Pyrosequencing revealed highly microbial phylogenetic diversity in ferromanganese nodules from farmland†
Min
Hu
a,
Fangbai
Li
*a,
Jing
Lei
b,
Yuan
Fang
b,
Hui
Tong
acd,
Weijian
Wu
acd and
Chengshuai
Liu
a
aGuangdong Key Laboratory of Agricultural Environment Pollution Integrated Control, Guangdong Institute of Eco-Environmental and Soil Sciences, Guangzhou 510650, P. R. China. E-mail: cefbli@soil.gd.cn; Fax: +86 20 87024123; Tel: +86 20 87024721
bAgricultural College of Guangxi University, Nanning 530004, P. R. China
cGuangzhou Institute of Geochemistry, Chinese Academy of Sciences, Guangzhou 510640, P. R. China
dGraduate University of The Chinese Academy of Sciences, Beijing 100039, P. R. China
First published on 11th December 2014
Abstract
There is renewed interest in the origin and makeup of ferromanganese nodules (FMNs), long known to soil mineralogists as unusual secondary minerals. However, new evidence suggests that microorganisms play a significant role in the generation of FMNs. The biogenic origin of nodules has remained elusive because until recently, little has been known about the overall microbial community structure in their microbiota. To learn more about the microbial community and to determine the relative abundance, diversity, and composition of the microbial communities present in FMNs and their surrounding soil, we used pyrosequencing to investigate 16S rRNA genes obtained from vertical soil profiles of both paddy fields and sugarcane fields. Using pyrotaq 16S rRNA gene sequencing, we show that the microbial phylogenetic diversity of nodules was higher than those reported in previous studies of this biosphere, and we identified many previously unidentified microorganisms. Here, we show that the microbial community of these nodules is dominated by Burkholderiales, Rhodocyclales, Acidobacteriales, Desulfuromonales, and Clostridiales, and there were no statistically significant differences found when comparing the microbial community structures of FMNs obtained from vertical soil sequences. Although the microbial composition was markedly different between the surrounding soil and the FMNs, the microbes found within the FMNs were very similar to other FMNs from both field types examined here. In addition to their geochemical properties and the microbial community composition of FMNs, we found that the levels of iron (Fe), manganese (Mn), and SiO2 greatly impact the microbial diversity among FMN communities. Our results indicate that the FMN microbial communities from different land-use types are very similar and suggest that natural selection of these microbes is based on the oligotrophic conditions and the high metal content. Researching FMNs in these two land-use patterns, which represent two different redox potentials, deepens our understanding of Fe–Mn biogeochemical cycling in these oligotrophic biospheres and suggests a biogenetical origin for these nodules.
Environmental impactAs unusual secondary minerals, ferromanganese nodules (FMNs) are ideal materials for researching the Fe–Mn element biogeochemical cycling in nature. The previous studies of FMNs were focused on their mineral composition or structure. However, new evidence suggests that microorganisms play a significant role in the generation of FMNs. In this study, we used pyrosequencing to investigate the relative abundance, diversity, and composition of the microbial communities present in FMNs and their surrounding soil, which were collected from vertical soil profiles of both paddy fields and sugarcane fields. We are also sure that this article will not only provide first-hand information of biogenetical origin for FMNs, but also generate a lot of interest among researchers involved in the interaction between microbes and minerals. |
1. Introduction
Abundant transition metals, iron and manganese, are widely distributed in nature and actively participate in electron-exchange reactions in the environment.1 Soil samples obtained from southern China often contain large quantities of iron and manganese, which undergo a rapid cycling between their reduced and oxidized states, which promote other element cycles in this environment.2 These alternative redox cycles are driven by the diffusion of soluble Fe(II)/Mn(II) in a reducing environment, followed by the precipitation of new solid Fe(III)/Mn(IV) minerals in the oxygenated regions.3 In addition, variations in oxygen diffusion and water content caused by differences in soil depth induce different redox potentials in the vertical horizon, which facilitates the Fe–Mn biogeochemical cycling in these soil profiles.4Because of the ion exchange reactions that precipitate ore components from water, ferromanganese nodules (FMNs) in the natural environment are often found at the boundary between the hydrosphere and the lithosphere (drying–wetting alternation region), where drastic oxidation and reduction cycles occur.5 FMNs were first discovered on the ocean floor, but plough soils such as paddy fields have a high level of anthropic activity, which provides a unique environment for the generation of FMNs. Here, the soil is consistently flooded during the main stages of rice growth and the fields are only left to drain after harvesting the rice.6 Flooding fields results in soil reduction of the plow layer, and the resultant Fe(II)/Mn(II) ions in the plow layer are leached by water percolation and accumulate in the subsoil depth, all of which are finally oxidized after the rice fields are drained.7 Therefore, FMNs from plowed soils such as paddy fields and sugarcane fields provide a unique opportunity to probe the mechanisms behind Fe–Mn biogeochemical cycling in an artificial environment.
In nature, FMNs are widely distributed and have a high redox activity; FMNs are considered a critical model to elucidate the mechanisms of Fe–Mn element cycling in natural environments. Previous studies have focused on the mineralogy8,9 or geochemistry10–13 of the FMNs. However, the unusual mineralogy of FMNs suggests that they are not simply created by the dissolution and leaching of iron/manganese minerals leaving insoluble materials behind. Many iron or manganese-oxidizing/reducing heterotrophic bacteria have been isolated from FMNs,14–16 demonstrating that microorganisms play significant roles in FMN genesis.17,18 Previous research studies suggest that a surprisingly phylogenetically diverse population of microorganisms are present on or inside the FMNs derived from terrestrial or aquatic biospheres.6,15,19,20 These pioneering studies demonstrate that Acidobacteria, Proteobacteria, Firmicutes and Nitrospira bacterium dominate the microbial phylotypes in FMNs.19,21 However, the rare archaeal biosphere of FMNs has not been explored in the previous studies, which is mainly due to the low through-put analytical methods applied. The isolation of Fe- or Mn- depositing microorganisms from the FMN environment also revealed that Burkholderia (e.g., Leptothrix) and Bacillus genus-affiliated bacteria widely participate in the Fe–Mn biogeochemicals of such biospheres.22 However, studies of the spatial distribution, diversity, and abundance of the microbial communities found in the FMNs derived from different land use patterns and soil profiles are still limited.
In this study, pyrosequencing of 16S rRNA genes (targeting bacteria and Archaea) was used to investigate the microbial phyla distribution in 13 FMN–soil pair samples, which were taken from the vertical soil horizon in both a paddy field and a sugarcane field in an Fe–Mn rich region in China. Then, the difference in microbial diversity between FMNs and the associated surrounding soil was compared using several diversity indices. More specifically, by comparing the profiles of the microbial community structure and the pertinent element geochemistry of FMNs in the vertical soil sequences, we determined which abiotic factors influenced the abundance, diversity, and composition of microbial communities in FMNs. Combined with similar analyses of the associated surrounding soil, this study provides an overall view of the microbial composition found in FMNs, including rare microbes, which led us to further elucidate the microbial community members that are involved in the Fe–Mn biogeochemical cycling of soil.
