Inactivation kinetics and mechanisms of viral and bacterial pathogen surrogates during urine nitrification†
Heather N.
Bischel
a,
Ariane
Schertenleib
a,
Alexandra
Fumasoli
b,
Kai M.
Udert
b and
Tamar
Kohn
*a
aLaboratory of Environmental Chemistry, School of Architecture, Civil and Environmental Engineering (ENAC), École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland. E-mail: tamar.kohn@epfl.ch
bEawag: Swiss Federal Institute of Aquatic Science and Technology, 8600 Dübendorf, Switzerland
First published on 4th December 2014
Abstract
This paper assesses the inactivation performance and mechanisms in urine nitrification reactors using bacteria and bacteriophages as surrogates for human pathogens. Two parallel continuous-flow moving bed biofilm reactors (MBBRs) were operated over a two-month period. One MBBR was used to conduct a continuous spike experiment with bacteriophage MS2. The second reactor provided the matrix for a series of batch experiments conducted to investigate the inactivation of Salmonella typhimurium, Enterococcus spp., MS2, Qβ, and ΦX174 during urine nitrification. The roles of aeration, biological activity, and solution composition in inactivation were evaluated. Whereas bacteriophages ΦX174 and MS2 remained infective following urine nitrification, partial inactivation of bacteriophage Qβ was observed. Qβ inactivation was attributed primarily to aeration with a potential additive effect of biological processes, i.e., processes that are attributable to the presence of other microorganisms such as sorption to biomass, predation or enzymatic activity. Tailing of Qβ inactivation to a plateau indicated a protective effect of the solution components in aerated nitrification reactors. In contrast to the bacteriophages, S. typhimurium and Enterococcus spp. were mainly affected by biological processes: they were inactivated in biologically active nitrification reactors while remaining stable in chemically equivalent filtered controls. The tested bacteria could, for example, be out-competed by other microbial communities or sorbed to biomass in the reactor. Microbial communities did not adapt to inactivate bacteriophage MS2 (e.g., via increased prevalence of virus predators) in the experimental time-scale evaluated, with no observed inactivation of MS2 during continuous input for 51 days in the flow-through MBBR. The compilation of these results suggests that biological nitrification as a fertilizer production process remains insufficient as a stand-alone technology for the sanitization of source-separated urine.
Water impactRecently, nitrification has been applied at pilot scales for the production of agricultural fertilizers from urine. While little is known regarding the inactivation of bacterial and viral pathogens during nitrification, such information is instrumental in evaluating the hygiene and safety of the nitrification end product. This study quantitatively assesses pathogen surrogate inactivation performance and mechanisms in urine nitrification reactors using two bacteria (Salmonella typhimurium and Enterococcus spp.) and three indicator viruses (MS2, ΦX174, and Qβ). Further, the influence of physical (i.e., aeration) and biological (i.e., microbial activity) processes on inactivation as well as the role of chemical solution characteristics of nitrified urine was assessed. Results indicate that biological nitrification is insufficient as a stand-alone technology for the sanitization of source-separated urine. |
Introduction
Nutrients excreted in urine have long been used as a fertilizer for agricultural applications of sewage sludge1 and wastewater2,3 or more directly through the application of source-separated urine.4,5 Urine contains the major fraction of nutrients found in human excreta: 80–90% of the nitrogen, 55–67% of the phosphorus and 50–80% of the potassium.4,6,7 Harvested urine not only provides value as an alternative fertilizer, but its use could also reduce pollution from unsafe excreta disposal, lower the costs of wastewater treatment, and lessen the ecological burden of fertilizer production and surplus use of chemical fertilizers.6,8Due primarily to the fecal contamination of source-separated urine,9 handling of urine and direct application as fertilizer in agriculture can pose microbial health risks.10 Storage of urine for 6 months at 20 °C is recommended prior to handling and usage in order to reduce or eliminate the risks.11 However, in the scale-up of urine nutrient recovery such storage time requirements are prohibitively long. Alternative urine treatment and nutrient stabilization technologies are now available to produce marketable fertilizers.12,13 A promising nutrient recovery process that yields a chemically stable solution from stored urine is nitrification.14 In configurations where nitrification is applied in combination with evaporation, a concentrated solution that preserves nearly all nutrients in the urine can be produced.15 Pilot reactors for combined nitrification/distillation systems are operated at eThekwini Water and Sanitation in Durban, South Africa and at the Swiss Federal Institute of Aquatic Science and Technology (Eawag) in Dübendorf, Switzerland as part of the Valorisation of Urine Nutrients in Africa (VUNA) project.13
Nitrification of urine, i.e., the oxidation of ammonia (NH3) to nitrate (NO3−), prevents ammonia volatilization, enhancing the recovery of nitrogen in urine. Two groups of nitrifying bacteria are involved in this process: ammonia oxidizing bacteria (AOB) and nitrite oxidizing bacteria (NOB). Nitrification typically oxidizes half of the NH3 content of urine. In addition, it reduces the pH of urine from 9 to around 6, which causes the remaining NH3 to become protonated to the nonvolatile NH4+ (pKa = 9.25).14 Concomitantly to nitrification, heterotrophic bacteria consume biologically degradable organic substances so that malodor is removed as well.
Refinement of urine nitrification processes presents several challenges. At a technical level, nitrifying bacteria are sensitive to environmental conditions and the intermediate product nitrite.15 From a public health perspective, the pathogen inactivation or removal efficacy during urine nitrification is unknown. Such information is instrumental in assessing the hygiene and safety of the nitrification end product (fertilizer) and to minimize health risks to reactor operators. The present study aimed to establish and operate bench-top urine nitrification reactors to evaluate the inactivation of viral and bacterial pathogen surrogates during this nutrient recovery process. Inactivation kinetics for five organisms were established and compared, and the main modes of inactivation were identified.
Two types of nitrification reactors were operated in this study: two 6.5 L continuous flow moving bed biofilm reactors (MBBR) and several smaller-volume batch MBBRs. In MBBRs, floating plastic carriers are used to provide surfaces for biofilms. The biofilm prevents washing out of slow growing bacteria such as nitrifiers and thereby allows high volumetric conversion rates without the need for membrane filtration or biomass recirculation.16 MBBRs are currently used for urine nitrification in Durban and Dübendorf.13 The use of several MBBRs in this study allowed evaluation of several bacteriophages and bacteria under field-relevant as well as varied experimental conditions.
