol, Biodegradation, 2008, 19, 683–693 CrossRef CAS PubMed.
- N. A. Zhou, A. C. Lutovsky, G. L. Andaker, H. L. Gough and J. F. Ferguson, Cultivation and characterization of bacterial isolates capable of degrading pharmaceutical and personal care products for improved removal in activated sludge wastewater treatment, Biodegradation, 2013, 24(6), 813–827 CrossRef CAS PubMed.
- F. Kurisu, M. Ogura, S. Saitoh, A. Yamazoe and O. Yaqi, Degradation of natural estrogen and identification of the metabolites produced by soil isolates of Rhodococcus sp. and Sphingomonas sp., J. Biosci. Bioeng., 2010, 109(6), 576–582 CrossRef CAS PubMed.
- J. P. Bagnall, A. Ito, E. J. McAdam, A. Soares, J. N. Lester and E. Cartmell, Resource dependent biodegradation of estrogens and the role of ammonia oxidising and heterotrophic bacteria, J. Hazard. Mater., 2012, 239, 56–63 CrossRef PubMed.
- Y. K. K. Koh, T. Y. Chiu, A. R. Boobis, M. D. Scrimshaw, J. P. Bagnall, A. Soares, S. Pollard, E. Cartmell and J. N. Lester, Influence of operating parameters on the biodegradation of steroid estrogens and nonylphenolic compounds during biological wastewater treatment processes, Environ. Sci. Technol., 2009, 43(17), 6646–6654 CrossRef CAS.
- D. T. Tan, W. A. Arnold and P. J. Novak, Impact of organic carbon on the biodegradation of estrone in mixed culture systems, Environ. Sci. Technol., 2013, 47(21), 12359–12365 CrossRef CAS PubMed.
- G. Boeije, R. Corstanje, A. Rottiers and D. Schowanke, Adaptation of the CAS test system and synthetic sewage for biological nutrient removal - Part I: development of a new synthetic sewage, Chemosphere, 1999, 38(4), 699–709 CrossRef CAS.
- Standard Methods for the Examination of Water and Wastewater. 20th Ed. American Public Health Association, American Water Works Association, Water Environment Federation.
- T. A. Ternes, M. Stumpf, J. Mueller, K. Haberer, R. D. Wilken and M. Servos, Behavior and occurrence of estrogens in municipal sewage treatment plants - I. investigations in Germany, Canada and Brazil, Sci. Total Environ., 1999, 225(1–2), 81–90 CrossRef CAS.
- M. Di Carro, C. Scapolla, C. Liscio and E. Magi, Development of a fast liquid chromatography-tandem mass spectrometry method for the determination of endocrine-disrupting compounds in waters, Anal. Bioanal. Chem., 2010, 398(2), 1025–1034 CrossRef CAS PubMed.
- G. Muyzer, E. C. de Wall and A. G. Uitterlinden, Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA, Appl. Environ. Microbiol., 1999, 59(3), 695–700 Search PubMed.
- A. K. Bartem, M. D. J. Lynch, J. C. Stearns, G. Moreno-Hagelsieb and J. D. Neufeld, Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads, Appl. Environ. Microbiol., 2011, 77(11), 3846–3852 CrossRef PubMed.
- J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Pena, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelly, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevnisky, P. J. Tumbaugh, W. A. Walters, J. Widmann, T. Yatsuneko, J. Zaneveld and R. Knight, QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 2010, 7(5), 335–336 CrossRef CAS PubMed.
- R. C. Edgar, Search and clustering orders of magnitude faster than BLAST, Bioinformatics, 2010, 26(19), 2460–2461 CrossRef CAS PubMed.
- D. McDonald, M. N. Price, J. Goodrich, E. P. Nawrocki, T. Z. DeSantis, A. Probst, G. L. Andersen, R. Knight and P. Hugenholtz, An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea, ISME J., 2012, 6(3), 610–618 CrossRef CAS PubMed.
- C. A. Lozupone, M. Hamady, S. T. Kelley and R. Knight, Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities, Appl. Environ. Microbiol., 2007, 73(5), 1576–1585 CrossRef CAS PubMed.
- C. A. Smith, K. T. O'Reilly and M. R. Hyman, Cometabolism of methyl tertiary butyl ether and gaseous n-alkanes by Pseudomonas mendocina KR-1 grown on C(5) to C(8) n-alkanes, Appl. Environ. Microbiol., 2003, 69(12), 7385–7394 CrossRef CAS.
- D. W. Connell, Basic Concepts of Environmental Chemistry, Lewis Publishers, New York, 2nd edn, 2005 Search PubMed.
- R. A. Deeb, K. M. Scow and L. Alvarez-Cohen, Aerobic MTBE biodegradation: an examination of past studies, current challenges, and future research directions, Biodegradation, 2000, 11, 171–186 CrossRef CAS.
- D. R. Johnson, D. E. Helbling, Y. Men and K. Fenner, Can meta-omics help to establish causality between contaminant biotransformations and genes or gene products?, Environ. Sci.: Water Res. Technol., 2015, 1 10.1039/C5EW00016E , Perspective.
- L. Racz, J. G. Muller and R. K. Goel, Fate of selected estrogens in two laboratory scale sequencing batch reactors fed with different organic carbon sources under varying solids retention times, Bioresour. Technol., 2012, 110, 35–42 CrossRef CAS PubMed.
- A. C. Layton, B. W. Gregory, J. R. Seward, T. W. Schultz and G. S. Sayler, Mineralization of steroidal hormones by biosolids in wastewater treatment systems in Tennessee USA, Environ. Sci. Technol., 2000, 34(18), 3925–3931 CrossRef CAS.
- C. P. Yu, H. Roh and K. H. Chu, 17β-estradiol-degrading bacteria isolated from activated sludge, Environ. Sci. Technol., 2007, 41(2), 486–492 CrossRef CAS.
- B. Horemans, J. Vandermaesen, L. Vankaecke, E. Smolders and D. Springael, Variovorax sp.-mediated biodegradation of the phenyl urea herbicide linuron at micropollutant concentrations and effects of natural dissolved organic matter as supplementary carbon source, Appl. Microbiol. Biotechnol., 2013, 97(22), 9837–9846 CrossRef CAS PubMed.
- J. Benner, D. E. Helbling, H. E. Kohler, J. Wittebol, E. Kaiser, C. Prasse, T. A. Ternes, C. N. Albers, J. Aamand, B. Horemans, D. Springael, E. Walravens and N. Boon, Is biological treatment a viable alternative for micropollutant removal in drinking water treatment processes?, Water Res., 2013, 47(16), 5955–5976 CrossRef CAS PubMed.
- D. T. Tan, H. R. Temme, W. A. Arnold and P. J. Novak, Estrone degradation: does organic matter (quality) matter?, Environ. Sci. Technol., 2015, 49(1), 498–503 CrossRef CAS PubMed.
- K. Chojnacka and A. Zielinksa, Evaluation of growth yield of Spirulina (Arthrospira) sp. In photoautotrophic, heterotrophic and mixotrophic cultures, World J. Microbiol. Biotechnol., 2012, 28(2), 437–445 CrossRef CAS PubMed.
- D. A. Lipson, R. K. Monson, S. K. Schmidt and M. N. Weintraub, The trade-off between growth rate and yield in microbial communities and the consequences for under-snow soil respiration in a high elevation coniferous forest, Biogeochemistry, 2009, 95(1), 23–35 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ew00014a |
| This journal is © The Royal Society of Chemistry 2015 |