Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Photo-induced uncaging of a specific Re(I) organometallic complex in living cells†
Anna
Leonidova‡
a,
Vanessa
Pierroz‡
ab,
Riccardo
Rubbiani‡
a,
Yanjun
Lan
a,
Anita G.
Schmitz
a,
Andres
Kaech
c,
Roland K. O.
Sigel
a,
Stefano
Ferrari
b and
Gilles
Gasser
*a
aDepartment of Chemistry, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland. E-mail: gilles.gasser@uzh.ch; Fax: +41-44-635-68-03; Tel: +41-44-635-46-30 Web: http://www.gassergroup.com
bInstitute of Molecular Cancer Research, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland
cCentre of Microscopy and Image Analysis, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland
First published on 9th June 2014
Abstract
In the last decades, a large number of organometallic complexes have shown promising anti-proliferative activity towards different cancer cell lines. However, these compounds generally had low cellular uptake and low selectivity towards cancer cells over healthy cells. The use of external triggers (e.g. light, ultra-sound, temperature, etc.) to modify the cytotoxic effect of a prodrug and the coupling of a targeting vector (e.g. peptides, antibodies, etc.) to a drug were found to be very successful techniques to tackle these drawbacks. Here, we envisioned combining these two methods, namely an external trigger (i.e. light activation) and a targeting vector, in an organometallic compound. More specifically, a Re(I) tricarbonyl N,N-bis(quinolinoyl) complex (Re-NH2) was derivatised with a photo-labile protecting group (PLPG) to cage Re-NH2 by formation of Re-PLPG. For organelle/cellular specificity, Re-PLPG was then further coupled to a nuclear localization sequence (NLS) or a bombesin peptide derivative to give Re-PLPG-NLS or Re-PLPG-Bombesin, respectively. Photolysis experiments in PBS buffer (pH 7.4) demonstrated that Re-NH2 was completely photo-released from Re-PLPG-NLS and Re-PLPG-Bombesin using a very low irradiation dose (1.2 J cm−2). To the best of our knowledge, these are the first two examples of the selective photo-release of an intact organometallic compound from a bioconjugate. Of high interest, both derivatives showed toxicity comparable to that of cisplatin towards cervical cancer cells (HeLa) upon light irradiation, although the phototoxic index (PTI) varied greatly with the targeting peptide. The cell death mechanism of Re-PLPG-NLS was explored using different techniques, including fluorescence microscopy, ICP-MS, gel electrophoresis, flow cytometry and transmission electron microscopy (TEM). It could be demonstrated that HeLa cells treated with Re-PLPG-NLS in the dark and upon irradiation showed severe cell stress (nucleolar segregation, pyknosis and vacuolation). The data obtained from an Annexin V/propidium iodide (PI) assay indicated that, after an early apoptotic stage, the onset induced by Re-PLPG-NLS led to cell death, with features ascribable to late apoptosis and necrosis, which were more marked for the treatment involving irradiation.