Inactivation was evaluated using gram negative and gram positive bacteria (Salmonella typhimurium and Enterococcus spp., respectively) as well as three bacteriophages (MS2, Qβ, and ΦX174) that served as surrogates for human enteric viruses. The Salmonella genus consists of rod-shaped flagellated facultative anaerobes of the family Enterobacteriaceae. S. typhimurium, one of thousands of non-typhoidal serotypes of the medically important species S. enterica, causes most cases of non-typhoidal Salmonella across Africa.17–19Enterococcus spp. is a common fecal indicator bacteria, selected because of its slightly longer persistence in urine relative to gram negative indicator organisms.20
The inclusion of virus surrogates is particularly important in this study as viruses are expected to persist much longer than gram negative and non-spore forming gram-positive bacteria in stored urine10,21 and are thus anticipated to be infective in the influent of field urine nitrification reactors. The selection of several bacteriophages also facilitates an evaluation of the effect of urine nitrification on viruses with a range of characteristics. MS2 and Qβ are positive-sense single-stranded RNA bacteriophages that infect Escherichia coli. MS2 and Qβ are structurally similar, small in size (21–29 nm diameter), and frequently used as models for enteric viruses.22 With an isoelectric point of 3.9 and 5.3, respectively,23 MS2 and Qβ are expected to be negatively charged in urine or nitrified urine. ΦX174 is a single-stranded DNA bacteriophage with a diameter of 27 nm and an isoelectric point of 6.6.23 Because of its low hydrophobicity and high stability against many environmental stressors,24 including stored urine,20 ΦX174 has been suggested as a model for more conservative virus inactivation.
Using the test organisms described above and varied continuous-flow and batch reactor conditions, the specific objectives of this study were to (1) assess the inactivation of gram negative and gram positive bacteria during urine nitrification, (2) similarly evaluate the inactivation of several bacteriophages as surrogates for human viruses, (3) elucidate the roles of physical and biological processes as well as the chemical solution composition of urine nitrification reactors in the inactivation of test organisms. Results are anticipated to inform the understanding of pathogen inactivation during urine nitrification and more broadly for other applications of nitrification for wastewater treatment.
Materials and methods
Materials
Approximately 25 L of nitrified urine and 10 L of Kaldnes® carriers with active biofilm were obtained from a 120 L urine nitrification reactor operating at Eawag and were stored at 4 °C until use in the laboratory reactors. Urine (100 L) to feed reactors was collected from the men's NoMix storage tank at Eawag and stored at 4 °C. Autoclaved phosphate-buffered saline (PBS; 5 mM PO42−, 10 mM NaCl, pH 7.5) was used for storage of bacteriophage stocks; PBS was adjusted to pH 6.1 using HCl for batch reactor experiments.Bacteriophage
Bacteriophages MS2 (DSMZ 13767), ΦX174 (DSMZ 4497), and Qβ (DSMZ 13768) and their respective bacterial hosts E. coli (DSMZ 5695 for MS2 and Qβ and DSMZ 13127 for ΦX174) were used. Bacteriophage stocks were prepared and enumerated by the double-layer agar method as described previously.25 The stock was conserved in PBS at 4 °C, and the volume of stock used to spike the batch reactors was <1% of the reactor volume. Phage concentrations are reported as plaque forming units (pfu) per mL. A PBS blank and an E. coli host blank were plated at each time point.Bacteria
An attenuated derivative of Salmonella typhimurium (strain SL1344) was generously provided by the Microbiology Department of Eawag. Isolates were grown to log phase in LB broth with 100 μg mL−1 ampicillin and stored in aliquots with 15% glycerol at −80 °C. S. typhimurium was grown overnight from storage in ampicillin-containing lysogeny broth immediately prior to use in experiments. Bacteria were pelleted and resuspended in PBS prior to reactor spiking (spike volume was <1% of reactor volume). The spread-plate method with 100 μL sample on ampicillin containing agar was used for enumeration and concentrations are reported as colony forming units (cfu) per mL. Spike concentrations were selected to be more than four orders of magnitude above low background concentrations of ampicillin resistant colonies detected in unspiked nitrified urine. Enterococcus spp. colonies were isolated from wastewater treatment plant influent (Vidy, Lausanne) on Bile Esculin Agar (Sigma-Aldrich, St. Louis, MO, USA) and subsequently grown in azide glucose broth (Sigma-Aldrich). Aliquots of log-phase growth Enterococcus spp. were stored with 15% glycerol at −80 °C. Enterococcus spp. from stored aliquots were grown overnight in azide glucose broth immediately prior to use in batch experiments. 100 μL of sample and 50 mL of PBS were filtered using the membrane filtration EPA method 1600 (ref. 26) for enumeration. A PBS blank was plated at each time point.Continuous flow MBBRs
Two bench-top 6.5 L MBBRs made of PVC were operated in parallel (Fig. 1). Material from reactor 1 served as the matrix for batch MBBR experiments described below, and reactor 2 was continuously spiked with MS2 to evaluate potential phage inactivation during nitrification. Reactors were seeded with the contents of an active urine nitrification reactor (i.e., nitrified urine and Kaldnes® K1 carriers with biofilm from Eawag) such that nitrifying organisms were in place at initiation. The filling ratio of the reactor was 50%, yielding a total approximate biofilm carrier surface area of 1.63 m2 per reactor, as determined by the specific surface area of the Kaldnes® (460 m2 m−3) carriers.Stored urine (9.0 ± 0.1) was the sole input to the reactor, serving as the ammonia source for nitrifying bacteria, the organic input for heterotrophic bacteria, and to balance the decrease in pH that occurs with ammonia nitrification. To control the urine input, a proportional-integral-derivative (PID) controller was configured for each reactor using a Liquisys M CPM 223 regulator (Endress+Hauser AG, Reinach, Switzerland) that regulated the inflow based on the pH in the reactor. pH was measured with a pH ISFET combination electrode (Endress+Hauser). The PID parameters (Kp, tmin) of the regulator were empirically determined to yield a rapid response to pH changes and to maintain the pH target of 6 to 6.1 (pH set-point, pH = 6.1; proportional gain, Kp = 3; minimal length of response, tmin = 0.1 s). The minimal length of response allowed at least one drop of urine input in the reactor per urine input event. The reactor was aerated with humidified air through a 125 mm Hobby Flexi Diffuser (Saint Vincent Group, Dubai, UAE). PVC tubing was used to deliver the urine input and moistened air within the reactor. To reduce evaporation and prevent escape of foam, the top of the reactors was plugged. Effluent from the reactor surface was continuously captured into a separate storage container.