2. Materials and methods
2.1. Sample collection and the analysis of geochemical properties
Several kilograms of soil blocks, which contained red-brown FMNs, were collected from two sites located in Laibin City, Guangxi Province, South China during October 2011. One soil profile sample was taken from paddy soil (23°20′N, 109°31′E) with a 0–200 cm horizon depth and the other was from a sugarcane field (23°42′N, 109°38′E) with 0–100 cm vertical horizon sequences. The paddy soil and sugarcane field samples used in this study are all belong to latosolic red soil, which are widely distributed in Guangxi province, China. There were 9 and 4 soil block samples collected from vertical horizons for analyses in both the paddy field and sugarcane field (20 cm depth interval). Totally, 26 samples (13 FMMs and 13 surrounding soil samples) were measured by the downstream geochemical and molecular analysis. Samples for biological analysis were collected into sterile 50 ml polypropylene tubes and shipped on ice to the laboratory where they were stored at −40 °C. In the laboratory, approximately 15 g FMNs from sugarcane fields (ca. 1 cm in diameter) and paddy soil (0.1–0.5 cm in diameter) (ESI Fig. S1†) were carefully picked up with autoclaved tweezers and the soil attached to the nodules was separated. The nodules were immersed into distilled water and sonicated for 1 min to disperse clinging soil particles from the nodules. This washing step was repeated three times to obtain clean FMNs. The air-dried FMNs and surrounding soil were ground using a mortar and pestle until the entire sample passed through a 100-mesh sieve. The following geochemical characteristics were determined for each sample and used in the subsequent statistical analyses: pH, OM (organic matter), Fed (dithionite extractable), Feo (oxalate extractable), Fep (pyrophosphate extractable), Fe(II), Fe(III), total Fe, Mnd (dithionite extractable), total Mn and weathering related materials (Fe2O3, SiO2, Al2O3). The geochemical composition of the FMNs and the surrounding soil was determined as described previously.2 The mineralogy of the FMNs was determined using powder X-ray diffraction (XRD) with a Bruker D8 diffractometer (Bruker Co., Ltd.) equipped with a Cu X-ray tube operated at 40 kV and 40 mA. Scans were collected from 10° to 90° 2θ-angles, with a step size of 0.02° and a counting time of 1 s per step. Phase identification was executed by matching XRD patterns with powder diffraction files (PDFs) in the database published by the International Centre for Diffraction Data (ICDD).2.2. DNA extraction from FMNs and the surrounding soil
Genomic DNA extraction from the FMNs and the surrounding soil was carried out, as previously described with slight modifications.19,23 Briefly, prior to genomic DNA extraction, the FMNs were intensively washed with distilled water and sterilized with a 0.1% NaClO solution for 1 min, rinsed three times with sterile distilled water, and then ground into a powder using a mortar and pestle. Then, genomic DNA was extracted from a 1.0 g sample of the ferromanganese powder using a PowerSoil DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA) according to the manufacturer's instructions. The DNA was eluted using 100 μL of TE buffer (10 mmol L−1 Tris–HCl (8.0), 1 mmol L−1 EDTA) and stored at −40 °C. This genomic DNA extraction was performed in triplicate for each sample. The extracts were run on a 1% agarose gel and the quantity of the eluted DNA was estimated using the Qubit® dsDNA BR (Broad Range) Assay Kit (Invitrogen, Merelbeke, Belgium) in combination with a Qubit® 2.0 Fluorometer (Invitrogen, Merelbeke, Belgium).2.3. 16S rRNA PCR and 454 pyrotag sequencing
Analysis of the 16S rRNA genes was performed as previously described.24 Briefly, the eubacterial sets of two primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACVSGGGTATCTAAT-3′) were applied to amplify the 16S rRNA gene that was designed to amplify the hypervariable V3–V4 region of the 16S rRNA gene from nearly all bacteria and archaea.25 The forward primer contained the Titanium A adaptor (5′-CGTATCGCCTCCCTCGCGCCATCAG-3′) and the reverse primer contained the Titanium B adaptor (5′-CTATGCGCCTTGCCAGCCCGCTCAG-3′). 8-bp Hamming barcodes that allow sample multiplexing during pyrosequencing were added to the 5′ end of primer 515F. Three replicates of DNA extract from each sample were amplified by PCR. The PCR was carried out with a PCR thermal cycler Model C1000 (Bio-rad Rad, Richmond, CA). The total volume of the reaction mixture was 50 μL containing 0.5 μL of each primer (50 pmol each), 5 μL of 2.5 mmol L−1 dNTP mixture, 5 μL of 10× Ex Taq buffer (20 mmol L−1 Mg2+; TaKaRa Inc., Dalian, China), 0.25 μL of Ex Taq DNA polymerase (TaKaRa), 1 μL of environmental DNA template and 37.75 μL Milli-Q water. Cycle conditions for the PCR amplification were as follows: initial denaturation at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s and extension at 72 °C for 30 s, and 5 min at 72 °C extension step after cycling was complete. All samples were amplified in triplicate and no template controls were included in all steps of the process. Next, 5 μl of each reaction mixture was analyzed on a 2% agarose gel and PCR products were gel-purified with a QIAquick Gel Extraction Kit (Qiagen). The purified PCR amplicons were combined in equimolar ratios into a single tube after the concentration of each amplicon was determined using the Quant-iT PicoGreen dsDNA reagent kit (Life Technologies, Merelbeke, Belgium) and sequenced by pyrosequencing using primer A on a 454 Life Sciences Genome Sequencer FLX (Roche Diagnostics, Indianapolis, IN, USA) machine following Titanium Chemistry at Macrogen Inc. (http://www.macrogen.com, Seoul, South Korea).2.4. Bioinformatics analyses
Firstly, all 454 community 16S rRNA sequences were denoised (homopolymer error-correction) using Denoiser V0.91 software26 according to the manual. Using QIIME-1.5.027, short reads were removed and only sequences longer than 200 bp were included in the downstream analysis, and low-quality reads were removed from the analysis using default settings (average quality value of > 20). Additionally, sequences containing >6 nt ambiguous nucleotides (“N”) in homopolymeric regions were removed. Sequences were then assigned to each sample by 8 bp barcode using a script derived from the QIIME pipeline. Secondly, the remaining sequences from all of the samples were clustered into Operational Taxonomic Units (OTUs) at 97% sequence similarity with the “uclust” model (search and clustering orders of magnitude faster than BLAST). The representative sequences in each OTU were assigned to taxonomic groups using the RDP classifier28 and comparing against the SILVA database29 within an 80% confidence threshold. Across all 13 nodule–soil pairs, we obtained 318![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 017 high quality sequences with an average read length of 285 bp after quality checking and denoising.26 The number of sequences per sample ranged from 10
017 high quality sequences with an average read length of 285 bp after quality checking and denoising.26 The number of sequences per sample ranged from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 293 to 22
293 to 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 161, with an average of 13
161, with an average of 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 827. Out of these sequences, 255, 251 (80.26%) were classified below the phylum level with 70% confidence or greater, using the RDP classifier28 and by comparing the sequences against the SILVA database.29 These sequences were affiliated with over 34 phyla across the entire dataset. When we grouped sequences into operational taxonomic units (OTUs) at the 3% dissimilarity level (roughly corresponding to the species level), there were 36
827. Out of these sequences, 255, 251 (80.26%) were classified below the phylum level with 70% confidence or greater, using the RDP classifier28 and by comparing the sequences against the SILVA database.29 These sequences were affiliated with over 34 phyla across the entire dataset. When we grouped sequences into operational taxonomic units (OTUs) at the 3% dissimilarity level (roughly corresponding to the species level), there were 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 018 OTUs in the complete dataset with an average of 1762 OTUs per sample.
018 OTUs in the complete dataset with an average of 1762 OTUs per sample.
Lastly, to estimate alpha diversity, alignment of the sequences was conducted by PyNAST.30 A random sub-sampling method from each sequence library was performed and used for microbial community diversity index calculations to control the effects of library size.31 Alpha diversity index (PD, Chao1 and observed species) analyses were performed for all samples with 100 repetitions using a step size of 5000 sequences per sample. For beta diversity analysis, all samples were also subsampled to 5000 sequences per sample to remove all possible side effects of sample size. Pairwise unweighted UniFrac distances calculated for the total community were analyzed in PRIMER v6.32 The principal coordinate analysis (PCoA) was performed on pairwise unweighted UniFrac distances33 using the QIIME software package. The DNA sequences were deposited in the MG-RAST database (http://metagenomics.anl.gov/)34 with access numbers: 4502145.3.