Introduction
Photodynamic therapy (PDT) is a medical technique that allows the destruction of target cells by irradiation of a photosensitiser (PS) with light. Offering important advantages, such as temporal and spatial control, PDT is currently used to treat certain cancers that are readily accessible with lamps or optic fibers (e.g. some types of head and neck, skin, bladder, lung and oesophagus cancers).2 Other applications of PDT – treatment of multi-drug resistant bacterial infections in particular – are also being actively explored.3–8 However, PDT suffers from a general drawback, namely its reliance on an oxygen-dependent mechanism. In fact, the two main PDT modes of action (type I which involves reactive oxygen species (ROS) production, and type II in which the damage is directly produced by 1O2) rely on the conversion of oxygen from its triplet ground state to its singlet excited state by a PS. This can be problematic since tumours, in their inner central part in particular, are often hypoxic.9,10 Hence, it would be extremely interesting to develop a light-mediated strategy that does not rely on the presence of oxygen. In this sense, singlet oxygen generation is not the only effect that light can induce. Chemical bonds can also be rearranged and/or cleaved upon light irradiation. Indeed, the activity of a compound may be modified by caging some of its functional groups with a photo-removable moiety. The release of the original compound can then be controlled by light irradiation. Such light-activated molecules have proven themselves useful tools in biological/chemical biology/medicinal chemistry studies, such as in the analysis of neurological and enzymatic processes, gene expression, cell signalling and many others.11,12 Several drugs and drug candidates have also been photo-caged and efficiently released upon light irradiation.13–19 Among them are aspirin,13 ibuprofen,13 ketoprofen,13 carbonic anhydrase II inhibitors,14 as well as a few anti-cancer agents such as fluxoridine,15 paclitaxel,16 tegafur,17 doxorubicin,18 and a photo-reactive DNA intercalator.19 Recently, this photo-caging strategy has also been combined with singlet oxygen generation: an anti-cancer drug candidate, combretastatin A-4, has been coupled to a porphyrin photosensitiser via a cleavable singlet oxygen linker.20 Surprisingly, although many studies reporting the use of photo-caged organic molecules have been published, examples of the specific release of intact (organo)metallic compounds using light as a trigger are very scarce.21 Surely, some complexes are known to undergo light-induced reactions involving the metal centre itself. This property was used not only to enhance their cytotoxicity,22–39 but also to design metal-based photo-cages.40–44 As shown by Franz and co-workers using a photo-labile o-nitrophenyl moiety, the release of a labile metal complex45 or the release of metal ions46,47 could also induce cytotoxicity. Of note, the photo-release of metal ions alone to explore neurological response and cell signalling is currently an intensively investigated research area.48–52 However, to the best of our knowledge, we were the first to report the photo-release of an intact cytotoxic substitutionally inert Ru(II) complex upon light irradiation.21 While most drugs on the market are organic molecules, the platinum(II)-based drug cisplatin and its derivatives oxaliplatin and carboplatin are playing a pivotal role in anti-cancer chemotherapy. In addition to purely metal coordination complexes, a significant number of organometallic compounds have been reported to be strongly toxic towards cancer cells in recent years.53–57 Among them, rhenium tricarbonyl complexes may not be the most numerous, but are particularly attractive, as their isostructural 99mTc analogues permit the use of imaging to observe the compound's behaviour in vivo. In this work, we are building on the information gained during a previous study on a Re(I) tricarbonyl bis(quinolinoyl) complex that showed a strong PDT profile1 to achieve multi-modal activity. We observed that synthetic modifications of the alkyl chain of these Re(I) complexes (see Scheme 1) strongly influenced both their dark and light-mediated cytotoxicities.1,58 Hence, photo-caging and peptide conjugation of these compounds could provide temporal and spatial control. Moreover, we envisaged boosting the uptake of our derivative by malignant tissues via conjugation of targeting peptides. Herein, we report the synthesis and characterization of photo-activated targeted Re(I) tricarbonyl N,N-bis(quinolinoyl) bioconjugates, as well as their photo-products. Their bioactivity towards cervical (HeLa) and prostate (PC-3) cancer cells, as well as towards normal lung fibroblasts (MRC-5), upon light activation is described. The possible modes of cytotoxic action are also explored in depth.