Monitoring of physiochemical parameters in continuous flow MBBRs
The pH and dissolved oxygen (DO) in the reactors were monitored and recorded once per minute using an Ecograph T RSG35 data logger (Endress+Hauser). The DO probes (Mettler-Toledo, Greifensee, Switzerland) were configured to separate COM223F regulators (Endress+Hauser). pH and DO probes were calibrated weekly, according to the manufacturer's instructions. The weights of influent and effluent tanks were recorded daily to determine the flow rate. When nitrified urine and biofilm carriers were removed from the reactor for batch experiments, the same volume of material was replaced using the nitrified urine stock stored at 4 °C and acclimated to room temperature. Temperature was recorded daily from the pH transmitter. Nitrite was monitored approximately once per week using Nitrite Test strips (Merck Millipore, Darmstadt, Germany) to avoid nitrite accumulation in the reactors. Hach-Lange cuvette tests were used to monitor ammonium (LCK 303), nitrite (LCK 342), total nitrogen (LCK 338) and chemical oxygen demand (COD; LCK 614) in reactor influent and effluent. For chemical analysis, 18 mL samples were taken and filtered through 0.45 μm cellulose nitrate filters (Albet LabScience, Dassel, Germany), discarding the first half of the filtrate prior to analysis according to the manufacturer's instructions. Samples were diluted with Milli-Q water to within the appropriate measurement range and measured with a DR 3900 spectrophotometer (Hach Lange GMBH, Düsseldorf, Germany). The volume, weight, date and time of samples were recorded. Measurements of the input stored urine (Table 2) were consistent with other analysis of urine from the NoMix men's storage tank at Eawag.15,27 Unless otherwise noted, results are presented as average values with standard deviations (SD).Continuous MS2 input to continuous flow MBBR
Reactor 2 was amended continuously with MS2 for 51 days. The initial MS2 concentration was forced to steady-state by first injecting a high concentration pulse followed by a reduced concentration continuous spike delivered via a syringe pump and a headspace-free syringe. This was achieved by first adding 1010 pfu mL−1 MS2 at 0.001 mL min−1 for 1 hour followed by a continuous input of 107 pfu mL−1 MS2 at 0.001 mL min−1. The syringe was refilled every 2–5 days. To monitor the inflow concentration, MS2 concentrations were measured in a side-by-side reference solution of the spike solution kept at the same room temperature as the syringe used to amend reactor 2. The reference solution MS2 concentration showed agreement with the syringe MS2 concentrations and allowed more frequent input concentration measurements (data not shown).The concentration of a microorganism with first order removal kinetics in a continuous flow reactor can be calculated assuming a perfectly mixed system of constant total reactor volume, V (6.5 L), according to the rate of change mass balance equation:
 | (1) |
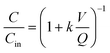 | (2) |
Batch and semi-batch MBBRs
To test the inactivation of bacteriophages and bacteria under varying experimental conditions, a series of 500 mL semi-batch and batch MBBRs were established in washed and autoclaved 1 L Pyrex glass bottles (Fisher Scientific, Reinach, Switzerland). Batch reactors refer to reactors with neither an influent flow nor an outflow, while semi-batch hereafter refers to reactors that received influent for the duration of the experiment but did not have an outflow. When biofilm carriers were added to the reactors, a filling ratio of 50% was used. Reactors were covered with parafilm to reduce loss by evaporation. Five types of reactors (Table 1) were evaluated for all target organisms, with Qβ and ΦX174 tested together in one series and MS2 tested with bacteria in a separate series. The main reactor conditions and parameters studied were given as follows: actively nitrifying urine reactors vs. filtered nitrified urine controls (to evaluate the effect of biological activity), aerated reactors vs. non-aerated reactors (effect of aeration), and urine reactors vs. PBS reactors (effect of matrix composition).| Reactor type | Description | Composition |
|---|---|---|
| Nitrified urine MBBR semi-batch | Biologically active system: conducted in duplicate for all test organisms | Reactor content: 500 mL of nitrified urine; 250 mL of active biofilm carriers |
| Reactor input: stored urine | ||
| Aeration: yes | ||
| Filtered nitrified urine semi-batch | Filtered control: to test the role of solution composition relative to biological activity | Reactor content: 500 mL of 0.45 μm-filtered nitrified urine; 250 mL of clean biofilm carriers |
| Reactor input: 0.45 μm-filtered nitrified urine | ||
| Aeration: yes | ||
| Nitrified urine batch | Aeration control: to test the role of aeration when compared with nitrified urine MBBR semi-batch | Reactor content: 500 mL of nitrified urine |
| Reactor input: none | ||
| Aeration: none | ||
| Phosphate-buffered saline (PBS) aerated batch | Clean aerated matrix control: to compare inactivation in aerated nitrified urine matrix to a clean PBS matrix and to evaluate the role of aeration in a clean matrix | Reactor content: 500 mL of PBS; 250 mL of clean biofilm carriers |
| Reactor input: none | ||
| Aeration: yes | ||
| PBS batch | Clean matrix control: to determine baseline inactivation at the experimental temperature, to compare inactivation in non-aerated nitrified urine matrix to a clean PBS matrix, and to evaluate the role of aeration in a clean matrix | Reactor content: 500 mL of PBS |
| Reactor input: none | ||
| Aeration: none |
The baseline data set was obtained from replicate actively nitrifying semi-batches that were seeded with the contents of the continuous flow reactor 1. Reactors were aerated and received urine input over 8 days. The influent flow rate was empirically determined to maintain a pH of 6.1 to 6.2. Reactor material for the ΦX174 and Qβ semi-batches was removed from reactor 1 on day 40 when the reactor 1 nitrification rate was 0.479 gN m−2 per day (day 39). Material for the MS2 and bacteria semi-batches was obtained on day 70; the nitrification rate on day 65 was 0.267 gN m−2 per day. The reactor nitrification rate per surface area (rn, gN m−2 per day) was calculated as:
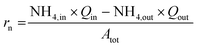 | (3) |
To disentangle the effect of biological activity from that of solution composition on inactivation, filter-sterilized controls with aeration were established for comparison with actively nitrifying reactors. These biologically inactive semi-batch reactors contained new biofilm carriers (cleaned with bleach and rinsed several times with sterile water) and were filled with 0.45 μm-filtered nitrified urine from the continuous flow MBBR. The same filtered nitrified urine also served as the influent at the rate established for the urine inflow to the active nitrification semi-batch.
