Inactivation mechanisms of cryptosporidium parvum oocysts by solar ultraviolet irradiation
Yuanyuan
Liu†
*ab,
Shengkun
Dong†
a,
Mark S.
Kuhlenschmidt
c,
Theresa B.
Kuhlenschmidt
c,
Jenny
Drnevich
d and
Thanh H.
Nguyen
a
aDepartment of Civil and Environmental Engineering, University of Illinois at Urbana-Champaign, Urbana IL 61801, USA. E-mail: Yuanyuan.Liu@pnnl.gov; sdong6@illinois.edu; thn@illinois.edu; Tel: +1 (509) 371 7370
bPacific Northwest National Laboratory, Richland WA 99352, USA
cDepartment of Pathobiology, University of Illinois at Urbana-Champaign, Urbana IL 61801, USA. E-mail: kuhlensc@illinois.edu; tkuhlens@illinois.edu
dHigh Performance Biological Computing Group and the Carver Biotechnology Center, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA. E-mail: drnevich@illinois.edu
First published on 5th January 2015
Abstract
Cryptosporidium parvum oocysts have been known to cause adverse health effects worldwide, and processes that contribute to their inactivation have gained wide attention in recent years. Solar inactivation is an important process that can improve surface water quality. Solar disinfection (SODIS) can be used to disinfect water as a point-of-use alternative, and disinfect wastewater in waste stabilization ponds. However, a clear understanding of the oocyst solar inactivation mechanisms is lacking. This study systematically investigated the oocyst solar inactivation mechanisms in the presence of a wide range of environmental factors and also provided an insight on the metabolic response of oocysts using the microarray analysis. The result revealed that oocyst inactivation by solar UVA/visible light was dominated by UVA-induced internal radical damages and was sensitive to both the temperature and the oocyst source. External radical producing sensitizers did not enhance the UVA/visible light inactivation of oocysts due to the protection by the oocyst wall. In contrast, UVB was found to directly damage the oocyst genome, ensuring an effective inactivation that correlated only with UV fluence after being corrected for light screening regardless of the oocyst source, temperature, and the presence of external sensitizers. Further microarray analysis suggested that the effective UVB inactivation could be explained by the down-regulation of most of the genes responsible for cellular metabolic activities and the lack of expression of stress protection mechanisms in oocysts after 30 minutes of UVB exposure. These results facilitate the understanding and design of water and wastewater treatment processes that involve natural sunlight exposure.
Water impactWe investigated solar inactivation mechanisms of Cryptosporidium parvum oocysts under a wide range of environmental parameters that control oocyst inactivation efficiency. For the first time, our findings revealed that UVA/visible light inactivated oocysts via endogenously produced radicals, and the presence of external radical producing sensitizers did not enhance oocyst inactivation. This indirect endogenous mechanism is sensitive to the oocyst source and temperature. In contrast, UVB directly damaged the oocyst genome, ensuring effective inactivation that correlated only with UV fluence and was independent of other environmental parameters. These findings contribute to the risk assessment for oocyst persistence in surface water and facilitate better solar disinfection design as a point-of-use water treatment method. |
1. Introduction
Water contamination with Cryptosporidium (C.) parvum oocysts is a serious threat to public health.1 More than 1000 outbreaks of human cryptosporidiosis, a severe diarrhea, have been recorded worldwide since the first human cryptosporidiosis was documented in 1976.2 Among the 199 waterborne disease outbreaks globally reported between 2004 and 2010, cryptosporidiosis was one of the most common diseases.3 Large-scale surface water surveys targeting rivers, streams, lakes, and springs from 17 states in the U.S. during the late 1980s' revealed that 39% to 100% of water samples were tested positive for Cryptosporidium.4–6 Because surface water is used as a drinking water source, EPA requires drinking water treatment plants to monitor their water source to determine the treatment requirement.7 Apart from drinking water, human exposure to contaminated surface water and wastewater treated by stabilization ponds is also possible through recreational and agricultural activities.8 Therefore, it is important to assess the persistence of oocyst infectivity in surface water. Among the factors that influence oocyst infectivity, solar inactivation by daily exposure to sunlight should be considered.9 Solar inactivation is already recommended as one of the point-of-use treatments for surface water when centralized water treatment facilities are not available,10 we argue that the knowledge of factors controlling oocyst inactivation by sunlight irradiation will allow a more accurate prediction of oocyst persistence in surface water that may be in contact with the public.Inactivation of C. parvum oocysts by sunlight irradiation is not readily predicted because oocysts are known to have a thick wall protecting the internal sporozoites from external damages, including sunlight. Previous studies have reported a 0.1 to 3 log10 inactivation of C. parvum oocysts by solar inactivation.11–20 This reported variation in inactivation efficiency could be attributed to the differences in the oocyst source, the UV transmission spectrum of reactor material, solution composition, temperature, and radiation spectrum used in these studies. The importance of solar spectrum was suggested by higher oocyst inactivation in UV transmitting containers such as borosilicate glass tubes11,12,15,16 compared to plastic containers that block solar UV transmission.13–15,17–20 In addition to UV blocking effect, the solution conditions also influenced oocyst inactivation due to light attenuation.14,16 For instance, an increase in temperature during sunlight exposure may induce the release of the sporozoites from the protective oocyst wall and subsequent inactivation.13,15,21 Although many environmental factors that influence solar inactivation of oocysts have been identified, the mechanisms of the impact of these environmental factors remain to be elucidated.
Despite the progress in understanding the influence of environmental factors on oocyst inactivation kinetics, few studies have investigated the mechanisms of solar inactivation of Cryptosporidium oocysts. Solar UVB, which caused direct genome damage to virus and bacteria,22–26 was also reported to be the most lethal portion of the solar spectrum for oocyst inactivation.16 Nevertheless, the mechanism of oocyst UVB inactivation remains unclear. In comparison to UVB, UVA/visible light is less powerful but more abundant in natural sunlight, which can penetrate deeper into water and transmit itself through bottles made from PET, a material that significantly attenuates UVB.27,28 It is reported that UVA/visible light damaged the oocyst wall after 10 h exposure20 and interfered with sporozoite exocytosis, a fundamental cellular process for sporozoites to attach and invade host cells.29 However, it is also unclear whether the production of reactive oxygen species (ROS), which are formed from excited external sensitizers such as natural organic matter (NOM),30,31 has a significant influence on oocyst inactivation. If this indirect exogenous mechanism dominated oocyst solar inactivation, the presence of NOM could stimulate the ROS production and enhance inactivation efficiency, rather than attenuate sunlight irradiation and reduce inactivation, as reported for the inactivation of some viruses.22,32–35 Besides, indirect endogenous inactivation initiated by ROS formed by internal sensitizers associated with the microorganisms themselves has been reported for viruses and bacteria36,37 but has not been studied on oocysts.