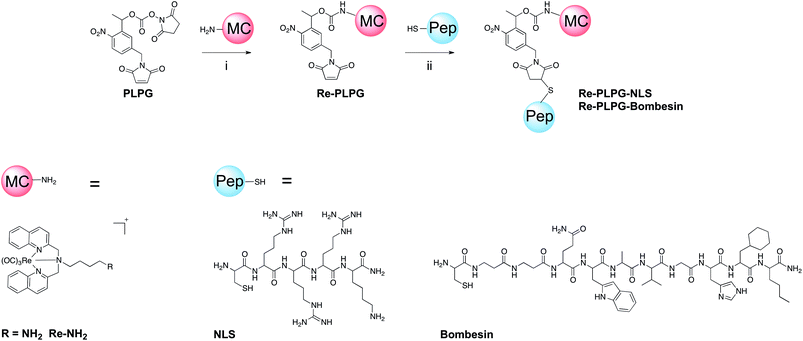 | ||
| Scheme 1 Synthesis of bioconjugated organometallic photo-caged derivatives Re-PLPG-NLS and Re-PLPG-Bombesin. (i) TEA, acetonitrile; (ii) H2O, MeOH. | ||
Results and discussion
Synthesis and characterization of the organometallic caged bioconjugates
Re-NH2 59 was caged with an o-nitrophenyl-based photo-linker, namely the Dmochowski photo-labile protecting group (PLPG).60 This type of photo-linker has been systematically studied and applied in numerous biochemical systems.12,61–64 Besides, it allows coupling of additional molecules – a property we used to attach targeting peptides to our caged Re(I) derivative. More specifically, the primary amino group of Re-NH2 was first reacted with PLPG via N-hydroxysuccinimide ester coupling to give Re-PLPG (Scheme 1).60 For this purpose, the original Dmochowski protocol was slightly modified. The phosphate buffer was replaced by organic solvents to solubilise the organometallic compounds, and a weak base was added to catch the protons liberated during the reaction. Although the presence of a base can induce maleimide ring opening,60 only a small amount of by-product was observed. The formation of Re-PLPG was confirmed by ESI-MS, which showed the expected mass of [M]+ at m/z 943.2, as well as the isotopic pattern characteristic of rhenium (Fig. S3 in the ESI†). In the 1H NMR spectrum, a new peak corresponding to the amide proton of the newly formed carbamate bond appeared at approximately 5.7 ppm. As expected, a shift of the signals of the protons in the immediate vicinity of the carbamate was observed. In Re-PLPG, the signals of the methyl group of the PLPG moiety and its neighbouring tertiary carbon proton shifted upfield compared to PLPG. Slight (both upfield and downfield) changes in chemical shifts were also observed for the protons of the organometallic part of Re-PLPG compared to Re-NH2 (Fig. S5–S7†).The photo-caged complex Re-PLPG was then conjugated to two different targeting peptides via Michael addition of the cysteine thiol of a peptide to the maleimide bond of the PLPG (Scheme 1). Re-PLPG was attached to (1) a nucleus localization signalling (NLS) peptide to target the compound to the nucleus in close proximity to DNA and (2) a bombesin derivative65,66 to improve the uptake of the compound by cancer cells over-expressing the gastrin-releasing peptide receptor (GRPR). A cysteine residue was added to each peptide to allow conjugation to the PLPG maleimide. After preparative HPLC purification and lyophilisation, light yellow solids of Re-PLPG-NLS and Re-PLPG-Bombesin were obtained. The expected masses at m/z 415.7 [M + 3H]4+, 553.9 [M + 2H]3+ and 830.3 [M + H]2+ (Re-PLPG-NLS) and at m/z 538.4 [M + 3H]4+, 717.4 [M + 2H]3+ and 1075.3 [M + H]2+ (Re-PLPG-Bombesin) were observed by ESI-MS (Fig. S9–S16†). The purity of the bioconjugates was unambiguously confirmed by LC-MS, in which two peaks in very close proximity with the same mass pattern were observed, corresponding to the two possible diastereomers formed during the Michael addition. Such behaviour was previously reported for biotin conjugated compounds photo-caged with a similar PLPG.18,67 Two small extra m/z peaks were sometimes observed in the mass spectra during LC-MS. They could be assigned to the photo-uncaged but not yet decarboxylated metal complex, as well as to the fully uncaged complex. This uncaging process clearly happens during the electrospray ionization process due to the harsh ionization conditions used for LC-MS, since the uncaged complex is eluted over a different time frame (Fig. S12 and S16–S18 in the ESI†).