2.5. Statistical analyses
Correlation between the microbial community diversity and environmental variables was estimated through Pearson correlation coefficients using the statistical software package SYSTAT 18 (SPSS, Inc.). Within each soil type, the divergence in microbial composition between ferromanganese nodules and surrounding soil was tested using the independent-samples t-test. Correlations between the community composition and environmental parameters were also analyzed by CCA (canonical correspondence analysis) using the program Canoco35 to determine whether the community structure is more strongly related to specific environmental variables than expected by chance.3. Results
3.1. The geochemistry of FMNs and their surrounding soil
The geochemical characteristics of all 13 nodule–soil pairs (4 pairs from the sugarcane field and 9 pairs from the paddy field in two vertical soil profiles) are presented in Table 1. The average pH value of the sugarcane field was 4.87, while that of the paddy field was 6.39. Not surprisingly, the mean Fed (dithionite extractable), total Fe and Fe2O3 contents of FMNs (average 228, 328 and 469 g kg−1, respectively, for the sugarcane field; 184, 308 and 440 g kg−1, respectively, for the paddy field) were significantly higher than those in the associated soil (average 84.2, 89.2 and 127 g kg−1, respectively, for the sugarcane field; 66.2, 86.1 and 123 g kg−1, respectively, for the paddy field); the contents of the oxalate extractable Fe, the pyrophosphate extractable Fe, Fe(II) and Fe(III) in both FMNs and their corresponding soils were in a similar range. The SiO2 and Al2O3 contents of FMNs (average 170 and 186 g kg−1, respectively, for the sugarcane field; 133 and 204 g kg−1, respectively, for the paddy field) were notably lower than the associated surrounding soils (average 367 and 276 g kg−1, respectively, for the sugarcane field; 497 and 210 g kg−1, respectively, for the paddy field). The total Mn was highly enriched in the FMNs (most of the Mn was dithionite extractable; average 0.17 g kg−1 for the sugarcane field and 1.87 g kg−1 for the paddy field) compared with the Mn in soil (average 0.08 g kg−1 for the sugarcane field and 0.34 g kg−1 for the paddy field). XRD studies revealed that the mineralogical makeup of these FMNs differs extensively between the sugarcane and paddy fields (ESI Fig. S2†): hematite [Fe2O3], kaolinite [Al2(OH)4Si2O5] and cronstedtite [Fe3FeSiO4(OH)5] were the predominated minerals in the sugarcane field samples, while quartz [SiO2], kaolinite, cronstedtite and goethite [α-FeO(OH)] were predominant in the paddy soil samples.| (A) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples | Sugarcane FMNs | Paddy FMNs | ||||||||||
| Mean | SD | Median | Min | Max | CV | Mean | SD | Median | Min | Max | CV | |
| a FMNs: ferromanganese nodules; SS: surrounding soil; OM: organic matter; Fed: dithionite extractable Fe; Feo: oxalate extractable Fe; Fep: pyrophosphate extractable Fe; Mnd: dithionite extractable Mn; NA: not available. Asterisks represent significant difference (P < 0.05) between ferromanganese nodules and corresponding surrounding soil examined by the independent t-test. The values of physicochemical characteristics are shown as mean, SD, median, min, max and CV in sugarcane field samples (N = 4) and paddy field samples (N = 9) from vertical soil profiles, respectively. | ||||||||||||
| pH | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| OM (g kg−1) | 2.68 | 0.22 | 2.72 | 2.39 | 2.89 | 0.08 | 3.56* | 0.53 | 3.67 | 2.26 | 4.03 | 0.15 |
| Fed (g kg−1) | 228* | 20.2 | 229 | 203 | 252 | 0.09 | 184* | 21.6 | 186 | 138 | 209 | 0.12 |
| Feo (g kg−1) | 8.07 | 0.15 | 0.89 | 0.69 | 1.02 | 0.18 | 0.83 | 0.21 | 0.84 | 0.58 | 1.14 | 0.25 |
| Fep (g kg−1) | 0.19 | 0.04 | 0.20 | 0.14 | 0.23 | 0.22 | 0.22 | 0.10 | 0.19 | 0.15 | 0.49 | 0.48 |
| Fe(II) (g kg−1) | 0.10 | 0.02 | 0.09 | 0.09 | 0.12 | 0.16 | 0.11 | 0.04 | 0.10 | 0.05 | 0.19 | 0.37 |
| Fe(III) (g kg−1) | 0.06 | 0.02 | 0.05 | 0.04 | 0.09 | 0.42 | 0.04 | 0.03 | 0.04 | 0.01 | 0.11 | 0.72 |
| Fe(II)/Fe(III) (g kg−1) | 1.86 | 0.64 | 2.14 | 0.91 | 2.26 | 0.34 | 3.7 | 2.95 | 3.32 | 0.95 | 10.83 | 0.80 |
| Total Fe (g kg−1) | 328* | 20.1 | 335 | 299 | 344 | 0.06 | 308* | 15.0 | 311 | 275 | 323 | 0.05 |
| Mnd (g kg−1) | 0.14 | 0.06 | 0.14 | 0.07 | 0.22 | 0.45 | 1.42* | 0.51 | 1.25 | 0.90 | 2.31 | 0.36 |
| Total Mn (g kg−1) | 0.17 | 0.06 | 0.14 | 0.07 | 0.21 | 0.40 | 1.87* | 0.46 | 1.74 | 1.42 | 2.76 | 0.24 |
| Fe2O3 (g kg−1) | 469* | 28.7 | 478 | 428 | 492 | 0.06 | 440* | 21.4 | 444 | 393 | 462 | 0.05 |
| SiO2 (g kg−1) | 170* | 5.60 | 169 | 165 | 178 | 0.03 | 133* | 7 | 136 | 121 | 144 | 0.05 |
| Al2O3 (g kg−1) | 186* | 18.3 | 184 | 171 | 207 | 0.10 | 204* | 11.2 | 203 | 190 | 225 | 0.05 |
| SiO2/Al2O3 | 0.92* | 0.08 | 0.93 | 0.81 | 0.99 | 0.09 | 0.66* | 0.05 | 0.65 | 0.60 | 0.74 | 0.08 |
| Al2O3/Fe2O3 | 0.4* | 0.06 | 0.39 | 0.35 | 0.48 | 0.16 | 0.46* | 0.03 | 0.45 | 0.43 | 0.52 | 0.06 |
| SiO2/(Al2O3 + Fe2O3) | 0.26* | 0.01 | 0.26 | 0.25 | 0.27 | 0.03 | 0.21* | 0.02 | 0.21 | 0.19 | 0.23 | 0.07 |