This paper systematically studied the dominant mechanisms of oocyst solar inactivation in the presence of a wide range of environmental parameters including the oocyst source, the presence of external sensitizer, temperature, and the UVA and UVB components of sunlight. We first determined the role of indirect exogenous inactivation mechanism for C. parvum oocysts, using organic matter isolates from a natural river and the effluent of a wastewater treatment plant. We differentiated the indirect endogenous and the direct UVB inactivation mechanisms based on inactivation results obtained at different temperatures because the bimolecular reactions associated with internal sensitizers are enhanced by temperature while direct DNA damages in non-cellular systems are independent of temperature.38 We established a correlation between the oocyst log10 removal and UV fluence for full-spectrum inactivation under a wide range of environmental factors. Finally, we used microarray analysis to determine the genetic response of oocysts to full-spectrum irradiation.
2. Materials and methods
2.1 Propagation of C. parvum oocysts
Two batches of oocysts were propagated from infected male Holstein calves using Cryptosporidium parvum AUCP-1 isolate to study the influence of oocyst sources on oocyst inactivation. Oocysts collection from calves were conducted in the Department of Pathobiology, UIUC, following protocols approved by the University of Illinois Institutional Animal Care and Use Committee (IACUC protocol 04070) as previously described.39 The collected oocysts were purified by sieve filtration, chloroform extraction, and Sheather's sugar floatation following a previous protocol within two weeks after the feces were collected.39 The purified infectious oocysts were stored in Tris-ethylenediamine-tetraacetic acid (Tris-EDTA) at 4 °C and used within one month. The Tris-EDTA was removed by centrifugation (Eppendorf) at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 2 min and rinsed with deionized water twice before experiments.
000 × g for 2 min and rinsed with deionized water twice before experiments.
2.2 Preparation of model natural and wastewater Organic Matter (OM)
Organic matter isolates from the Suwannee River (SRNOM) and from a wastewater treatment plant in Saudi Arabia (EfOM) were selected as model external sensitizers. Information on these isolates can be found in a previous study.37 Briefly, 50 mg of each isolate were added into 100 mL deionized water, and the pH of the solution was adjusted to 8 with NaOH. These solutions were stirred for 24 h and then filtered through 0.22 μm Millipore filters. The total organic carbon (TOC) concentrations of the SRNOM and EfOM stock solutions were 73.4 and 166.2 mg C L−1, respectively.2.3 Oocyst inactivation by simulated sunlight
Sunlight irradiation at a spectrum from 280 to 700 nm was simulated with an Atlas Suntest® XLS+ photosimulator (Chicago, IL) equipped with a xenon arc lamp (280–800 nm).22 The spectrum was shown in a previous study.37 For experiments with only UVA/visible portion of the sunlight, the radiation below 320 nm was cut off by a secondary UVB filter (Newport, FSQ-WG320), which was placed on top of the reactor. Oocyst inactivation experiments were conducted using a 10 mL beaker with the wall painted black to reduce light scattering. Two mL of the oocyst suspension at a concentration of 1 × 108 oocysts mL−1 in 1 mM NaHCO3 at pH 8 were exposed to simulated UVA/visible light or full-spectrum irradiation (referred to as UVB) for 10 h or 1.5 h, respectively. The solar simulator intensities were set at 400 W m−2 for UVA/visible light irradiation and 250, 400, and 750 W m−2 for full-spectrum irradiation. During the experiment, the solution was mixed by a magnetic stirrer at 130 rpm. The sample temperature was controlled by a water bath and monitored by a thermometer. To investigate the role of exogenously formed radicals produced by sunlight radiation, 20 mg C L−1 NOM was added to a selected set of solutions. At each sampling point, the sample was drawn from one of the beakers and was immediately used in the oocyst infectivity analysis. For any given solution condition, dark control experiments were also conducted. Duplicate experiments were conducted due to limited oocysts quantity.Infectivity of oocysts was analyzed by in vitro culture infection of the HCT-8 line.40,41 In brief, the oocysts were rinsed with deionized water and disinfected with 40% bleach for 10 min. The samples were handled under sterile conditions from then on. The bleach was removed by rinsing with cold sterile PBS five times, and the oocyst pellet was resuspended in 0.5 mL HCT-8 cell culture medium. The oocyst sample was then serially diluted and inoculated to the confluent monolayer HCT-8 cells in 96-well plates. After a 48 hour incubation, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol/acetic acid was used to fix and permeabilize the HCT-8 cells and to inactivate the C. parvum sporozoites. The sporozoites were then marked by immunodetection with sporo-glo (Waterborne, Inc.) and recognized under a fluorescence microscope. Each cluster of sporozoites was deemed as being from infectious oocysts, and single sporozoites were regarded as being from noninfectious oocysts. The oocyst sample was diluted until no infectious oocysts could be detected. Then, a most probable number calculator was used to determine the number of infectious oocysts in the original oocyst sample.
1 methanol/acetic acid was used to fix and permeabilize the HCT-8 cells and to inactivate the C. parvum sporozoites. The sporozoites were then marked by immunodetection with sporo-glo (Waterborne, Inc.) and recognized under a fluorescence microscope. Each cluster of sporozoites was deemed as being from infectious oocysts, and single sporozoites were regarded as being from noninfectious oocysts. The oocyst sample was diluted until no infectious oocysts could be detected. Then, a most probable number calculator was used to determine the number of infectious oocysts in the original oocyst sample.
2.4 UVA/visible light irradiated radical concentration determination
The concentrations of hydroxyl radicals (˙OH) and singlet oxygen (1O2) formed from excited NOM by UVA/visible light irradiation were measured using phenol (Acros Organics)22,42 and furfuryl alcohol (FFA, Acros Organics)22,43 as probes, respectively. Forty mL of solutions containing 20 mg C L−1 SRNOM or EfOM and the corresponding probes (10 μM phenol for ˙OH or 50 μM FFA for 1O2) were buffered to pH 8 with 1 mM NaHCO3 and exposed to simulated UVA/visible light at 400 W m−2 for 10 h. Samples were taken at different times and centrifuged at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 min. The supernatant was taken to analyze the concentrations of phenol and FFA by a reverse-phase HPLC 1200 Series using an Eclipse XDB-C18 column.22 The concentrations of ˙OH and 1O2 were calculated from decay rates of phenol and FFA, following previous publications.22,43,44 Duplicate experiments were conducted due to limited sample quantity.
000 × g for 30 min. The supernatant was taken to analyze the concentrations of phenol and FFA by a reverse-phase HPLC 1200 Series using an Eclipse XDB-C18 column.22 The concentrations of ˙OH and 1O2 were calculated from decay rates of phenol and FFA, following previous publications.22,43,44 Duplicate experiments were conducted due to limited sample quantity.