Photo-release of the organometallic complex
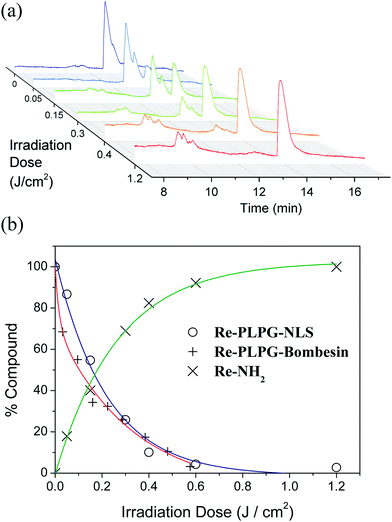
In order to verify the photo-uncaging efficiency of our system, the photo-release of Re-NH2 from the two prepared bioconjugates, namely Re-PLPG-NLS and Re-PLPG-Bombesin, was then investigated in phosphate buffer solution (PBS, pH 7.4). To determine the light dose required to photo-release Re-NH2 from Re-PLPG-NLS and Re-PLPG-Bombesin in cells, the two compounds were irradiated using a Rayonet UV reactor (350 nm) that was later used in the cell culture experiments. Both Re-PLPG-NLS and Re-PLPG-Bombesin completely photo-released Re-NH2 using a very low irradiation dose of 1.2 J cm−2 – four times lower than the dose used to completely photolyze the caged Ru complex previously reported by our group.21 During the irradiation, the composition of the solutions was monitored by LC-MS. As shown in Fig. 1 for Re-PLPG-NLS, prior to irradiation, only two peaks at 12.6–13.3 min, corresponding to the two diastereomers, were present. As the caged complex was irradiated, new peaks arose in the chromatograms. The first small peaks at 9.8–10 min were assigned to the by-products of the UV decomposition of the PLPG (Fig. S17 and S19†) that have been reported for o-nitrobenzyl photo-linkers.12 The released Re-NH2 complex is detected at 14.5 min. In the case of Re-PLPG-Bombesin, the order of peak appearance changes, as bombesin is more lypophilic than NLS (Fig. S18†). The released Re-NH2 is eluted first, followed by o-nitrobenzyl moiety by-products, while the remaining Re-PLPG-Bombesin peak is eluted last. Photolysis quantum yields were then measured using a 355 nm laser setup. As expected, the targeting peptide had no influence, and both Re-PLPG-NLS and Re-PLPG-Bombesin achieved a good photolysis quantum yield of 10 ± 2% (value obtained using azobenzene actinometry using the azobenzene quantum yield of 0.15, Fig. S20 and S21†).68(Photo-)toxicity
The cytotoxicity of the photo-caged bioconjugates Re-PLPG-NLS and Re-PLPG-Bombesin, as well as of the Re-NH2 complex, towards cancerous (HeLa) and non-cancerous (MRC-5) cells was evaluated in the presence and the absence of light. Cisplatin was used as a positive control (although it is not a PDT agent), in order to compare the cytotoxicity of our compounds to that of a successful metal-based drug. Re-PLPG-Bombesin was additionally tested on prostate cancer cells (PC-3), as the analogue of bombesin used in this study has been optimised on this cell line, which over-expresses GRPR.65,66 HeLa cells also over-express receptors belonging to the bombesin receptor family, namely the bombesin receptor subtype 3 (BSR-3), for which our bombesin derivative should also have some affinity.69 As shown in Table 1, 48 h of incubation, photo-caging and peptide conjugation strongly affected both the dark and light toxicity of the compounds. In general, the bioconjugates with NLS peptide (Re-NLS and Re-PLPG-NLS) showed a marked increase in dark cytotoxicity compared to Re-NH2 (up to 13-fold), while the bombesin derivatives displayed only moderate toxicity towards MRC-5 cells and remained inactive up to the highest concentration used in this study (100 μM) towards HeLa and PC-3. While the presence of the PLPG rendered Re-PLPG-NLS almost 2.5 times more toxic towards HeLa cells than Re-NLS, it did not affect the cytotoxicity of the bombesin derivative Re-PLPG-Bombesin towards HeLa and PC-3, and actually attenuated (up to 2-fold) the effect of the compounds on the non-cancerous cell line MRC-5.| Compound | HeLa darka (μM) | HeLa UV (μM) | MRC-5 darka (μM) | MRC-5 UV (μM) | PC-3 darka (μM) | PC-3 UV (μM) |