| (B) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples | Sugarcane SS | Paddy SS | ||||||||||
| Mean | SD | Median | Min | Max | CV | Mean | SD | Median | Min | Max | CV | |
| pH | 4.87 | 0.53 | 4.645 | 4.53 | 5.67 | 0.11 | 6.39 | 1.58 | 7.05 | 4.45 | 8.22 | 0.25 |
| OM (g kg−1) | 11.7 | 8.09 | 8.89 | 5.99 | 23.2 | 0.69 | 9.41* | 3.87 | 8.11 | 5.71 | 18.9 | 0.41 |
| Fed (g kg−1) | 84.2* | 6.06 | 86.7 | 75.2 | 88.3 | 0.07 | 66.1* | 9.66 | 67.4 | 55.5 | 80.1 | 0.15 |
| Feo (g kg−1) | 0.80 | 0.58 | 0.55 | 0.45 | 1.67 | 0.73 | 1.11 | 0.49 | 0.98 | 0.59 | 1.89 | 0.44 |
| Fep (g kg−1) | 0.11 | 0.11 | 0.07 | 0.04 | 0.27 | 0.97 | 0.17 | 0.12 | 0.13 | 0.08 | 0.45 | 0.71 |
| Fe(II) (g kg−1) | 0.09 | 0.04 | 0.08 | 0.06 | 0.15 | 0.44 | 0.11 | 0.06 | 0.09 | 0.07 | 0.26 | 0.52 |
| Fe(III) (g kg−1) | 0.02 | 0.01 | 0.02 | 0.01 | 0.04 | 0.57 | 0.11 | 0.12 | 0.06 | 0.02 | 0.40 | 1.10 |
| Fe(II)/Fe(III) (g kg−1) | 4.36 | 1.60 | 4.43 | 2.39 | 6.20 | 0.37 | 1.56 | 0.78 | 1.41 | 0.65 | 3.23 | 0.50 |
| Total Fe (g kg−1) | 89.1* | 5.03 | 90.6 | 82 | 93.5 | 0.06 | 86.1* | 6.50 | 86.1 | 74.9 | 96.8 | 0.08 |
| Mnd (g kg−1) | 0.11 | 0.03 | 0.10 | 0.08 | 0.15 | 0.32 | 0.24* | 0.09 | 0.20 | 0.17 | 0.47 | 0.39 |
| Total Mn (g kg−1) | 0.08 | 0.02 | 0.07 | 0.06 | 0.11 | 0.29 | 0.34* | 0.09 | 0.21 | 0.13 | 0.43 | 0.37 |
| Fe2O3 (g kg−1) | 127* | 7.19 | 129 | 117 | 134 | 0.05 | 123* | 9.29 | 123 | 107 | 138 | 0.08 |
| SiO2 (g kg−1) | 367* | 11 | 366 | 358 | 381 | 0.03 | 497* | 59.3 | 517 | 430 | 590 | 0.10 |
| Al2O3 (g kg−1) | 276* | 24.1 | 282 | 246 | 297 | 0.09 | 209* | 38.8 | 193 | 139 | 239 | 0.17 |
| SiO2/Al2O3 | 1.3* | 0.10 | 1.31 | 1.25 | 1.46 | 0.07 | 2.49* | 0.79 | 2.67 | 1.80 | 4.24 | 0.30 |
| Al2O3/Fe2O3 | 2.2* | 0.20 | 2.24 | 1.91 | 2.30 | 0.08 | 1.79* | 0.29 | 1.79 | 1.13 | 1.87 | 0.16 |
| SiO2/(Al2O3 + Fe2O3) | 0.91* | 0.04 | 0.91 | 0.86 | 0.96 | 0.04 | 1.53* | 0.37 | 1.69 | 1.16 | 2.25 | 0.22 |
As shown in Fig. 1, the Fe and Mn found deep within the soil were more stable in FMNs than in the corresponding surrounding soil samples. Fed was mostly iron in the FMNs and the soil, but was found in larger concentrations deep in the soil of paddy fields. In addition, the content of Mnd accounted for the vast majority of the total amount of Mn. The Fe(II)/Fe(III) ratio was 2.3 times higher in soil than in the associated FMNs from the sugarcane field, which indicated a higher oxidation capability of the surrounding soil. However, the Fe(II)/Fe(III) ratio had an adverse pattern, indicating that the oxidation ability was stronger in FMNs than in the corresponding soil from the paddy field samples. The enrichment ratio36 of weathering related materials in nodules from the paddy field and the sugarcane field exhibits similar patterns (Fig. 2) and showed the highest enrichment of Fe2O3 (∼4), followed by Al2O3 and SiO2 (<1). Generally, the enrichment ratio of Fe2O3 and Al2O3 was decreased with the vertical soil depth in the paddy soil. However, data from the sugarcane field did not show such trends. The contrasting form and enrichment ratio of the Fe element in FMNs from these two samples indicate that the iron cycle increased with vertical soil depth was more prominent in the paddy field than in the sugarcane field (Fig. 1 and 2).
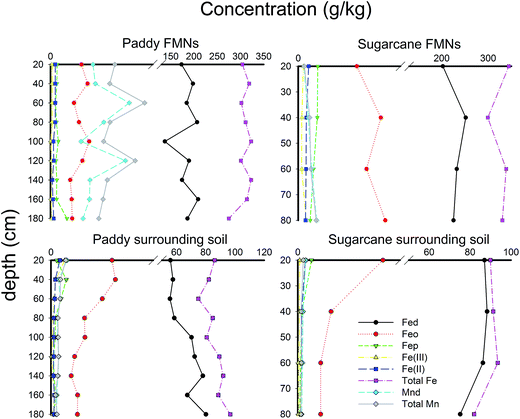 | ||
| Fig. 1 Key iron and manganese profile characteristics in vertical soil depth sequences. Sample designation refers to Table 1. Complete physicochemical characteristics are available in Table 1. | ||
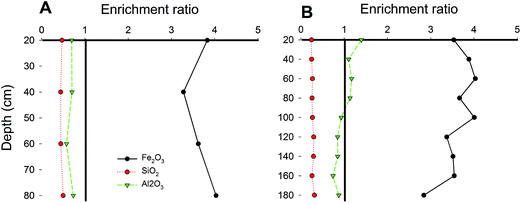 | ||
| Fig. 2 Plot of enrichment factors for major weathering materials with soil depth profiles. Line plots showing the enrichment ratio of weathering related materials in FMNs compared to soils with vertical depth profiles in sugarcane (A) and paddy (B) field samples. The enrichment ratio is the concentration of the materials in the ferromanganese nodules divided by the concentration of the materials in the associated surrounding soil.36 Data are the same as those in Table 1. | ||
As shown in ESI Tables S1 and S2,† the correlation between geochemical properties exhibited different patterns in FMNs and the surrounding soil. Not surprisingly, the pH had a prominent correlation with other geochemical characteristics in the soil. The content of Fed and Mnd also had a significant negative correlation in the surrounding soil (Pearson coefficient −0.699, P < 0.01). Furthermore, the content of the weathering-related material had a significant correlation between the Fe- or Mn-content in the soil (see ESI Tables S1 and S2 for details†). However, we did not detect a significant correlation between the geochemical parameters in the FMN samples and the parameters from the surrounding soil. The most notable correlation was observed in the SiO2 content with Fed (Pearson coefficient = 0.591, P < 0.05) and Mnd (Pearson coefficient −0.730, P < 0.01), which showed an adverse pattern in the surrounding soil (SiO2 had a negative relationship with Fed and a positive relationship with Mnd).