To assess the physical role of gas bubbles on inactivation (i.e., aeration), a nitrified urine control without inflow, biofilm carriers or aeration was tested for comparison with the actively nitrifying, aerated semi-batch. Some biological activity was expected to be sustained in these reactors, although at lower rates than the aerated nitrifying semi-batch.
Finally, PBS batch controls either with clean biofilm carriers and aeration or without both were tested to compare inactivation in nitrified urine to that in a simple matrix (represented by PBS).
In the tests containing nitrifying bacteria, pH and nitrite concentrations were measured daily to verify the batch reactor stability. For the remaining reactors, the pH and temperature were measured at the beginning of the experiment, and pH stability was verified at the end of the experiment. For ΦX174 and Qβ, additional PBS batch tests without biofilm carriers were conducted with and without aeration to verify observations. For Qβ, additional replicate nitrified urine batch tests without urine input or biofilm carriers were conducted with and without aeration. Material for these tests was obtained from reactor 1 when the nitrification rate was 0.218 gN m−2 per day.
Data analysis
First-order inactivation rate constants (k, per day) were determined from a least-square linear regression of data from each batch and semi-batch experimental condition according to the equation: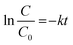 | (4) |
Results and discussion
The operation stability of laboratory urine nitrification reactors established in this study is first reported for comparison with field nitrification activities, followed by a discussion of the inactivation of pathogen surrogates observed in batch, semi-batch and continuous reactors.Continuous flow MBBR performance
The pH, DO, temperature and nitrification rates of the two continuous-flow urine nitrification reactors are presented in Fig. S1 (reactor 1) and S2 (reactor 2).† Average measured physical and chemical parameters are shown in Table 2. Approximately half of the total ammonia in urine was oxidized under optimum reactor conditions, indicating active biological nitrification. This is consistent with the literature: a maximum of 50% of ammonia in urine was converted to nitrate in either a MBBR, a continuous flow stirred reactor, or a sequencing batch reactor.14 Heterotrophic bacteria degraded approximately 90% of the COD in the influent. The average pH in the reactors was stable at 6.05 (SD < 0.01) throughout the operating and experimental period, except for short periods of electronic malfunction and during the first several days of operation when urine input was limited to prevent nitrite accumulation in the reactor. pH is an important parameter influencing AOB and NOB activity.14 DO was also relatively stable throughout operation.| Reactor 1 | Reactor 2 | |||
|---|---|---|---|---|
| Parameter | Influent (Avg. ± SD) | Reactor content (Avg. ± SD) | Influent (Avg. ± SD) | Reactor content (Avg. ± SD) |
| a Averages for pH, temperature, nitrification rate (rn), hydraulic retention time (HRT) and volume losses for the reactor contents are calculated from the data presented in Fig. S1–S3. The average (Avg.) and standard deviation (SD) are based on n = 8 measurements for the influent and other reactor content measurements. b NM = not measured; nitrite concentrations were below detection in the initial tests of the urine influent and are usually negligible in stored urine. c Higher apparent Ntot in the effluent can be explained in part by water loss in addition to measurement accuracy. d N/A = not applicable. | ||||
| NO2 [mg L−1] | NMb | 1.37 ± 0.43 | NM | 1.27 ± 0.30 |
| NH4,tot [mg L−1] | 3760 ± 180 | 1980 ± 130 | 3570 ± 300 | 1850 ± 250 |
| N tot [mg L−1]c | 3970 ± 440 | 4270 ± 330 | 3780 ± 170 | 4250 ± 150 |
| COD [mg L−1] | 3870 ± 61 | 419 ± 72 | 3890 ± 260 | 401 ± 70 |
| COD/N [mg O2 mg−1 N] | 1.04 ± 0.12 | 0.09 ± 0.01 | 1.06 ± 0.08 | 0.09 ± 0.01 |
| pH [−] | 9.04 ± 0.09 | 6.05 ± < 0.01 | 9.04 ± 0.1 | 6.05 ± < 0.01 |
| Temperature [°C] | NM | 19.1 ± 1.8 | NM | 20.2 ± 1.3 |
| r n [gN m−2 per day] | N/Ad | 0.56 ± 0.14 | N/A | 0.50 ± 0.10 |
| HRT [day] | N/A | 20.7 ± 8.2 | N/A | 17 ± 3.7 |
| Losses [vol%] | N/A | 1.1 ± 0.9 | N/A | 1.6 ± 0.9 |
The average nitrification rate in continuous flow reactors over the course of operation was 0.56 ± 0.14 gN m−2 per day in reactor 1 and 0.49 ± 0.10 gN m−2 per day in reactor 2, lower than the reported maximum nitrification rate of 1.7 gN m−2 per day in a 2.8 L reactor.14 Over time the nitrification rate also decreased. The lower nitrification rate may be explained by temperature: the average temperature in both MBBRs was approximately 5 °C lower than the reported temperature of operating reactors in the literature.15 Temperature is an important factor influencing the growth of nitrifiers; a 5 °C decrease in temperature could reduce the biomass activity of AOB by 25%.28 However, temperature measurements were taken only once per day, limiting a detailed analysis of this effect. Furthermore, the salt concentrations were higher than in the reactor from which the inoculum originated. High salt concentrations are known to inhibit nitrifying bacteria.29 The greatest decrease in nitrification rate appeared in reactor 1, where a substantial fraction of the reactor material (approximately 2 L of nitrified urine and 1 L of biofilm carriers) was removed for batch experiments and replaced by stored material. As the nitrification rate decreased in the reactors, a lower urine input flow rate was required to maintain the set pH. Therefore the input and output flow rates decreased over time in both reactors, and changes were more pronounced in reactor 1 than those in reactor 2 (Fig. S3†).