|---|---|---|---|---|---|---|
| a IC50 after 48 h treatment and incubation; n.d. = not determined. | ||||||
Re-NH2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
187.1 ± 17.9 | 17.3 ± 2.9 | >100 | 40.3 ± 5.4 | >100 | >100 |
| Re-NLS 1 | 35.1 ± 1.8 | 18.3 ± 1.4 | 17.8 ± 1.8 | 13.0 ± 2.5 | n.d. | n.d. |
| Re-PLPG-NLS | 14.5 ± 5.2 | 9.3 ± 0.8 | 36.2 ± 0.6 | 20.5 ± 5.5 | n.d. | n.d. |
| Re-Bombesin 1 | >100 | 5.3 ± 1.0 | 44.1 ± 9.9 | 41.6 ± 15.9 | >100 | 13.6 ± 1.7 |
| Re-PLPG-Bombesin | >100 | 9.7 ± 4.4 | 72.3 ± 3.6 | 23.3 ± 0.6 | >100 | 19.2 ± 2.4 |
| Cisplatin | 9.2 ± 0.6 | 26.8 ± 1.7 | 10.5 ± 2.8 | 47.8 ± 1.5 | 15.7 ± 3.5 | 74.8 ± 14.8 |
The influence of light on the activity of Re-PLPG-NLS and Re-PLPG-Bombesin was studied by incubating cells with the compounds for 4 h. This incubation time frame has been shown to allow sufficient uptake of Re-NH2 for its phototoxic effect to be observed.1 After incubation, the medium was replaced with a compound-free fresh medium, the cells were irradiated at 350 nm (UV reactor) with a low light dose of 2.58 J cm−2, and the viability of the cells was quantified 44 h later. Light irradiation increased the toxicity of almost all tested compounds towards HeLa, PC-3 and MRC-5. The UVA irradiation by itself did not affect cell viability (Fig. S22†). Interestingly, although light significantly increased the cytotoxicity of the Re-NH2 complex towards HeLa (10-fold) and MRC-5 cells (2.5-fold), irradiated PC-3 cells remained unaffected at a Re-NH2 concentration of up to 100 μM. Derivatization of Re-NH2 with bombesin allowed much more efficient targeting of this particular cancer cell line (5–7 phototoxic index (PTI)). In addition, the bombesin conjugates Re-Bombesin and Re-PLPG-Bombesin showed good light-induced increases in cytotoxicity towards the HeLa cell line (10–20 fold). However, no significant difference (within the error range) in phototoxicity between Re-Bombesin and Re-PLPG-Bombesin – i.e. no influence of the PLPG – was observed. These results suggest a predominant singlet oxygen-dependent mode of cytotoxic action that, together with an acceptable PTI towards cancer cell lines and moderate toxicity towards healthy MRC-5 cells, makes these two bombesin derivatives interesting for PDT applications. In fact, some commercially available PSs have comparable PTIs (at a similar light dose of 1.5 J cm−2), e.g. Photofrin (PTI 3), Hypercin (PTI 27) and ALA-induced PPIX.70 Only Foscan and Fospeg achieve PTIs of 268 and 4695 respectively.70 Compared with our previously reported caged Ru(II) species that use a similar photo-caging moiety, Re-Bombesin and Re-PLPG-Bombesin possess better phototoxicity characteristics (higher PTI and higher light toxicity).21 However, in the area of photoactive metal compounds acting via singlet oxygen production or ligand lability, similar or higher (typically in the 100–200 range) PTIs with both UVA and visible light have already been achieved,34,35,44,71–78 although often with higher light doses (up to 100 J cm−2) and longer irradiation times.