2.5 Kinetic data analysis
The data from oocyst inactivation kinetic experiments at a given condition were combined into one dataset and fit with a pseudo-1st-order kinetics model. The pseudo-1st-order inactivation rate constant, kobs (h−1), was determined by the slope of ln(C/C0) versus time (h), where C is the infectious oocyst concentration and C0 is the initial infectious oocyst concentration. To account for light attenuation, kobs was adjusted by a light screening correction factor based on the concept discussed in a previous publication.22 To compare oocyst inactivation kinetics under different conditions, such as dark controls with irradiated experiments, the intercepts and the slopes of the kinetics plots were statistically compared using ANOVA with 95% confidence interval.45The UV fluence (kJ m−2) was determined by multiplying the measured fluence rate (kW m−2) by the irradiation time (s):46L × t. The fluence rate at each solar simulator intensity setting was calculated from the 1 × 10−4 M p-nitroanisole (PNA)/0.01 M pyridine (pyr) actinometer system under the full-spectrum or the UVA/visible light irradiation following the equations:47–49
 | (1) |
 | (2) |
 | (3) |
 of the PNA/pyr actinometer system was first calculated. Then, (Lλ)a was estimated at each wavelength λ by the assumption that
of the PNA/pyr actinometer system was first calculated. Then, (Lλ)a was estimated at each wavelength λ by the assumption that  . The values of ελ can be found in Leifer,47
. The values of ελ can be found in Leifer,47 and (Lλ)m can be determined by the spectroradiometer measurement. Eventually, the fluence rate (kW m−2) of the actinometer system, (L)a, was calculated by (Lλ)a at each wavelength λ from eqn (3).
and (Lλ)m can be determined by the spectroradiometer measurement. Eventually, the fluence rate (kW m−2) of the actinometer system, (L)a, was calculated by (Lλ)a at each wavelength λ from eqn (3).
2.6 Microarray analysis
Three samples (2 mL of 1 × 108 oocysts L−1) were exposed to full-spectrum irradiation for 30 min at 25 °C, and another three samples were kept in the dark as controls. After 30 min, the samples were frozen and thawed in liquid nitrogen for three cycles, and the RNA of the oocysts was extracted following the instructions from RNeasy Mini Kit (Qiagen, CA). The RNA quality was evaluated using an Agilent Bioanalyzer (Santa Clara, CA) and the RNA quantity was determined using a Qubit® RNA HS Assay Kit (Grand Island, NY). 200 ng of total RNA per sample were reverse transcribed and linearly amplified according to manufacturer's instructions using the Agilent low input QuickAmp single color labelling kit (Agilent Technologies, Santa Clara, CA).An 8 × 15 K microarray for Iowa-II Cryptosporidium parvum was custom-made by Agilent based on the design of a previous study50 and interrogated a total of 3805 genes. Gene annotations for each probe, including the Gene Ontology terms, were downloaded from CryptoDB.org on December 5, 2014. Major functional classifications for the genes were taken from the same previous study.50 The number of unique 60-mer probes per gene was dependent on the length of the gene: 650 genes had a single detection probe, 106 had two, and 3049 had three. Of these, 6218 were spotted once (1× probes) and 3790 were spotted twice (2× probes). In addition, 47 probes of these were replicated for an additional 30 times (30× probes) for quality control. An additional 459 positive and 77 negative controls were added to the array for a total of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 744 spots. RNA samples, including 6 samples from two independent triplicate experiments and 2 technical replicates, were hybridized to the microarray overnight at 65 °C and washed according to the standard Agilent protocols. Slides were scanned using an Axon 4000B scanner and images were analysed with GENEPIX 6.1 software (Molecular Devices, Sunnyvale, CA).
744 spots. RNA samples, including 6 samples from two independent triplicate experiments and 2 technical replicates, were hybridized to the microarray overnight at 65 °C and washed according to the standard Agilent protocols. Slides were scanned using an Axon 4000B scanner and images were analysed with GENEPIX 6.1 software (Molecular Devices, Sunnyvale, CA).
Microarray data pre-processing and statistical analyses were done in R (v 3.1.2)51 using the limma package (v 3.22.1).52 Median foreground and median background values from the 8 arrays were read into R, and any spots that had been manually flagged (−100 values) were given a weight of zero.53 The background values were ignored because investigations showed that trying to use them to adjust for background fluorescence added more noise to the data. One of the sunlight-treated samples had a poor hybridization and was removed from the analysis. The single-channel foreground values from the remaining 7 samples were not normalized before log2-transformation because most methods of normalization assume that 1) most genes are not changing and 2) most of the changes are not in the same direction.53 Preliminary investigations using quantile-normalized data showed an asymmetric distribution of fold-changes with a large skew towards negative fold-changes, suggesting that normalization assumptions had been violated (data not shown). Given that wide-spread down-regulation of gene expression upon full-spectrum treatment was expected, we performed hierarchical clustering on log2-transformed fluorescence values without normalization (i.e., “raw” expression values) and found that the two treatments clustered separately and that the replicates treated with full-spectrum radiation had the same amount of variation as the control replicates (Fig. 1). Therefore, normalization was not needed and the statistical analysis was done on the log2-transformed raw expression data.
A separate statistical model was fit on each set of 1×, 2×, and 30× probes; the 1× probes were fit with a standard limma statistical model54 to estimate the expression difference between full-spectrum treated and dark control samples, while the 2× and 30× probes were fit with a similar model that additionally accounted for the multiple spots per probe.52 After fitting the models, the results for the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 055 unique probe IDs were combined back together before adjusting the raw p-values using the False Discovery Rate method.55 In this study, the probes were recognized as having a significant fold-change if the FDR p-value was less than 0.05. All probes were categorized into three groups: having a significant up-regulated fold-change, having a significant down-regulated fold-change, and not having a significant fold-change. Consistency among the multiple probes per gene was high with the Pearson correlation coefficients ranging from 0.905 to 0.933. Over- and under-representation analysis of the major functional classifications in the up- and down-regulated genes was done using the goseq package (v 1.18.0),56 which assesses and corrects for any biases in gene length among the significant genes.
055 unique probe IDs were combined back together before adjusting the raw p-values using the False Discovery Rate method.55 In this study, the probes were recognized as having a significant fold-change if the FDR p-value was less than 0.05. All probes were categorized into three groups: having a significant up-regulated fold-change, having a significant down-regulated fold-change, and not having a significant fold-change. Consistency among the multiple probes per gene was high with the Pearson correlation coefficients ranging from 0.905 to 0.933. Over- and under-representation analysis of the major functional classifications in the up- and down-regulated genes was done using the goseq package (v 1.18.0),56 which assesses and corrects for any biases in gene length among the significant genes.