Bacteria and bacteriophage inactivation in semi-batch and batch reactors
Inactivation of bacteriophages MS2, ΦX174 and Qβ as well as the bacteria Enterococcus spp. and S. typhimurium was tested over 7 to 10 days in semi-batch MBBRs containing either active nitrifying biofilm carriers or filtered nitrified urine with clean biofilm carriers (Table 1, Fig. 2). Enterococcus spp. and S. typhimurium underwent 4–5 log inactivation over 7 days in the active biological batch, compared to no inactivation in the filtered control. While ΦX174 and MS2 were not or only minimally inactivated in either the active nitrification system or the filtered nitrified urine, infective Qβ concentrations decreased by 3–4 logs in both reactor types.Inactivation of target organisms was also tested in aerated batch reactors containing only PBS and clean biofilm carriers, without any input (Fig. 3). Only Qβ was inactivated with k > 0.5 per day in repeat aerated PBS batch tests. For an unknown reason, the concentration of ΦX174 decreased substantially in one aerated PBS control. In 10 other batch or semi-batch tests for ΦX174, including duplicate aerated PBS control batches (Table S1†), inactivation of ΦX174 was minimal. This outlier was therefore excluded from the subsequent rate calculation and discussion.
Finally, inactivation of all test organisms was monitored in nitrified urine and PBS batch reactors without aeration (Fig. 3). In biologically active nitrified urine held without aeration, Enterococcus spp. and S. typhimurium exhibited inactivation similar to aerated semi-batches. The bacteria were stable or exhibited comparably low inactivation rates in PBS without aeration. Qβ also exhibited modest inactivation in nitrified urine without aeration and was relatively stable or slowly inactivated in PBS without aeration. MS2 and ΦX174 were stable in nitrified urine or only slowly inactivated in PBS without aeration.
First-order inactivation rate constants (k) were calculated for all semi-batch and batch systems (Tables 3 and S1†). Reproducibility between replicate active nitrification semi-batch reactors was high; for each of the three bacteriophages and two bacteria, there was no significant difference within 95% confidence limits between first-order inactivation rates measured in replicate reactors. The inactivation curve observed for Qβ in aerated nitrified urine reactors consisted of an initial exponential decrease and a secondary plateau. The first-order rate constants (k) were therefore calculated for the initial decrease that occurred in the first 4 days of the experiment.
| Biologically active nitrified urine semi-batches [per day]b | Filtered nitrified urine semi-batch, aerated [per day] | Nitrified urine semi-batch, not aerated [per day] | PBS batch, aerated with biofilm carriers [per day] | PBS batch, not aerated, without biofilm carriers [per day] | |
|---|---|---|---|---|---|
| a Standard error of regression for the slope coefficient (k) determined from the log-transformed culturable fraction versus time, based on n data points. b Average of semi-batch results reported with range and R2 for each replicate experiment. c NS = not significantly different from zero at a 95% confidence level. d Calculated k for the first 4 days of the biologically active nitrified urine semi-batches and the filtered nitrified urine semi-batch. | |||||
| MS2 | NSc | 0.17 ± 0.04 (n = 6; R2 = 0.79) | NS | NS | 0.13 ± 0.04 (n = 6; R2 = 0.69) |
| Qβd | 1.77 (1.46–2.04) (n = 6, 5, 3, 3; R2 = 0.62, 0.74, 0.97, 0.996) | 1.88 ± 0.32 (n = 6; R2 = 0.90) | 1.50 ± 0.07 (n = 6; R2 = 0.99) | 1.82 ± 0.24 (n = 8; R2 = 0.91) | 0.39 ± 0.09 (n = 7; R2 = 0.81) |
| ΦX174 | NS | NS | NS | 0.38 ± 0.01 (n = 3; R2 = 0.999) | 0.39 ± 0.01 (n = 7; R2 = 0.99) |
| S. typhimurium | 1.42 (1.31–1.52) (n = 5, 6; R2 = 0.83, 0.85) | NS | 1.50 ± 0.30 (n = 5; R2 = 0.89) | NS | NS |
| Enterococcus spp. | 0.92 (0.91–0.92) (n = 5, 6; R2 = 0.91, 0.83) | NS | 0.71 ± 0.15 (n = 6; R2 = 0.85) | 0.35 ± 0.09 (n = 6; R2 = 0.79) | 0.27 ± 0.02 (n = 6; R2 = 0.98) |
A comparison of the inactivation kinetics observed in the different reactors allowed us to identify the main modes of bacteria and virus inactivation during nitrification. Specifically, we could assess (1) the physical role of the air–water interface, (2) the role of a biologically active bacterial community, and (3) the role of chemical solution conditions. These mechanisms are discussed below, and a recapitulation of the primary observed modes of inactivation is presented in Table 4.
| Mode of inactivation | MS2 | ΦX174 | Qβ | S. typhimurium | Enterococcus spp. |
|---|---|---|---|---|---|
| a A protective effect of the chemical matrix was observed for Qβ during aeration, leading to tailing of the inactivation curve. This effect could also be relevant for MS2 and ΦX174 but could not be observed due to the overall lack of inactivation of these two bacteriophages. | |||||
| (1) Physical effect of the air–water interface | No | No | Yesa | No | No |
| (2) Presence of biologically active community | No | No | Possible | Yes | Yes |
| (3) Chemical matrix effects | No | No | No | No | No |
Inactivation at the air–water interface
Aeration of the MBBRs is essential in order to maintain dissolved oxygen levels, as well as to provide mixing of the solution. Previous studies have found that viruses can become inactivated at the air–water interface.30–32 In a study of bacteriophages MS2 and ΦX174 at the air–water interface (AWI), Thompson et al.30 propose that loss of infectivity occurs when hydrophobic regions of the virus capsid partition out of solution into the gas phase via reconfiguration of the capsid proteins. More precisely, inactivation was shown to occur at the triple-phase-boundary at the interface of air, liquid and solid, and inactivation is influenced by both the hydrophobicity of the solid phase30,31 and the surface properties of the virus.32 In the present study, aerated reactors in glass bottles also contained polyethylene biofilm carriers, PVC tubing and air diffusers, providing several surface characteristics at which the liquid–air–solid interface may be formed. Aeration, therefore, may lead to inactivation by increasing the air–water–solid interface compared to non-aerated reactors.This assumption was evaluated by comparing inactivation in aerated and non-aerated nitrified urine semi-batches or PBS control batches (Fig. 3). Little to no difference in Enterococcus spp. or S. typhimurium inactivation was observed between aerated and non-aerated batches of similar solution and biological conditions, indicating that the physical presence of air bubbles did not affect bacteria viability. Similarly, little to no difference in the infectivity of ΦX174 and MS2 was observed between aerated and unaerated nitrified urine or PBS reactors.