3.2. The microbial diversity indices are affected by soil depth and geochemical characteristics
We examined the microbial communities of FMNs and their associated soil at similar levels (5000 randomly chosen 16S rRNA gene sequences per sample were used in our analyses). The results showed that in almost all nodule–soil pairs at an identical soil depth, the predicted microbial community diversity (PD, Chao1 and observed species, at a genetic distance of 3%) in the surrounding soil exceeded that of the corresponding FMNs (ESI Table S3†). However, the different microbial community diversity indices used (PD, Chao1 and observed species) did not show consistent trends between the FMNs and the surrounding soils with horizon depth, but the highest microbial indices were found at a depth of 20–40 cm in the sugarcane field sample and at a depth of 60–80 cm in the paddy field sample (ESI Table S3†). Of those FMNs whose geochemical characteristics were considered, Fed and SiO2 had a significantly positive correlation (P < 0.01) with either phylogenetic distance (PD) (Pearson coefficient 0.67 and 0.70 for Fed and SiO2, respectively). But, there was a negative relationship observed between the Mn content (Mnd and total Mn) and species diversity indices (Chao1 and observed species) (Table 2). In the associated surrounding soil, we only detected that the Fe(II)/Fe(III) had a significantly positive correlation with PD (Pearson coefficient 0.60, P < 0.05) and Chao1(Pearson coefficient 0.87, P < 0.01) diversity index.| Ferromanganese nodules | Surrounding soil | |||||
|---|---|---|---|---|---|---|
| PD* | Chao1 | Observed species | PD | Chao1 | Observed species | |
| a PD: phylogenetic distance. Marked correlations are significant at 0.05 (*) and 0.01 (**) levels of probability. NP: not possible to calculate. | ||||||
| pH | NP | NP | NP | 0.280 | 0.497 | 0.035 |
| OM | −0.42 | −0.519 | −0.430 | −0.240 | 0.137 | 0.017 |
| Fed | 0.673* | 0.696** | 0.635* | −0.307 | −0.369 | −0.083 |
| Feo | 0.242 | 0.347 | 0.305 | 0.057 | 0.271 | 0.007 |
| Fep | −0.339 | −0.286 | −0.328 | 0.121 | 0.234 | 0.095 |
| Fe(II) | −0.178 | −0.094 | −0.124 | 0.092 | 0.284 | 0.125 |
| Fe(III) | 0.073 | 0.145 | 0.090 | 0.241 | 0.381 | 0.250 |
| Fe(II)/Fe(III) | −0.121 | −0.615* | −0.199 | −0.595* | −0.887** | −0.384 |
| Total Fe | 0.341 | 0.399 | 0.408 | −0.341 | −0.446 | −0.074 |
| Mnd | −0.371 | −0.587* | −0.460 | 0.155 | 0.214 | 0.054 |
| Total Mn | −0.511 | −0.714** | −0.584* | 0.164 | 0.231 | 0.190 |
| Fe2O3 | 0.341 | 0.399 | 0.408 | −0.341 | −0.446 | −0.074 |
| SiO2 | 0.702** | 0.890** | 0.767* | 0.160 | 0.275 | −0.052 |
| Al2O3 | 0.017 | 0.242 | −0.052 | −0.090 | −0.248 | 0.111 |
| SiO2/Al2O3 | 0.501 | 0.736** | 0.577* | 0.229 | 0.393 | 0.059 |
| Al2O3/Fe2O3 | −0.149 | −0.353 | −0.228 | 0.008 | −0.129 | 0.129 |
| SiO2/(Al2O3 + Fe2O3) | 0.596* | 0.815** | 0.653** | 0.225 | 0.383 | 0.020 |
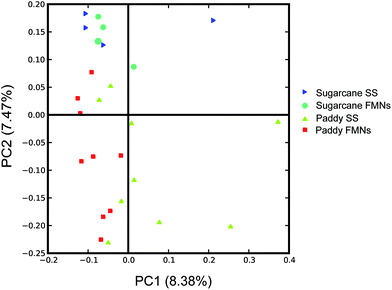
The average unweighted UniFrac distance33 was computed (a measure of differences in microbial community structures) from OTU abundance patterns among all category pairs (including FMN individuals with vertical soil depth in the identical site, nodule–soil pair in the same depth and FMNs from different sites) to access the microbial community differences in this system. To control the differences in coverage, all analyses were performed on an equal number of 5000 randomly selected sequences. Overall, the microbial community structures of FMN samples from the same soil vertical depth were much more similar to one another (average unweighted UniFrac distance 0.40 for paddy field samples and 0.33 for sugarcane field samples, respectively) than the FMN samples from different sites (average unweighted UniFrac distance 0.42, respectively), while the surrounding soil from the vertical horizon had the most divergence in community structure (average unweighted UniFrac distance 0.70 for paddy field samples and 0.45 for sugarcane field samples, respectively) (Table 3). These results demonstrated that the microbiota inside the FMNs were stable in different land-use patterns and depth, although the surrounding soil exhibited large variance. As expected, PCoA analysis (Principal Coordinate Analysis) using pairwise unweighted UniFrac distance showed that the FMN samples from the same land-use pattern clustered together, and that FMN samples or associated soils from different locations did not (Fig. 4). This suggested a phylogenetic dissimilarity between the microbial community structures of FMNs and surrounding soils in vertical soil depth rather than FMNs from different land-use patterns.
| Paddy FMNs | Paddy SS | Sugarcane FMNs | Sugarcane SS | |
|---|---|---|---|---|
| a Mean (±SD) unweighted UniFrac distance for pair-wise comparisons between FMN individuals with soil depth in identical place, nodule–soil pair in the same depth and FMNs from different sites. Sample designations are included in Table 1. | ||||
| Paddy FMNs | 0.40 ± 0.11 | |||
| Paddy SS | 0.59 ± 0.23 | 0.70 ± 0.24 | ||
| Sugarcane FMNs | 0.42 ± 0.09 | 0.58 ± 0.24 | 0.33 ± 0.05 | |
| Sugarcane SS | 0.51 ± 0.16 | 0.54 ± 0.21 | 0.44 ± 0.16 | 0.45 ± 0.17 |
3.3. The taxonomic compositions in FMNs and the corresponding surrounding soils
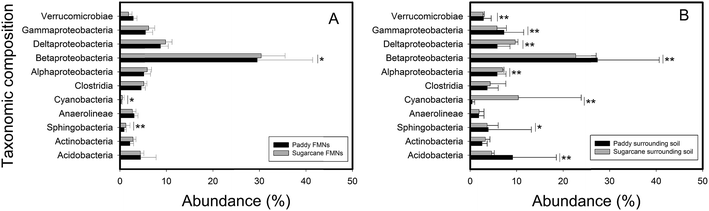
Phylogenetic diverse eubacterial phylotypes were detected across all nodule–soil pair samples; β-proteobacterial sequences were most abundant (Fig. 3, ESI Tables S4 and S5†). This phylum was dominated by the members of Burkholderiales and Rhodocyclales (ESI Fig. S3†). Most of the α-proteobacterial sequences were classified as Rhizobiales taxa while the Pseudomonadales taxon dominated the δ-proteobacteria phylum. The Desulfuromonales were the most abundant groups within the γ-proteobacteria phyla. Less predominant bacteria in FMNs and associated soils were Acidobacteria, Antinobacteria, Verrucomicrobiae, Clostridia, Sphingobacteria, Cyanobacteria (had a high abundance in topsoil of sugarcane field) and Anaerolineae (ESI Tables S4 and S5†). The relative abundance of archaea was low both in FMNs (average of 1.78%) and surrounding soil (average of 3.36%). And about half of the archaeal sequences (average of 46.5% for FMNs and 53.5% for surrounding soil) were classified to unclassified archaea. Definitely rare biospheres, but still detected in this system (<1%), included Thermoprotei, Methanobacteria, Methanomicrobia, Thermoplasmata, Bacteroidetes, Chlamydiae, Chloroflexi and Nitrospira. Comparative analysis of pyrosequencing data indicated that no significant differences were observed in the microbial composition in order level between the nodule–soil pairs from the same type of plough (ESI Fig. S3†). Furthermore, the distribution of high abundance phyla exhibited no difference inside these two types of FMNs. But we identified several phyla (including β-proteobacteria, γ-proteobacteria, α-proteobacteria, δ-proteobacteria, Cyanobacteria, etc.) with significantly different levels of abundance in these two FMN associated surrounding soils (Fig. 3).3.4. Correlations among selected geochemical properties and bacterial phyla distribution in the nodule–soil system
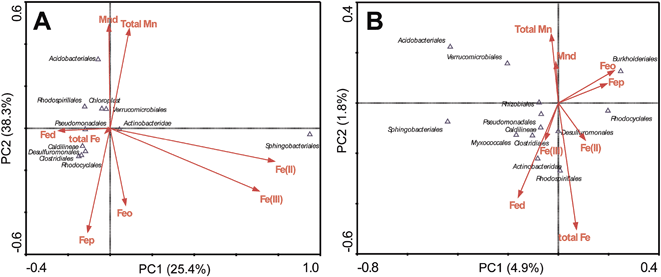