2.7 RT-qPCR confirmation
RT-qPCR of three selected genes was conducted to confirm the microarray data. These genes were cgd3_3770, a 2135 bp gene associated with the chaperone heat shock protein, cgd7_3430, a 1913 bp gene coding for the protein kinase, and cgd5_1290, a 1640 bp gene coding for the membrane-associated adenyl cyclase. The primer sets used for these genes were adapted from a previous study.50 iTaq™ Universal SYBR® Green One-Step Kit and a MiniOpticon™ real-time PCR system (Bio-Rad, CA) were used to amplify these genes from 0.5 μl of extracted total RNA. The relative gene expression from the RT-qPCR data was determined using the 2−ΔΔCt method as described previously,57 using 18 s rRNA gene of C. parvum as the internal control.58 The fold-change data derived from the microarray analysis for these three genes were compared with the relative gene expression obtained by the RT-qPCR method. Three biological replicates were conducted for both the dark controls and the full-spectrum radiation treated samples.3. Results and discussion
3.1 The effect of UVA/visible light of simulated solar radiation on oocyst inactivation
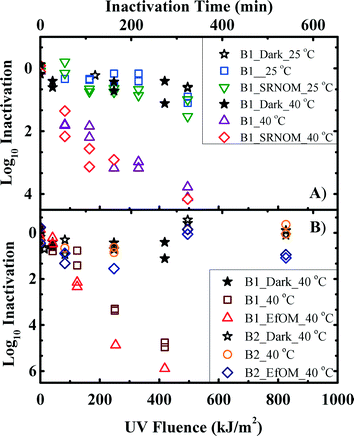
UVA/visible light inactivation kinetics of oocysts in the presence and absence of organic matter sensitizers was determined for two batches of oocysts from two different calves. The results of oocyst log10 inactivation from batch 1 with/without SRNOM at temperatures of 25 or 40 °C are shown in Fig. 2A. At 25 °C, the inactivation rate constants for dark controls were statistically similar compared with the irradiated experiments with or without 20 mg L−1 SRNOM (p = 0.58 and 0.06, respectively). At 40 °C, 4 log10 inactivation was achieved after the oocysts were exposed to a UV fluence of 500 kJ m−2. The oocyst inactivation kinetics with and without SRNOM were statistically similar after kobs was adjusted by a light screening correction factor (p = 0.58), though all were different from that of the dark controls (p = 7 × 10−5). This observation indicated the negligible effect of this exogenous sensitizer on oocyst inactivation, despite the formation of 1O2 and ˙OH radicals as reported in Table 1.| T (°C) | ˙OH (fM)c | 1O2 (pM)c | |
|---|---|---|---|
| a Sensitizer-free buffer solution containing only oocysts. b SRNOM and EfOM solution without oocysts. c The two values of ˙OH and 1O2 concentrations are replicate values from duplicate experiments. | |||
| Sensitizer-freea | 25 ± 1 | 0.26 | 0.004, not detected |
| 40 ± 1 | 0.35, 0.28 | 0.012, 0.008 | |
| SRNOMb | 25 ± 1 | 0.33, 0.22 | 0.15, 0.15 |
| 40 ± 1 | 0.62, 0.48 | 0.13, 0.14 | |
| EfOMb | 25 ± 1 | 0.25, 0.28 | 0.13, 0.11 |
| 40 ± 1 | 0.60, 0.60 | 0.13, 0.11 | |
Because no oocyst inactivation was observed at 25 °C, oocyst inactivation in the presence and absence of wastewater organic matter (EfOM) was studied only at 40 °C, and the results are shown in Fig. 2B. The result of UVA/visible light inactivation of oocysts from batch 1 in the presence and absence of EfOM was similar (p = 0.17), indicating that the EfOM did not change oocyst inactivation significantly. Different inactivation rate constants obtained for the dark controls and the irradiated experiments (p = 10−9) suggested that UVA/visible light inactivate oocyst when the solution temperature reaches 40 °C.
The two organic matter isolates have been shown to produce reactive radicals (Table 1). The radicals can further cause MS2 bacteriophage and rotavirus inactivation through the indirect exogenous inactivation mechanism,35,37 which is initiated by light absorption via external sensitizers.35,59 If indirect exogenous inactivation dominates, the oocyst inactivation rate should be dependent on the presence of the external sensitizers.22 At 25 °C, we detected 0.004 pM of 1O2 in one experiment containing only oocysts in the solution. For the other experiment with the oocyst only solution, the 1O2 was below the detection limit. At 40 °C, we found 0.008 and 0.012 pM of 1O2 in oocyst only solution. For SRNOM or EfOM solutions free of oocysts, the 1O2 concentration ranged from 0.11 to 0.15 pM and 0.11 to 0.14 pM for 25 °C and 40 °C, respectively. This range of 1O2 concentration is similar to the 1O2 concentrations (0.01 to 0.25 pM) reported by previous studies,22,60 in which bacteriophage MS2 were inactivated by exogenously produced 1O2. In addition, to preserve oocyst infectivity and to use oocysts in environmentally relevant conditions, extensive purification using bleach to eliminate all organic substances from the stool samples of the infected calves was not applied. This residue of organic matter in the oocyst solution was found to produce ˙OH concentrations of 0.26 fM at 25 °C and 0.28 or 0.35 fM at 40 °C. In solutions that contained both the oocysts and the sensitizer, the concentration of ˙OH should be the summation of the ˙OH concentrations produced by both the oocysts and the sensitizer. Thus, the ˙OH concentrations in the inactivation experiments should be 0.5 fM at 25 °C and 0.9 fM at 40 °C. This range of ˙OH concentrations found here is again similar to the previously reported ˙OH concentrations produced by organic matter isolates.60 The absence of enhanced oocyst inactivation in solutions containing organic matter isolates suggested that oocysts cannot be inactivated by exogenously produced radicals. This observation may be attributed to the protection of the oocysts' thick wall. Furthermore, oocysts were inactivated by UVA/visible light at a higher temperature (40 °C) even in the absence of added dissolved organic matter (DOM), indicating the occurrence of internal damages caused by excited internal sensitizers such as RNA and proteins. Those bimolecular reactions are likely enhanced by temperature.38,59 In summary, the dependence of oocyst inactivation by UVA/visible light irradiation on temperature implied the dominant role of indirect endogenous inactivation.