In contrast, aeration did cause inactivation of Qβ in both nitrified urine and PBS (with or without biofilm carriers present). In PBS, 4- to 5-log inactivation was observed over 6 days in reactors with aeration, compared to 1-log inactivation in PBS without aeration. In nitrified urine, significant inactivation was observed both in the presence and absence of aeration (Fig. 3 and Table S1†). Inactivation of Qβ can therefore not be explained by aeration alone. However, aeration caused the initial inactivation to proceed at a markedly faster rate, leading to a 3- to 4-log decrease in infective Qβ over the course of 4 days. Interestingly, the inactivation rate slowed down after approximately 4 days in the aerated reactors that contained nitrified urine (either unfiltered or filtered), leading to a tailing inactivation curve. This feature was not observed in any other Qβ experiments. A tailing inactivation curve has been observed previously in phage disinfection kinetics and, for disinfection of MS2 by ClO2, was recently attributed to deposition of protective material on the phage exterior.33 Therefore, while the aeration of nitrified urine is initially the primary cause of Qβ inactivation, it may also facilitate the creation and deposition of protective material for Qβ or other viruses, protecting them from complete inactivation.
To rationalize why Qβ was susceptible to aeration whereas MS2 and ΦX174 were not, their surface properties must be considered. From the literature, it is known that viruses containing hydrophobic regions on the capsid are more sensitive to AWI inactivation.31,32 While highly similar in structure to MS2, Qβ is more hydrophobic than MS2,34 a characteristic likely contributing to its susceptibility to inactivation at the triple-phase boundary. In the context of urine nitrification, aeration may thus contribute to the inactivation of viruses with hydrophobic capsids, although protection from inactivation may limit the extent of inactivation. In contrast, more hydrophilic viruses and bacteria appear to be resistant to AWI inactivation.
Role of biological treatment in the inactivation of pathogen surrogates
In biological treatment systems, several physiochemical and biological processes can lead to pathogen inactivation. In activated sludge wastewater treatment, viruses and bacteria can be adsorbed on sludge flocs.35,36 Pathogens may also be out-competed by active biological communities. For example, the regrowth of S. typhimurium has been suppressed by indigenous microflora in biologically active compost relative to sterilized compost.37 Predation of viruses or bacteria by protozoa or other sludge microbes as well as enzymatic activity can also inactivate pathogens.38–40 A comparison of actively nitrifying semi-batches with semi-batches containing filtered nitrified urine (i.e., no bacteria larger than 0.45 μm) facilitated evaluation of the role of microbial activity in the inactivation of target organisms during nitrification.The concentrations of Enterococcus spp. and S. typhimurium were stable in the absence of bacteria (i.e., in filtered nitrified urine). On the contrary, inactivation was observed in nitrification batches, reaching 3-log reduction for Enterococcus spp. and 5-log inactivation for S. typhimurium over 6 days (Fig. 2). Biological activity therefore had an effect on the survival of bacteria. Because of the similarities between temperature, aeration, pH and other solution conditions between the two systems, the resulting difference in inactivation is likely attributable to biological processes relevant within the experimental time frame, such as competition for nutrients with the indigenous organisms, sorption to Kaldnes® biofilms and predation. Inactivation of both S. typhimurium and Enterococcus spp. was similar between aerated and non-aerated (unfiltered) nitrified urine, despite the expectation of reduced biological activity in the non-aerated batch. This indicates that adsorption to biomass or enzymatic activity, rather than competition in growth, may have played important roles. Additionally, because the HRT of field nitrification reactors is expected to be shorter than the duration of batch studies conducted, competition is expected to be less important. Further study is required to evaluate the relative contribution of different biological processes to inactivation.
Concentrations of infective MS2, Qβ and ΦX174 followed the same evolution in both aerated biologically active and filtered systems (Fig. 2). This result indicates that the biological activity in the nitrification reactors did not cause bacteriophage inactivation, and inactivation of Qβ was largely attributable to aeration. However, inactivation of Qβ was also observed in non-aerated nitrified urine held without biofilm carriers (Fig. 3), indicating an additional inactivating effect of biological activity on Qβ. This finding was surprising because biological activity was expected to be reduced in the absence of aeration. Qβ inactivation reached 4–5 logs in 8 days for the unaerated system with a starting nitrification rate of 0.5 gN m−2 per day, while inactivation reached only 2–3 logs in 7 days in the unaerated system with a starting nitrification rate of 0.2 gN m−2 per day, suggesting that higher microbial activity may lead to more inactivation. This corresponded to an inactivation rate constant in the batch with higher microbial activity (Table 3) that was approximately twice that of the lower microbial activity batch (Table S1†). Therefore, while aeration appears to be a primary mode of inactivation for Qβ, inactivation in unaerated nitrified urine batch controls is likely due to biological processes of suspended microbial communities not attached to biofilm carriers or to degradation by proteolytic enzymes present in the unaerated nitrified urine batch.
To evaluate the ability of the biological community to adapt to inactivate persistent bacteriophage, MS2 was continuously spiked into the continuous MBBR over 51 days. Measured MS2 concentrations in the continuous-flow MBBR mirrored the expected concentration of a modeled conservative tracer added with equivalent influent concentrations and no degradation (k = 0 in eqn (1), Fig. 4). The difference between measured MS2 and modeled tracer concentrations was less than 0.5 log over the course of the experiment, indicating that little to no MS2 was lost due to adsorption to the reactor or to inactivation. This is consistent with little to no inactivation of MS2 observed in all batch and semi-batch reactors. Because inactivation of MS2 was not enhanced through time, the biological community facilitating nitrification and organic degradation in the MBBR did not adapt to alter MS2 infectivity within the experimental time scale. It was postulated that the microbial community could adapt with MS2 as a continuously added substrate. Bacteria, protozoa or other organisms can engulf viruses or release virucidal agents, so the long-term input of MS2 could favor the growth of these organisms and lead to increased MS2 inactivation. However, the time-scale over which microbial communities change in response to new substrates is highly variable. This effect could be further evaluated after months or even several years of exposing an operating nitrification reactor to different (pathogen surrogate) substrates.