The correlations between bacterial phyla and selected soil properties such as Fed, total Fe, Feo, Fep, Fe(II), Fe(III), Mnd and total Mn were evaluated using ordination plots, according to the CCA (canonical correspondence analysis) (Fig. 5), in which the first two CCA axes explained 25.4% and 8.3% of the total variance in the FMN microbial community data, comparing only explained 4.9% and 1.8% in the associated surrounding soils. CCA analyses showed, in FMN biospheres, Sphingobacteriales were centered along a gradient with relatively high Fe(II) or Fe(III) content. In contrast, Acidobacteria was associated with higher values of Mnd and total Mn. However, the pattern in surrounding soils exhibited different trends, in that the total Fe and Fed contributed most on the microbial taxa distribution. Correlations between weathering related materials and bacterial phyla were revealed by the linear regression analyses (ESI Table S6†). In FMNs, we found only Myxococcales had significant correlation with Fe2O3 (Pearson coefficient 0.629, P < 0.05) and SiO2 (Pearson coefficient 0.700, P < 0.01), while in the associated surrounding soils, Rhodocyclales were associated with values of Fe2O3 (Pearson coefficient −0.563, P < 0.05) and Al2O3 (Pearson coefficient 0.557, P < 0.05), but had a negative relationship with SiO2 (Pearson coefficient −0.703, P < 0.01).4. Discussion
4.1. Land-use patterns seriously impact the microbial community structure of soil
This investigation compared the difference between the microbial compositions, the diversity, and the geochemistry of soil from paddy fields and sugarcane fields, which are the primary land-use patterns of fields in South China. Paddy fields experience an alternation between reducing and oxidizing states during the stages of rice growth and harvesting. In contrast, sugarcane fields possess a higher oxygen content where the oxidative state remains dominated during crop growth. The Fe(II)/Fe(III) ratios of FMNs and the surrounding soil from the two types of land use studied here (Table 1) indicate that the oxidation capacity of sugarcane soil (mean Fe(II)/Fe(III) ratio 4.36) is much higher than that of paddy soil (mean Fe(II)/Fe(III) ratio 1.56). The higher ratio of Fe(II)/Fe(III) in FMNs than that of the corresponding surrounding soil in paddy fields suggests that Fe(II) oxidation occurs inside the nodules, while Fe(III) reduction occurs in the corresponding surrounding soil. The high oxygen levels and low pHs may be a response to the predominantly oxidized state of the sugarcane fields, while the relatively low ratio of Fe(II)/Fe(III) within FMNs reveals that the reduction of iron is active on the surface of FMNs from the sugarcane field. Previous research has demonstrated that the composition of the soil microbial community is closely correlated with land use patterns.37 The relatively high abundance of Burkholderiales, Rhodocyclales, Acidobacteriales and Clostridiales in these two types of plagues is consistent with previous studies.38,39 The pH and oxidizing/reducing capacity (generated by different land use patterns) are responsible for the markedly different microbial community structures between paddy and sugarcane fields.4.2. Iron- and manganese-redoxing bacteria
Microorganisms are active and may involve direct enzymatic interventions or the metabolic production of specific chemical reactants that cause precipitates to generate FMNs; this biogenic theory of FMNs is supported by the detection of respiring cells on the surface of FMNs using the in situ metabolic dye (2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyltetrazolium chloride).40 Nonetheless, by using ultra-deep sequencing on the microbial community (>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sequences per sample), we successfully detected many iron- and manganese-reducing/oxidizing bacteria. Previous research using oxygenic gradient tube enrichments of circumneutral iron- or manganese-oxidizing bacteria from FMNs revealed functional groups including the Rhizobiaceae group in α-Proteobacteria, the Actinobacteria/Bacillus group in Firmicutes and the Xanthomonas group in the γ-Proteobacteria.21 Our results demonstrate that these phylotypes are present in the FMNs and in the surrounding soil samples, but not the predominated species. The well-studied Fe-reducing anaerobic bacteria, Geobacteraceae,41 were recovered in large numbers using pyrosequencing data, and represented 1.1–4.6% of FMNs, compared with 0.1–4.5% in the surrounding soil (ESI Tables S4 and S5†). However, another facultative anaerobe, Shewanellaceae, which reduces iron and manganese metabolically,42 was not detected in this large 16S rRNA sequence dataset. The relatively high abundance of Geobacteraceae suggests that this species is very much involved in the Fe–Mn biogeochemical cycling in the FMN environment, and may reduce insoluble minerals such as iron- or manganese oxides and promote the mobility of the iron–manganese elements in the soil biosphere. In the presence of Fe(II), Geobacter is able to reduce hematite (the primary minerals in FMNs) better than Shewanella bacteria,43 thus the physiology of Geobacteraceae is more suitable for the FMN habitat than Shewanellaceae in the FMN habitat.
000 sequences per sample), we successfully detected many iron- and manganese-reducing/oxidizing bacteria. Previous research using oxygenic gradient tube enrichments of circumneutral iron- or manganese-oxidizing bacteria from FMNs revealed functional groups including the Rhizobiaceae group in α-Proteobacteria, the Actinobacteria/Bacillus group in Firmicutes and the Xanthomonas group in the γ-Proteobacteria.21 Our results demonstrate that these phylotypes are present in the FMNs and in the surrounding soil samples, but not the predominated species. The well-studied Fe-reducing anaerobic bacteria, Geobacteraceae,41 were recovered in large numbers using pyrosequencing data, and represented 1.1–4.6% of FMNs, compared with 0.1–4.5% in the surrounding soil (ESI Tables S4 and S5†). However, another facultative anaerobe, Shewanellaceae, which reduces iron and manganese metabolically,42 was not detected in this large 16S rRNA sequence dataset. The relatively high abundance of Geobacteraceae suggests that this species is very much involved in the Fe–Mn biogeochemical cycling in the FMN environment, and may reduce insoluble minerals such as iron- or manganese oxides and promote the mobility of the iron–manganese elements in the soil biosphere. In the presence of Fe(II), Geobacter is able to reduce hematite (the primary minerals in FMNs) better than Shewanella bacteria,43 thus the physiology of Geobacteraceae is more suitable for the FMN habitat than Shewanellaceae in the FMN habitat.
Generally, microorganisms oxidize Mn(II) much faster than oxidation via abiotic means, suggesting that biological oxidation is dominant in nature,44 and the manganese-oxidizing bacteria actively function in the formation of FMNs. Pseudomonas, a genus which contains well-studied manganese-oxidizing species,45 is relatively abundant in our data, indicating its importance in manganese biogeochemical cycles of the microbial ecosystem within the FMNs. Leptothrix belong to Burkholderiales and are known for their oxidation of both Fe(II) and Mn(II) in their sheath.46 In this study Leptothrix was present in 0.7–2.9% of the total 16S rRNA sequences from the FMNs. Another iron-oxidizing chemolithotrophic bacterium, Gallionela, oxidizes Fe(II) to Fe(III) for energy conservation,47 but was not detected in the FMN microbial community in this study. All of these data indicate that Geobacteraceae and Leptothrix play important roles in FMN mineral formation during the process of biological precipitation or the dissolution of Fe–Mn elements.