In contrast to the results obtained from batch 1, oocysts from batch 2 experienced no inactivation at 40 °C after exposure to UV fluence as high as 800 kJ m−2, as shown in Fig. 2B. The inactivation kinetics for the dark controls were not significantly different from those obtained from neither the sensitizer-free solution (p = 0.88) nor the solution containing the EfOM (p = 0.38). The observed different susceptibility of the same strain of oocysts from batch 1 and batch 2 indicated that oocyst inactivation by UVA/visible light irradiation could be unpredictable and dependent on the oocyst hosts.
3.2 The effect of the UVB component of solar radiation on oocyst inactivation
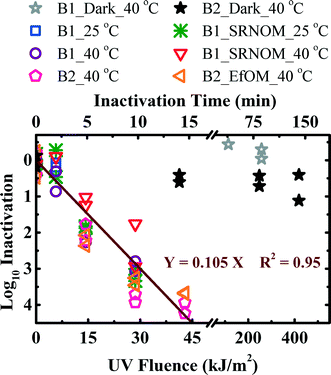
Full-spectrum inactivation was studied in the absence or presence of the organic matter isolates at temperatures of 25 and 40 °C to reveal oocyst inactivation mechanisms by UVB irradiation (Fig. 3). Oocysts from both batch 1 and batch 2 were used in full-spectrum inactivation experiments. In all cases, the oocyst inactivation kinetics were significantly different from the dark control (p ranging from 10−7 to 10−5). The inactivation rate constants (kobs) for all experimental conditions under full-spectrum inactivation varied from 33.92 to 51.25 h−1 (Table 2). The inactivation kinetics of oocysts with and without NOM as an external sensitizer were statistically similar after kobs was adjusted by a light screening correction factor (p = 0.08 and 0.24 for 25 and 40 °C, respectively). Similar to the observation with UVA/visible light irradiation, the negligible effect of NOM on the full-spectrum inactivation of oocysts suggests the dominant role of endogenous inactivation for oocysts. There were two key differences between oocyst inactivation with/without UVB components. First, full-spectrum inactivation was independent of temperature (p = 0.68), while UVA/visible light inactivation was dependent on temperature (p = 10−10 for batch 1, which was sensitive to irradiation). Second, the two batches of oocysts were similarly inactivated under full-spectrum irradiation (p = 0.64) but differently under UVA/visible light irradiation (p = 10−10). Those differences indicated that direct mechanism, which is not dependent on temperature due to the direct damages of DNA,38 is the most important inactivation mechanism when the UVB portion of sunlight is present.| Irradiation | T (°C) | Sensitizer | Batch | k obs (h−1) | |
|---|---|---|---|---|---|
| Without sensitizer | With sensitizer | ||||
| The two values in the table are replicate values from duplicate experiments. All kobs for dark controls were statistically similar (p > 0.14), therefore the average of all kobs for dark controls at the same temperature were listed in this table for simplicity. The kobs for dark controls was averaged from four (a) and six (b) measurements, followed by the standard deviation. | |||||
| Dark controla | 25 ± 1 | — | 1 | −0.01 ± 0.45 | — |
| Dark controlb | 40 ± 1 | — | 1 and 2 | 0.02 ± 0.23 | — |
| UVA/visible | 25 ± 1 | SRNOM | 1 | 0.26, 0.34 | 0.52, 0.47 |
| UVA/visible | 40 ± 1 | SRNOM | 1 | 1.32, 1.43 | 1.32, 1.52 |
| UVA/visible | 40 ± 1 | EfOM | 1 | 2.32, 2.16 | 2.88, 2.85 |
| UVA/visible | 40 ± 1 | EfOM | 2 | −0.05, −0.24 | 0.03, 0.02 |
| Full-spectrum | 25 ± 1 | SRNOM | 1 | 46.15, 50.33 | 44.41, 49.37 |
| Full-spectrum | 40 ± 1 | SRNOM | 1 | 41.38, 42.36 | 42.77, 35.29 |
| Full-spectrum | 40 ± 1 | EfOM | 2 | 42.27, 33.92 | 51.25, 38.33 |
3.3 Role of UV fluence for oocyst inactivation under full-spectrum irradiance
The correlation between the UV fluence and oocyst log10 removal for solar simulator intensity ranging from 250 to 750 W m−2 was established and shown in Fig. 4. The experiments were conducted only at 25 °C, since full-spectrum inactivation was independent of temperature. In addition, irradiance was also concentrated by wrapping aluminum foil around the reactors, as suggested by a previous study.15 The actinometer results from experiments using reactors with and without foil wrapping showed that the aluminum foil concentrated the average irradiance 1.5 to 1.7 times. As shown in Fig. 4, the oocyst log10 inactivation was linearly correlated to UV fluence (log10 inactivation = 0.105 × UV fluence) with R2 = 0.88. The correlation between the log10 inactivation and UV fluence in variable light intensity experiments is similar to the correlation obtained from oocyst inactivation in the absence and presence of external sensitizers, as shown in Fig. 3 (log10 inactivation = 0.105 × UV fluence with R2 = 0.95). In addition, another correlation (log10 inactivation = 0.107 × UV fluence with R2 = 0.90) was also found between oocyst inactivation and UV fluence for all experimental conditions. The positive correlations between oocyst log10 inactivation and UV fluence were obtained for all experiments under full-spectrum irradiation, suggesting that higher UV fluence directly led to greater oocyst inactivation. The direct UVB inactivation mechanism of oocysts in the presence of the UVB component implied that the impact of environmental factors such as material of the container, the presence of NOM, and turbidity on oocyst inactivation may be evaluated by measuring the UV fluence with an actinometer system.3.4 Transcript level response of oocysts to full-spectrum irradiation
The inactivation results described above suggest that direct UVB inactivation dominated under full-spectrum solar irradiation. We further used microarray analysis to determine the response of oocyst physiological regulations on full-spectrum sunlight irradiation. The fold-change in gene expression obtained by the microarray analysis was also confirmed by RT-qPCR analysis. The gene expression fold-change of cgd3_3770, cgd7_3430, and cgd5_1290, after full-spectrum irradiation, were 0.46 (±0.02), 0.39 (±0.02), and 0.36 (±0.06), as determined by the microarray analysis. These fold-change values obtained with the microarray analysis were not significantly different (p > 0.05) from the gene expression fold-change determined by RT-qPCR for cgd3_3770 (0.57 ± 0.12), cgd7_3430 (0.45 ± 0.11), and cgd5_1290 (0.49 ± 0.18).UVB exposure caused overall massive down-regulation in expression, with 1518 genes (41%) significantly down-regulated versus only 36 genes (1%) significantly up-regulated. One explanation for this result is that there is no active transcriptional regulation by the oocysts, but instead it is due to random DNA damage from the UVB, resulting in blocked transcription.22–26 If this were the case, then we would expect the UVB-treated replicates to have larger variation in expression values than the dark controls due to the random nature of the DNA damage. However, we saw similar levels of expression variation in each of the treatments, suggesting that there is active transcription regulation in response to the UVB (Fig. 1). Another prediction of the random DNA damage theory is that longer genes will be more likely to be damaged and thus down-regulated. We did see a significant relationship between gene length and the likelihood of down-regulation, although the data suggest a quadratic rather than a linear relationship (Fig. 5). Specifically, a ~5000 bp gene is 10 times as long as a ~500 bp gene, but less than twice as likely to be down-regulated (~52% down-regulated vs. ~31.5% down-regulated, based on the regression line). Therefore, while random DNA damage from UVB appears to have some effect on down-regulation, it is not the sole cause and active regulation by the oocysts is supported.