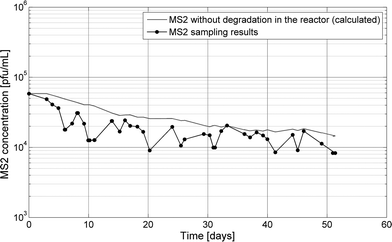 | ||
| Fig. 4 MS2 was spiked continuously in a continuous flow MBBR for 51 days. A tracer was modeled in the reactor using the measured MS2 input concentrations and reactor flow rates. | ||
Solution chemistry matrix effects
In stored urine, the three key parameters governing pathogen inactivation are free ammonia (NH3) activity, pH and temperature.9,20,21,27,41 NH3 is a known biocide for most organisms, as is high pH.42 The survival time of bacteria and viruses in urine declines with increasing temperatures.20,21 Gram-negative bacteria are generally more rapidly inactivated in stored urine than gram-positive bacteria, and viruses are typically more persistent than both. In this study, NH3 concentrations were reduced by microbial oxidation and pH was lowered from that of the influent, yielding less detrimental conditions for the test microorganisms following nitrification than during urine storage at high pH.To evaluate the role of the bulk nitrified urine solution composition on target organism inactivation, aerated batches containing PBS were compared to aerated nitrified urine and filtered nitrified urine. In the chemically complex solutions (i.e., nitrified urine and filtered nitrified urine, Table 3), the inactivation of bacteriophage was either comparable to or less pronounced than in the buffer (Tables 3 and S1†). As was observed for Qβ, the solution could also provide a protective coating for the other phage, but this was not further evaluated due to the lack of overall inactivation of MS2 and ΦX174.
The concentrations of Enterococcus spp. and S. typhimurium decreased substantially in active nitrification reactors but were unchanged in filtered nitrified urine and relatively stable in PBS. This suggests that the sole mode of inactivation during nitrification was biological processes, and there was no additional effect of the matrix composition. Further, the solution did not provide protection for bacteria as observed for Qβ.
While the temperature was not specifically controlled in the continuous or batch MBBRs (laboratory temperature, ~20 °C), batch controls in PBS conducted at the same temperature as urine batch tests indicated little additional inactivation effect of temperature for the duration of the experiment. Phage and bacteria remained relatively stable in unaerated PBS (Fig. 3). In summary, the nitrified urine solution composition and experimental temperature had little biocidal effect on bacteria or bacteriophage and, in the case of Qβ, may instead contribute to protection of the virus from complete inactivation during aeration.
Implications for urine nitrification applications
While results of batch and semi-batch MBBRs cannot be extrapolated directly to the continuous flow MBBRs because several parameters were different (e.g., aeration rate, reactor dimensions and material), they permit evaluation of several inactivation mechanisms for the bacteria and bacteriophage and can inform further research with continuous flow MBBRs (Table 4). Bacteriophages ΦX174 and MS2 were more resistant to inactivation during urine nitrification than Qβ or tested bacteria. The presence of active nitrification relative to controls inactivated the tested bacteria but did not directly affect bacteriophages. Conversely, bacteriophages may be protected by macromolecules or particles generated during aeration in nitrified urine, as observed for Qβ. This protective or tailing effect was evident for Qβ only and not for the tested bacteria. Qβ was sensitive to aeration in batch reactors, while MS2, ΦX174 and bacteria were not.In further development of nitrification for the production of fertilizers from source-separated urine, it is anticipated that nitrification will provide inactivation capacity for bacterial pathogens but viruses may remain infective following treatment. For example, field-scale nitrification reactors established in the VUNA project have a HRT of 3 to 6 days. Under these conditions, assuming steady state of the reactor has been reached (eqn (2)), and applying first-order inactivation rates presented in Table 3, S. typhimurium and Enterococcus spp. are expected to undergo 0.7 to 1-log and 0.6 to 0.8-log inactivation, respectively. Bacteriophage Qβ could reach 0.8 to 1.1-log removal if no protective effect of the matrix is assumed, while no treatment benefit is expected for MS2 or ΦX174. The persistence of these viruses raises concern for the treatment capacity of urine nitrification for human viruses and therefore its ability to improve the hygiene of urine fertilizer production.
Nitrification of urine removes a significant amount of the biocidal effect afforded by ammonia in stored urine. The inactivation of bacteria and viruses could be enhanced via longer storage of urine prior to nitrification, but stabilization of the urine for nutrient recovery remains important. Additionally, because some viruses as well as spore-forming bacteria are known to persist in stored urine, even with extended storage times, downstream treatment of nitrified urine would be necessary to inactivate such pathogens. Distillation for example, although energy intensive, provides the production benefit of concentrating the liquid nitrified urine fertilizer and producing a clean water by-product, while also conferring a pathogen treatment benefit.
Acknowledgements
We thank our funding sources: the US National Science Foundation International Research Fellowship Program (grant no. 1159225), the Swiss National Science Foundation (grant no. 200021_146829/1), and the Bill and Melinda Gates Foundation. This study was part of the research project VUNA – Promoting sanitation and nutrient recovery through urine separation.References
- Sewage sludge: Land utilization and the Environment, ed. C. E. Clapp, W. E. Larson and R. H. Dowdy, American Society of Agronomy, Inc., Madison, WI, 1994 Search PubMed.
- H. N. Bischel, G. L. Simon, T. M. Frisby and R. G. Luthy, Environ. Sci. Technol., 2012, 46, 180–188 CrossRef CAS PubMed.
- Water reuse for irrigation: agriculture, landscapes, and turf grass, ed. V. Lazarova and A. Bahri, CRC Press, 2004 Search PubMed.
- T. Karak and P. Bhattacharyya, Resour., Conserv. Recycl., 2011, 55, 400–408 CrossRef PubMed.
- M. Johansson, H. Jönsson, C. Höglund, A. Richert Stintzing and L. Rodhe, Final Rep. R&D Proj. Source-Separated Hum. Urin. – a Futur. source Fertil. Agric. Stock. Reg., 2001 Search PubMed.