4.3. The impact of minerals weathering on the FMN microbial communities
Increasing evidence suggests that microbes play a significant role in mediating the dissolution and mineral oxidation, which modifies the rate and mechanism of chemical and physical weathering and clay growth.48,49 Fe(II) is produced on the subsurface under anoxic conditions by dissimilatory iron(III) reducing bacteria (DIRB) coupled with biotic/abiotic weathering of minerals.50 Herein, we observed the relationship between weathering minerals (Fe2O3, SiO2, Al2O3), microbial diversity, and the composition to investigate the link between the weathering process and the presence of certain microbial populations. Researching the subsurface environments has revealed that silicate weathering caused by bacteria is dependent on the trace nutrient contents of each aquifer mineral.51 Our results show that the SiO2 content has a significantly positive correlation with microbial diversity within FMNs (PD diversity, Pearson coefficient 0.702, P < 0.01), but the associated surrounding soil did not exhibit such patterns (Table 2). Silicon, a beneficial nutrient, can limit microbial growth and metabolism.51 In the nutrient-limited FMN biosphere, the progression of weathering may be influenced by the SiO2 nutritional potential, which stimulates microorganisms to accelerate the dissolution of clay minerals and enhance the release of SiO2 and other nutrients from FMN minerals, ultimately increasing the diversity of the microbial community. These results support the idea that the microbial activity is a major driving force behind mineral weathering even in the isolated environment of FMNs. The biochemistry of SiO2 utilization by microbes is not well understood.17 However, the significant positive correlation between the SiO2 content and the presence of Myxococcales bacterium (ESI Table S2†) suggests that these bacteria may participate in silicon's biogeochemical cycling in FMN's ecosystem. Furthermore, a dissimilatory iron-reducing member of the Anaeromyxobacter subgroup of Myxococcales has been isolated,52 demonstrating the importance of these bacteria in Fe–Si coupled element cycling in nature and in the process of nodule formation.4.4. The microbial community structure is stable in FMNs from different land-use patterns and from vertical soil profiles
The sugarcane soil had a lower pH (average pH 4.9) than the paddy soil (average pH 6.4), which accounts for the high divergence of microbial compositions in the FMNs associated with the surrounding soil from both sugarcane and paddy fields (average unweighted Unifrac distance 0.54), which coincides with the idea that pH has a significant influence on the diversity of soil microorganisms.53–55 Likewise, the pH of the soil in the vertical horizon in paddy fields (from 4.81 to 7.97) was responsible for the marked discrepancy between the microbial community structure in surrounding soil and the content from different vertical horizon depths (average unweighted Unifrac distance 0.70). Although the physical and chemical properties of FMNs from different soil profiles have been investigated,56–58 the microbial community of nodules in vertical soil depth has only been addressed by Cahyani et al., revealing that the bacterial communities in the manganese nodules were markedly different from those in the plowed layer of the rice fields and their subsoil.6 Our study demonstrated that the community structures were significantly different in the FMNs associated with the surrounding soil from these two types of plowed fields or in vertical soil sequences, however, the microbiota in FMNs were very similar despite the different land-use patterns (average unweighted Unifrac distance 0.42, Table 3) or the different soil depths (average unweighted Unifrac distance 0.40 and 0.33 for paddy field and sugarcane field samples, respectively). This disequilibrium suggests that the ecological and evolutionary forces behind the development of FMNs (typical high metal content and low organic matter content, Table 1) may shape their microbial diversity and community structure in this oligotrophic and isolated environment, which results in a particular structure of microbial composition. Without available carbon and nitrogen sources, FMN microbes derive energy mainly by transferring electrons between iron and manganese oxides and hydroxides, to sustain the reproduction of microorganisms. Due to this “function-selection”,59 the microbial community structure is stable in FMNs regardless of the community divergence observed in their surrounding soil biogeochemical properties. The dispersal limitation primarily drives the biogeographical patterns in soil, and the geographic distance should be the best predictor of genetic divergence between communities and habitats.60,61 This is not the case in the FMN environment; the geographic factors that determine the spatial pattern of microbial communities were not demonstrated in our analyses, and can only be evaluated in further studies on more systematical samples and record data from various distances and soil types.5. Conclusion
We used pyrosequencing to analyze the microbial communities present in ferromanganese nodules, which were collected from vertical soil profiles of paddy and sugarcane fields. We found a high phylogenetic diversity among the microbes from these oligotrophic biospheres, including many phylotypes, which have not been reported previously. Our studies also suggest the presence of an iron- and manganese-oxidizing or reducing microorganism in the FMN environment, and Geobacteraceae and Leptothrix are the major functional groups active in Fe–Mn biochemical cycling. Biomineralization plays an important role in secondary mineral and soil formation, and weathering-related components (such as Fe2O3, SiO2 and Al2O3) determined the degree and extent of weathering. Our study assessed the correlation between weathering indices and the diversity of the microbial community or the taxonomic composition, and found that the SiO2 content was correlated with increased microbial diversity in the FMN environment, which increases our understanding of the role that microorganisms play in the weathering process. Only certain organisms can survive in the presence of oligotrophic forces and a high-metal environment; the natural selection of organisms that thrive in this environment can explain the similar microbial communities we observed in FMNs from different land-use patterns and vertical soil sequences.Currently, many researchers are dedicated to the isolation, characterization, and quantification of iron- and manganese-oxidizing bacteria from the environment. Other studies are focused on determining the rates of iron and manganese oxidation and on finding key species or genes that respond to this process using genomic, transcriptomic, and proteomic methods. This research elucidates the significant role that microbes play in the mechanism and generation of ferromanganese nodules. However, to determine the biogenic mechanisms of FMNs in natural ecosystems, further studies using metagenomic and metranscriptomic techniques in combination with iron- and manganese-isotope fractionation and systematic sampling are required. Further studies are also necessary to investigate the genes or species that respond to the Fe–Mn biochemical cycling in these environments to ultimately answer the question: “How do biotic factors impact the production of these unusual secondary minerals?”
Abbreviations
| FMNs | Ferromanganese nodules |
| qPCR | Quantitative real-time PCR |
| T-RFLP | Terminal restriction fragment length polymorphism |
| ARDRA | Amplified ribosomal DNA restriction analysis |
| DGGE | Denaturing gradient gel electrophoresis |
| OTUs | Operational taxonomic units |
| OM | Organic matter |
| Fed | Dithionite extractable Fe |
| Feo | Oxalate extractable Fe |
| Fep | Pyrophosphate extractable Fe |
| Mnd | Dithionite extractable Mn |
| PCoA | Principal coordinate analysis |
| CCA | Canonical correspondence analysis |
Acknowledgements
This work was supported by grants from the National Science Foundation of China (41025003 and 41201253), and Science and Technology Planning Project of Guangdong Province, China (S2011030002882).References
- K. H. Nealson and D. Saffarini, Annu. Rev. Microbiol., 1994, 48, 311–343 CrossRef CAS PubMed.
- W. F. Tan, F. Liu, Y. H. Li, H. Q. Hu and Q. Y. Huang, Pedosphere, 2006, 16, 72–81 CrossRef CAS.
- A. Kappler and K. L. Straub, Rev. Mineral. Geochem., 2005, 59, 85–108 CrossRef CAS.
- D. J. Burdige, Earth-Sci. Rev., 1993, 35, 249–284 CrossRef CAS.
- R. G. Burns and V. Mee Burns, Nature, 1975, 255, 130–131 CrossRef CAS.
- V. R. Cahyani, J. Murase, E. Ishibashi, S. Asakawa and M. Kimura, Soil Sci. Plant Nutr., 2007, 53, 575–584 CrossRef CAS PubMed.
- Anaerobic Microbiology in Waterlogged Rice Fields, ed. M. Kimura, Marcel Dekker, New York, 2000 Search PubMed.
- C. A. Asikainen and S. F. Werle, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 17579–17581 CrossRef CAS PubMed.
- J. E. Post, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 3447–3454 CrossRef CAS.
- S. E. Calvert, B. L. Cousens and M. Y. S. Soon, Chem. Geol., 1985, 51, 9–18 CrossRef CAS.
- H. L. Ehrlich, Miner. Metall. Process., 2000, 17, 121–128 CAS.
- H. Elderfield, C. J. Hawkesworth, M. J. Greaves and S. E. Calvert, Geochim. Cosmochim. Acta, 1981, 45, 513–528 CrossRef CAS.
- A. Manceau, N. Tamura, R. S. Celestre, A. A. MacDowell, N. Geoffroy, G. Sposito and H. A. Padmore, Environ. Sci. Technol., 2002, 37, 75–80 CrossRef.
- W. C. Ghiorse, Annu. Rev. Microbiol., 1984, 38, 515–550 CrossRef CAS PubMed.
- L. Y. Stein, M. T. La Duc, T. J. Grundl and K. H. Nealson, Environ. Microbiol., 2001, 3, 10–18 CrossRef CAS.
- A. S. Templeton, H. Staudigel and B. M. Tebo, Geomicrobiol. J., 2005, 22, 127–139 CrossRef CAS.
- H. L. Ehrlich, Geomicrobiol. J., 1999, 16, 135–153 CrossRef.
- J. Greenslate, Nature, 1974, 249, 181–183 CrossRef CAS.