Major functional classification is only available for 957 of the 3805 genes on the array (27.3%). These genes account for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 662
662![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 056 bp of length out of the total of 6
056 bp of length out of the total of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 847
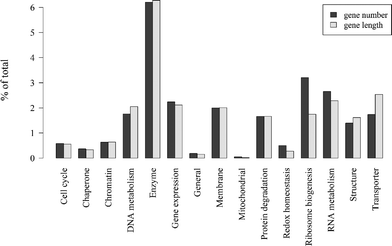
847![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 724 bp across all genes (24.3%), suggesting that having annotation or not is independent of gene length. A breakdown of the major functional classifications based on the proportion of the total number of genes and the proportion of total gene length (Fig. 6) shows that enzyme is the most prevalent functional category by both percentage of gene number (6.2%) and gene length (6.3%). Ribosome biogenesis is the next most abundant category by number (3.2%), but these genes are shorter on average because the proportion of total gene length is lower (1.7%). Conversely, transporter genes are slightly longer on average (1.7% of gene number and 2.5% of gene length). The rest of the functional categories have fairly similar proportions of gene numbers and gene lengths.
724 bp across all genes (24.3%), suggesting that having annotation or not is independent of gene length. A breakdown of the major functional classifications based on the proportion of the total number of genes and the proportion of total gene length (Fig. 6) shows that enzyme is the most prevalent functional category by both percentage of gene number (6.2%) and gene length (6.3%). Ribosome biogenesis is the next most abundant category by number (3.2%), but these genes are shorter on average because the proportion of total gene length is lower (1.7%). Conversely, transporter genes are slightly longer on average (1.7% of gene number and 2.5% of gene length). The rest of the functional categories have fairly similar proportions of gene numbers and gene lengths.
If we divide the genes by significance level and direction of change, those that are non-significant (n = 2251) and those that are significantly down-regulated (n = 1518) have similar proportions of genes with unknown functions (74% and 76%, respectively). Up-regulated genes (n = 36) have a slightly lower percentage of genes with unknown function (64%), which after correcting for gene length is marginally significant (Wallenius non-central hypergeometric test,56p = 0.060). A breakdown of the major functional categories by significance level and direction of change is in Fig. 7. We assessed whether each of the functional categories was over- or under-represented in the up- and down-regulated genes after removing the biases due to gene length noted above. Among the genes that were up-regulated, only those coding for RNA metabolism (p = 0.011) and ribosome biogenesis (p = 0.043) were over-represented, and no functional categories were under-represented. The results for all down-regulated genes are presented in Table 3. Genes coding for transporter and cell cycle were significantly over-represented while genes coding for redox homeostasis and ribosome biogenesis were significantly under-represented (all p < 0.05). Genes coding for RNA metabolism and gene expression were marginally over-represented (0.05 < p < 0.1). The rest of the down-regulated genes were not significantly over- or under-represented.
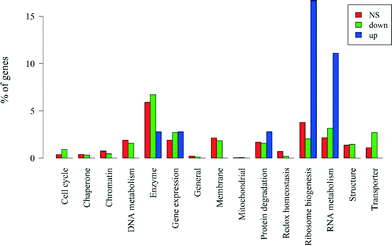 | ||
| Fig. 7 Percentages of the number of non-significant (NS), down-regulated and up-regulated genes in each functional category, including genes with unknown functions. | ||
| Category | Over-represented P-value | Under-represented P-value | Number of genes significantly down-regulated in each category |
|---|---|---|---|
| Transporter | 0.001 | 0.999 | 41 |
| Cell cycle | 0.024 | 0.992 | 14 |
| RNA metabolism | 0.072 | 0.952 | 48 |
| Gene expression | 0.092 | 0.940 | 41 |
| Enzyme | 0.348 | 0.700 | 102 |
| Mitochondrial | 0.525 | 0.903 | 1 |
| Structure | 0.561 | 0.550 | 22 |
| Protein degradation | 0.637 | 0.464 | 24 |
| Chaperone | 0.711 | 0.497 | 5 |
| Membrane | 0.748 | 0.334 | 28 |
| Unknown | 0.767 | 0.256 | 1125 |
| General | 0.800 | 0.484 | 2 |
| Chromatin | 0.878 | 0.234 | 7 |
| DNA metabolism | 0.885 | 0.173 | 24 |
| Redox homeostasis | 0.988 | 0.044 | 3 |
| Ribosome biogenesis | 0.989 | 0.019 | 31 |
Several important stress-related protective agents such as the superoxide dismutase (cgd5_3230) and NADPH (cgd8_2710), which are believed to participate in relieving the oxidative stress, were all down-regulated significantly, suggesting the oocysts were not able to recover from the oxidative stress. This agrees with the observed oocyst inactivation under full-spectrum simulated sunlight.
Almost half of the genes coding for enzymes were down-regulated (40 out of 98); while they were not over- nor under-represented as a group, they included genes involved in the post-translational modification of proteins, including protein kinase, peptidase, and protein phosphatase. This observation suggests that C. parvum AUCP-1 was not able to utilize the protein degradation pathways to reuse amino acids for metabolic purposes, which is an important mechanism for the oocysts to survive in such a low-nutrient environment. About 67 genes related to DNA metabolism were tracked, among which 24 genes were down-regulated and the rest kept the same expression level after solar exposure. It is important to note that 54.2% of these 24 genes are responsible for DNA repair, and 33.3% are associated with DNA replication. This may explain why a mass scale down-regulation was observed, as the expression for DNA repair machinery was not active. In summary, our data showed that while a large proportion of core metabolic pathways were down-regulated in oocysts, a larger amount of genes for enzymes retained their original expression level after the solar exposure.