- H. Heinonen-Tanski and C. van Wijk-Sijbesma, Bioresour. Technol., 2005, 96, 403–411 CrossRef CAS PubMed.
- T. A. Larsen and W. Gujer, Water Sci. Technol., 1996, 34, 87–94 CrossRef CAS.
- K. M. Udert and M. Wächter, Water Res., 2011, 46, 453–464 CrossRef PubMed.
- C. Schönning, R. Leeming and T. A. Stenström, Water Res., 2002, 36, 1965–1972 CrossRef.
- C. Höglund, T. A. Stenström and N. Ashbolt, Waste Manage. Res., 2002, 20, 150–161 CrossRef PubMed.
- WHO, Guidelines for the safe use of wastewater, excreta and greywater. Volume 4 excreta and greywater use in Agriculture, World Health Organization, 2006, vol. 4 Search PubMed.
- M. Maurer, W. Pronk and T. A. Larsen, Water Res., 2006, 40, 3151–3166 CrossRef CAS PubMed.
- K. M. Udert, C. A. Buckley, M. Wächter, C. S. McArdell, T. Kohn, L. Strande, H. Zöllig, A. Hug, A. Oberson and B. Etter, in WISA Biennial Conference, Mbombela, Mpumalanga, South Africa, 2014 Search PubMed.
- K. M. Udert, C. Fux, M. Münster, T. A. Larsen, H. Siegrist and W. Gujer, Water Sci. Technol., 2003, 48, 119–130 CAS.
- K. M. Udert and M. Wächter, Water Res., 2012, 46, 453–464 CrossRef CAS PubMed.
- E. Morgenroth, in Biological Wastewater Treatment, Principles, Modelling and Design, ed. M. Henze, M. C. M. van Loosdrecht and G. A. Ekama, WA Publishing, London, UK, 2008 Search PubMed.
- E. L. Hohmann, Food Saf., 2001, 32, 263–269 CAS.
- M. Mcclelland, K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, N. Grewal, E. Mulvaney, E. Ryan and H. Sun, Nature, 2001, 413, 852–856 CrossRef CAS PubMed.
- N. A. Feasey, G. Dougan, R. A. Kingsley, R. S. Heyderman and M. A. Gordon, Lancet, 2012, 379, 2489–2499 CrossRef.
- B. Vinneras, A. Nordin, C. Niwagaba and K. Nyberg, Water Res., 2008, 42, 4067–4074 CrossRef PubMed.
- C. Höglund, N. Ashbolt, T. A. Stenström and L. Svensson, Adv. Environ. Res., 2002, 6, 265–275 CrossRef.
- J. Langlet, F. Gaboriaud, J. F. L. Duval and C. Gantzer, Water Res., 2008, 42, 2769–2777 CrossRef CAS PubMed.
- S. E. Dowd, S. D. Pillai, S. Wang and M. Yavuz, Appl. Environ. Microbiol., 1998, 64, 405–410 CAS.
- J. F. Schijven and S. M. Hassanizadeh, Crit. Rev. Environ. Sci. Technol., 2000, 30, 49–127 CrossRef CAS.
- B. M. Pecson, L. V. Martin and T. Kohn, Appl. Environ. Microbiol., 2009, 75, 5544–5554 CrossRef CAS PubMed.
- USEPA, Method 1600: Enterococci in Water by Membrane Filtration Using membrane-Enterococcus Indoxyl-β-D-Glucoside Agar (mEI), 2009 Search PubMed.
- L. Decrey, K. M. Udert, E. Tilley, B. M. Pecson and T. Kohn, Water Res., 2011, 45, 4960–4972 CrossRef CAS PubMed.
- C. Hellinga, M. C. M. van Loosdrecht and J. J. Heijnen, Math. Comput. Model. Dyn. Syst., 1999, 5, 351–371 CrossRef.
- J. H. Hunik, H. J. G. Meijer and J. Tramper, Appl. Microbiol. Biotechnol., 1992, 37, 802–807 CrossRef CAS.
- S. S. Thompson, M. Flury, M. V. Yates and W. A. Jury, Appl. Environ. Microbiol., 1998, 64, 304–309 CAS.
- S. S. Thompson and M. V. Yates, Appl. Environ. Microbiol., 1999, 65, 1186–1190 CAS.
- T. Trouwborst, S. Kuyper, J. C. de Jong and A. D. Plantinga, J. Gen. Virol., 1974, 24, 155–165 CrossRef CAS PubMed.
- T. Sigstam, A. Rohatschek, Q. Zhong, M. Brennecke and T. Kohn, Water Res., 2014, 48, 82–89 CrossRef CAS PubMed.
- C. Dika, M. H. Ly-Chatain, G. Francius, J. F. L. Duval and C. Gantzer, Colloids Surf., A, 2013, 435, 178–187 CrossRef CAS PubMed.
- T.-D. Kim and H. Unno, Water Sci. Technol., 1996, 33, 243–250 CrossRef CAS.
- S. Soda, M. Ike and M. Fujita, J. Biosci. Bioeng., 1999, 87, 513–518 CrossRef CAS.
- J. Sidhu, R. A. Gibbs, G. E. Ho and I. Unkovich, Water Res., 2001, 35, 913–920 CrossRef CAS.
- R. L. Ward, Appl. Environ. Microbiol., 1982, 43, 1221–1224 CAS.
- C. van der Drift, E. van Seggelen, W. Hol and J. Tuinte, Appl. Environ. Microbiol., 1977, 34, 315–319 CAS.
- M. D. Sobsey and J. S. Meschke, World Health Organization Meeting September 23–25, 2003, Rome, Italy, 2003 Search PubMed.
- A. Chandran, S. K. Pradhan and H. Heinonen-Tanski, J. Appl. Microbiol., 2009, 107, 1651–1657 CrossRef CAS PubMed.
- W. N. Cramer, W. D. Burge and K. Kawata, Appl. Environ. Microbiol., 1983, 45, 760–765 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Contains results from the monitoring of physiochemical parameters in continuous flow MBBRs as well as inactivation rate constants calculated for supplementary batch tests for ΦX174 and Qβ. See DOI: 10.1039/c4ew00065j |
| This journal is © The Royal Society of Chemistry 2015 |