- J. Z. He, L. M. Zhang, S. S. Jin, Y. G. Zhu and F. Liu, Geomicrobiol. J., 2008, 25, 14–24 CrossRef CAS.
- S. Nitahara, S. Kato, T. Urabe, A. Usui and A. Yamagishi, FEMS Microbiol. Lett., 2011, 321, 121–129 CrossRef CAS PubMed.
- D. E. Northup, S. M. Barns, L. E. Yu, M. N. Spilde, R. T. Schelble, K. E. Dano, L. J. Crossey, C. A. Connolly, P. J. Boston, D. O. Natvig and C. N. Dahm, Environ. Microbiol., 2003, 5, 1071–1086 CrossRef.
- R. B. Trimble and H. L. Ehrlich, Appl. Microbiol., 1968, 16, 695–702 CAS.
- L. M. Zhang, F. Liu, W. F. Tan, X. H. Feng, Y. G. Zhu and J. Z. He, Soil Biol. Biochem., 2008, 40, 1364–1369 CrossRef CAS PubMed.
- S. T. Bates, D. Berg-Lyons, J. G. Caporaso, W. A. Walters, R. Knight and N. Fierer, ISME J., 2011, 5, 908–917 CrossRef CAS PubMed.
- Z. Liu, C. Lozupone, M. Hamady, F. D. Bushman and R. Knight, Nucleic Acids Res., 2007, 35, 120 CrossRef PubMed.
- J. Reeder and R. Knight, Nat. Methods, 2010, 7, 668–669 CrossRef CAS PubMed.
- J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Pena, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelley, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevinsky, P. J. Turnbaugh, W. A. Walters, J. Widmann, T. Yatsunenko, J. Zaneveld and R. Knight, Nat. Methods, 2010, 7, 335–336 CrossRef CAS PubMed.
- J. R. Cole, Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity and J. M. Tiedje, Nucleic Acids Res., 2009, 37, 141–145 CrossRef PubMed.
- E. Pruesse, C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, J. Peplies and F. O. Glöckner, Nucleic Acids Res., 2007, 35, 7188–7196 CrossRef CAS PubMed.
- J. G. Caporaso, K. Bittinger, F. D. Bushman, T. Z. DeSantis, G. L. Andersen and R. Knight, Bioinformatics, 2010, 26, 266–267 CrossRef CAS PubMed.
- T. M. Gihring, S. J. Green and C. W. Schadt, Environ. Microbiol., 2012, 14, 285–290 CrossRef CAS PubMed.
- PRIMER v5: User manual/tutorial, ed. K. R. Clarke and R. N. Gorley, Plymouth, UK, 2001 Search PubMed.
- C. Lozupone and R. Knight, Appl. Environ. Microbiol., 2005, 71, 8228–8235 CrossRef CAS PubMed.
- F. Meyer, D. Paarmann, M. D'Souza, R. Olson, E. Glass, M. Kubal, T. Paczian, A. Rodriguez, R. Stevens, A. Wilke, J. Wilkening and R. Edwards, BMC Bioinf., 2008, 9, 386 CrossRef CAS PubMed.
- CANOCO reference manual and CanoDraw for windows user's Guide: Software for canonical community ordination (Version 4.5), ed. C. J. F. Ter Braak and P. Smilauer, Microcomputer Power, Ithaca, NY, USA, 2002 Search PubMed.
- D. Gasparatos, Environ. Chem. Lett., 2013, 11, 1–9 CrossRef CAS.
- K. L. Steenwerth, L. E. Jackson, F. J. Calderón, M. R. Stromberg and K. M. Scow, Soil Biol. Biochem., 2002, 34, 1599–1611 CrossRef CAS.
- V. Acosta-Martínez, S. Dowd, Y. Sun and V. Allen, Soil Biol. Biochem., 2008, 40, 2762–2770 CrossRef PubMed.
- E. B. Hollister, A. S. Engledow, A. J. M. Hammett, T. L. Provin, H. H. Wilkinson and T. J. Gentry, ISME J., 2010, 4, 829–838 CrossRef CAS PubMed.
- M. N. Spilde, D. E. Northup, P. J. Boston, R. T. Schelble, K. E. Dano, L. J. Crossey and C. N. Dahm, Geomicrobiol. J., 2005, 22, 99–116 CrossRef CAS.
- B. Lin, M. Braster, B. M. van Breukelen, H. W. van Verseveld, H. V. Westerhoff and W. F. M. Röling, Appl. Environ. Microbiol., 2005, 71, 5983–5991 CrossRef CAS PubMed.
- C. R. Myers and K. H. Nealson, J. Bacteriol., 1990, 172, 6232–6238 CAS.
- H. A. Crosby, E. E. Roden, C. M. Johnson and B. L. Beard, Geobiology, 2007, 5, 169–189 CrossRef CAS PubMed.
- B. M. Tebo, J. R. Bargar, B. G. Clement, G. J. Dick, K. J. Murray, D. Parker, R. Verity and S. M. Webb, Annu. Rev. Earth Planet. Sci., 2004, 32, 287–328 CrossRef CAS.
- C. A. Francis and B. M. Tebo, Appl. Environ. Microbiol., 2001, 67, 4272–4278 CrossRef CAS.
- F. C. Boogerd and J. P. de Vrind, J. Bacteriol., 1987, 169, 489–494 CAS.
- R. Hallberg and F. G. Ferris, Geomicrobiol. J., 2004, 21, 325–330 CrossRef CAS.
- J. F. Banfield, W. W. Barker, S. A. Welch and A. Taunton, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 3404–3411 CrossRef CAS.
- M. Skidmore, S. P. Anderson, M. Sharp, J. Foght and B. D. Lanoil, Appl. Environ. Microbiol., 2005, 71, 6986–6997 CrossRef CAS PubMed.
- Soils: Genesis and Geomorphology, ed. R. Schaetzl and S. Anderson, Cambridge University Press, Cambridge, UK, 2005 Search PubMed.
- P. C. Bennett, J. R. Rogers, W. J. Choi and F. K. H. Hiebert, Geomicrobiol. J., 2001, 18, 3–19 CrossRef CAS.
- N. Treude, D. Rosencrantz, W. Liesack and S. Schnell, FEMS Microbiol. Ecol., 2003, 44, 261–269 CrossRef CAS.
- C. L. Lauber, M. Hamady, R. Knight and N. Fierer, Appl. Environ. Microbiol., 2009, 75, 5111–5120 CrossRef CAS PubMed.
- J. Rousk, E. Baath, P. C. Brookes, C. L. Lauber, C. Lozupone, J. G. Caporaso, R. Knight and N. Fierer, ISME J., 2010, 4, 1340–1351 CrossRef PubMed.
- J. Rousk, P. C. Brookes and E. Bååth, Soil Biol. Biochem., 2010, 42, 926–934 CrossRef CAS PubMed.
- A. Manceau, C. Tommaseo, S. Rihs, N. Geoffroy, D. Chateigner, M. Schlegel, D. Tisserand, M. A. Marcus, N. Tamura and Z. S. Chen, Geochim. Cosmochim. Acta, 2005, 69, 4007–4034 CrossRef CAS PubMed.
- B. Palumbo, A. Bellanca, R. Neri and M. J. Roe, Chem. Geol., 2001, 173, 257–269 CrossRef CAS.
- Y. Timofeeva, Geochem. Int., 2008, 46, 260–267 CrossRef.
- R. E. Ley, D. A. Peterson and J. I. Gordon, Cell, 2006, 124, 837–848 CrossRef CAS PubMed.
- S. Dequiedt, J. Thioulouse, C. Jolivet, N. P. A. Saby, M. Lelievre, P. A. Maron, M. P. Martin, N. C. Prévost-Bouré, B. Toutain, D. Arrouays, P. Lemanceau and L. Ranjard, Environ. Microbiol. Rep., 2009, 1, 251–255 CrossRef PubMed.
- N. Fierer and R. B. Jackson, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 626–631 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4em00407h |
| This journal is © The Royal Society of Chemistry 2015 |