Differences in gene expression were found between oocysts exposed to full-spectrum simulated solar radiation and to UVC. After either 30 min solar exposure, as studied here, or UVC exposure studied previously,50 genes coding for enzymes and RNA metabolism comprised the largest group among the genes with significant changes in expression levels. In our study, 6 of 13 up-regulated genes with known functions are responsible for RNA metabolism (e.g., RNA binding, RNA modification, RNA splicing), suggesting that RNA repair-associated machinery was activated.50 For oocysts after UVC exposure in a previous study,50 41 genes with known functions were up-regulated; among these genes only 2 were responsible for RNA metabolisms. While we observed 41% (388 of 948 genes) of the targeted genes with known biological functions becoming down-regulated after solar irradiation, only 5.2% (42 of 811 genes) of the targeted genes with known biological functions were down-regulated after UVC irradiation. Most of stress-related genes coding for redox homeostasis remained unchanged after UVC irradiation, while these genes were mostly down-regulated after full solar spectrum irradiation. We also found that only three genes related to thioredoxin became down-regulated after irradiation. We observed that 41 of 66 transporter genes were down-regulated, and none became up-regulated. ABC-transporter and ATPase genes were found among the down-regulated genes. In contrast to our observation, Zhang et al.50 found at least 15 up-regulated transporter genes after 30 min of UVC irradiation. Thus, no significant stress protection mechanisms in oocysts were engaged after 30 min of exposure to full-spectrum radiation, while some repairing machinery was activated shortly after the UVC exposure.
4. Environmental implications
This study systematically investigated the solar inactivation mechanisms for C. parvum oocysts under a wide range of environmental conditions. Oocyst inactivation was controlled by different mechanisms in the presence or absence of the UVB component. UVA/visible light inactivation was dominated by indirect endogenous mechanism, while full-spectrum inactivation involving UVB was dominated by direct UVB inactivation mechanism. Unlike viruses and bacteria, oocyst solar inactivation was independent of the external sensitizer (NOM), likely due to the presence of the oocysts' thick wall. We also found that UVA/visible light inactivation of oocysts was sensitive to the oocyst source and temperature, suggesting the unpredictable efficiency of solar inactivation by UVA/visible light irradiation. In contrast, full-spectrum inactivation was more efficient and independent of the oocyst source, temperature, and the presence of NOM. Because UVA/visible light inactivation is unpredictable, using containers that block the UVB may not ensure oocyst inactivation in SODIS treatment. Increasing the UV fluence of full-spectrum solar irradiation, such as wrapping the bottom of the container with aluminum foil or allowing for longer exposure time, can proportionally improve the efficiency of oocyst inactivation. The role of irradiation on oocyst solar inactivation found here implies that oocysts residing at depth may not be reached by solar UVB due to light-screening by DOM and other water constituents, thus reducing the potential of solar inactivation.Acknowledgements
This work was supported by NSF Career grant #0954501. We acknowledge Dr. Mark Band and the Functional Genomics Unit of the Carver Biotechnology Center at the University of Illinois for microarray hybridizations. Professor Zhu from Texas A& M and Professor Zhou from UC Davis are acknowledged for allowing us to obtain the array design from Agilent. Ofelia Romero-Maraccini is acknowledged for her intellectual input.References
- M. C. Hlavsa, J. C. Watson and M. J. Beach, Morb. Mortal. Wkly. Rep., 2005, 54, 1–8 Search PubMed.
- R. Fayer and L. Xiao, Cryptosporidium and Cryptosporidiosis, CRC Press, New York, 2007 Search PubMed.
- E. Dreelin, R. Ives, S. Molloy and J. Rose, Int. J. Environ. Res. Public Health, 2014, 11, 10480–10503 CrossRef PubMed.
- J. B. Rose, C. P. Gerba and W. Jakubowski, Environ. Sci. Technol., 1991, 25, 1393–1400 CrossRef CAS.
- M. W. LeChevallier, W. D. Norton and R. G. Lee, Appl. Environ. Microbiol., 1991, 57, 2610–2616 CAS.
- M. W. LeChevallier, W. D. Norton, J. E. Siegel and M. Abbaszadegan, Appl. Environ. Microbiol., 1995, 61, 690–697 CAS.
- U.S. Environmental Protection Agency (EPA), Washington, D.C., National primary drinking water regulations: long term 2 enhanced surface water treatment rule. Final rule, Federal Register, 06-00004, 2006.
- P. M. Oates, P. Shanahan and M. F. Polz, Water Res., 2003, 37, 47–54 CrossRef CAS.
- B. King and P. Monis, Parasitology, 2007, 134, 309–323 CrossRef CAS PubMed.
- K. G. McGuigan, R. M. Conroy, H.-J. Mosler, M. D. Preez, E. Ubomba-Jaswa and P. Fernandez-Ibañez, J. Hazard. Mater., 2012, 235–236, 29–46 CrossRef CAS PubMed.
- S. J. Connelly, E. A. Wolyniak, C. E. Williamson and K. L. Jellison, Environ. Sci. Technol., 2007, 41, 7101–7106 CrossRef CAS.
- F. Méndez-Hermida, E. Ares-Mazás, K. G. McGuigan, M. Boyle, C. Sichel and P. Fernández-Ibáñez, J. Photochem. Photobiol., B, 2007, 88, 105–111 CrossRef PubMed.
- H. Gómez-Couso, M. Fontán-Sainz and E. Ares-Mazás, Am. J. Trop. Med. Hyg., 2010, 82, 35 CrossRef PubMed.
- H. Gómez-Couso, M. Fontán-Saínz, C. Sichel, P. Fernández-Ibáñez and E. Ares-Mazás, Trop. Med. Int. Health, 2009, 14, 620–627 CrossRef PubMed.
- M. Fontán-Sainz, H. Gómez-Couso, P. Fernández-Ibáñez and E. Ares-Mazás, Am. J. Trop. Med. Hyg., 2012, 86, 223–228 CrossRef PubMed.
- B. King, D. Hoefel, D. Daminato, S. Fanok and P. Monis, J. Appl. Microbiol., 2008, 104, 1311–1323 CrossRef CAS PubMed.
- F. Méndez-Hermida, J. Castro-Hermida, E. Ares-Mazas, S. Kehoe and K. G. McGuigan, Appl. Environ. Microbiol., 2005, 71, 1653–1654 CrossRef PubMed.
- W. Heaselgrave and S. Kilvington, Acta Trop., 2011, 119, 138–143 CrossRef PubMed.
- H. Gómez-Couso, M. Fontán-Sainz, K. G. McGuigan and E. Ares-Mazás, Acta Trop., 2009, 112, 43–48 CrossRef PubMed.
- K. G. McGuigan, F. Méndez-Hermida, J. A. Castro-Hermida, E. Ares-Mazás, S. C. Kehoe, M. Boyle, C. Sichel, P. Fernández-Ibáñez, B. P. Meyer, S. Ramalingham and E. A. Meyer, J. Appl. Microbiol., 2006, 101, 453–463 CrossRef CAS PubMed.
- H. Gómez-Couso, M. Fontán-Sainz, P. Fernández-Ibáñez and E. Ares-Mazás, Acta Trop., 2012, 124, 235–242 CrossRef PubMed.
- O. C. Romero, A. P. Straub, T. Kohn and T. H. Nguyen, Environ. Sci. Technol., 2011, 45, 10385–10393 CrossRef CAS PubMed.
- D. S. Goodsell, Oncologist, 2001, 6, 298–299 CrossRef CAS PubMed.
- A. Favre, C. Saintomé, J.-L. Fourrey, P. Clivio and P. Laugâa, J. Photochem. Photobiol., B, 1998, 42, 109–124 CrossRef CAS.
- H. Görner, J. Photochem. Photobiol., B, 1994, 26, 117–139 CrossRef.
- M. H. Patrick and J. M. Snow, Photochem. Photobiol., 1977, 25, 373–384 CrossRef CAS PubMed.
- M. B. Fisher, M. Iriarte and K. L. Nelson, Water Res., 2012, 46, 1745–1754 CrossRef CAS PubMed.
- A. R. Marques, F. D. C. O. Gomes, M. P. P. Fonseca, J. S. Parreira and V. P. Santos, Sol. Energy, 2013, 87, 158–167 CrossRef CAS PubMed.
- B. J. King, D. Hoefel, P. E. Wong and P. T. Monis, PLoS One, 2010, 5, e11773 Search PubMed.
- D. Vione, G. Falletti, V. Maurino, C. Minero, E. Pelizzetti, M. Malandrino, R. Ajassa, R.-I. Olariu and C. Arsene, Environ. Sci. Technol., 2006, 40, 3775–3781 CrossRef CAS.
- N. V. Blough and R. G. Zepp, in Active oxygen in chemistry, Springer, 1995, pp. 280–333 Search PubMed.
- A. B. Boehm, K. M. Yamahara, D. C. Love, B. M. Peterson, K. McNeill and K. L. Nelson, Environ. Sci. Technol., 2009, 43, 8046–8052 CrossRef CAS PubMed.
- D. C. Love, A. Silverman and K. L. Nelson, Environ. Sci. Technol., 2010, 44, 6965–6970 CrossRef CAS PubMed.
- T. Kohn and K. L. Nelson, Environ. Sci. Technol., 2007, 41, 192–197 CrossRef CAS.
- A. I. Silverman, B. M. Peterson, A. B. Boehm, K. McNeill and K. L. Nelson, Environ. Sci. Technol., 2013, 47, 1870–1878 CrossRef CAS PubMed.
- R. G. Zepp, T. Callaghan and D. Erickson, J. Photochem. Photobiol., B, 1998, 46, 69–82 CrossRef CAS.
- O. C. Romero-Maraccini, N. J. Sadik, S. L. Rosado-Lausell, C. R. Pugh, X.-Z. Niu, J.-P. Croué and T. H. Nguyen, Environ. Sci. Technol., 2013, 47, 11004–11012 CrossRef CAS PubMed.
- S. Li, M. Paulsson and L. O. Björn, J. Photochem. Photobiol., B, 2002, 66, 67–72 CrossRef CAS.
- J. K. Johnson, J. Schmidt, H. B. Gelberg and M. S. Kuhlenschmidt, J. Parasitol., 2004, 90, 980–990 CrossRef CAS PubMed.
- F. M. Schets, G. B. Engels, A. During and A. A. D. Husman, Appl. Environ. Microbiol., 2005, 71, 6793–6798 CrossRef CAS PubMed.
- P. C. Davidson, T. B. Kuhlenschmidt, R. Bhattarai, P. K. Kalita and M. S. Kuhlenschmidt, Water, Air, Soil Pollut., 2014, 225, 1–12 CrossRef CAS.
- Z. Alexieva, H. Yemendzhiev and P. Zlateva, Biodegradation, 2010, 21, 625–635 CrossRef CAS PubMed.
- W. R. Haag, E. Gassman and A. Braun, Chemosphere, 1984, 13, 631–640 CrossRef CAS.
- J. Kochany and J. R. Bolton, J. Phys. Chem., 1991, 95, 5116–5120 CrossRef CAS.
- J. Neter, W. Wasserman and M. H. Kutner, Applied Linear Statistical Models: Regression, Analysis of Variance, and Experimental Designs, CRC Press, Boston, MA, 1990 Search PubMed.
- J. L. Zimmer, R. M. Slawson and P. M. Huck, Water Res., 2003, 37, 3517–3523 CrossRef CAS.
- A. Leifer, The kinetics of environmental aquatic photochemistry: theory and practice, American Chemical Society, 1988 Search PubMed.
- D. Dulin and T. Mill, Environ. Sci. Technol., 1982, 16, 815–820 CrossRef CAS PubMed.
- R. G. Zepp, Environ. Sci. Technol., 1978, 12, 327–329 CrossRef CAS.
- H. Zhang, F. Guo, H. Zhou and G. Zhu, BMC Genomics, 2012, 13, 647 CrossRef CAS PubMed.
- The R Project for Statistical Computing, Institute for Statistics and Mathematics of WU (Wirtschaftsuniversität Wien). 2013 Search PubMed.
- G. K. Smyth, J. Michaud and H. S. Scott, Bioinformatics, 2005, 21, 2067–2075 CrossRef CAS PubMed.
- G. K. Smyth and T. Speed, Methods, 2003, 31, 265–273 CrossRef CAS.
- G. K. Smyth, Stat. Appl. Genet. Mol. Biol., 2004, 3, 3 Search PubMed.
- Y. Benjamini and Y. Hochberg, J. R. Stat. Soc. B, 1995, 57, 289–300 Search PubMed.
- M. Young, M. Wakefield, G. Smyth and A. Oshlack, Genome Biol., 2010, 11, R14 CrossRef PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- U. o. I. a. C. RRC Core Genomics Facility, Endogenous Control Primer Sequences:, http://www.uic.edu/depts/rrc/cgf/realtime/primerseq.html.
- R. J. Davies-Colley, A. M. Donnison and D. J. Speed, Water Sci. Technol., 2000, 42, 149–158 CAS.
- S. L. Rosado-Lausell, H. Wang, L. Gutiérrez, O. C. Romero-Maraccini, X.-Z. Niu, K. Y. H. Gin, J.-P. Croué and T. H. Nguyen, Water Res., 2013, 47, 4869–4879 CrossRef CAS PubMed.
Footnote |
| † Author contributions: these authors contributed equally. Liu, Y. contributed to oocyst solar inactivation kinetic study and Dong, S. contributed to damage analysis of oocysts on the genetic level. |
| This journal is © The Royal Society of Chemistry 2015 |