Upon light irradiation, the NLS derivatives behaved quite differently to the bombesin bioconjugates. Re-NLS and Re-PLPG-NLS possess lower PTIs (2 or less), and show a more pronounced difference between the PLPG-peptide conjugate (Re-PLPG-NLS) and the peptide-only derivative (Re-NLS). In this case, the presence of the PLPG confers a certain advantage. Re-PLPG-NLS had a stronger (2-fold) anti-proliferative effect on malignant HeLa cells compared with Re-NLS. In addition, Re-PLPG-NLS affected non-cancerous MRC-5 cells less, compared with Re-NLS. To explore the origin of these differences, the effect of Re-PLPG-NLS on cells and cellular components was studied in-depth.
Cellular localization
To investigate the biological behaviour of Re-PLPG-NLS in more detail, its cellular localisation was assessed. The known luminescence1,59,79 of the Re-NH2 complex allowed us to visualize the sub-cellular localization of its bioconjugates (Fig. 2). While the complex by itself1 and its photo-caged version Re-PLPG were observed exclusively in the cytoplasm (Fig. 2B and C), conjugation to an NLS peptide (Re-PLPG-NLS) brought the compound inside the nucleus, as designed (Fig. 2D). However, the compound did not show homogeneous distribution inside the nucleus, but accumulated in a particular sub-nuclear domain – the nucleolus. We previously reported this cellular distribution for Re-NH2 directly conjugated to NLS peptide.1 Responsible mainly for ribosomal RNA transcription, processing and assembly, nucleoli also play an important role in cell cycle regulation and stress response.80 As up-regulated ribosomal RNA synthesis and the consequent enlargement of nucleoli are characteristic of many cancer cells, these organelles have gained attention as promising drug targets in the field of anti-cancer research.81 | ||
| Fig. 2 Fluorescence microscopy of HeLa cells: (A) control; (B) Re-NH2; (C) Re-PLPG and (D) Re-PLPG-NLS (50 μM, 2 h). | ||
Quantification of Re-PLPG-NLS uptake in whole cells and nucleoli
Due to possible quenching effects inside the cells, fluorescence microscopy is not always reliable when it comes to quantifying sub-cellular accumulation of a compound. Such effects with this class of compounds were recently observed by our group.58,82 The cellular distribution of Re complexes can be quantitatively determined by isolating the cellular compartments of interest and measuring the Re content by ICP-MS. According to this method, we incubated HeLa cells with 20 μM of Re-NH2, Re-NLS and Re-PLPG-NLS for 2 h, and collected their pellets (whole cell samples) or isolated their nucleoli through differential centrifugation (nucleoli samples), following an established method with opportune modifications (Fig. S23 in the ESI†).83 Both nucleoli and whole cell extracts were then lysed, the protein content was measured, and the obtained samples were lyophilized and analysed by ICP-MS. The Re content of the whole cell samples followed the order of accumulation Re-NH2 ≪ Re-NLS < Re-PLPG-NLS. Interestingly, these data demonstrated very efficient uptake of Re-PLPG-NLS by HeLa. Indeed, in just 2 h, the cells internalised almost half of all Re available in solution. Moreover, evaluation of the accumulation of the target Re compound in the nucleoli indicated that approximately 25% of Re-PLPG-NLS taken up by the cells was concentrated in these organelles (Fig. S23†). The results obtained from the quantitative determination are in good agreement with the microscopic experiments and emphasize the fast and significant uptake of the Re complex.Effect of Re-PLPG-NLS on nucleic acids
In order to achieve a better understanding of the possible mode of action of our Re compound, we investigated the possible damage to DNA or RNA induced by Re-PLPG-NLS in the dark and upon irradiation. Given the high positive charge of Re-PLPG-NLS (5+) at physiological pH and the fact that the NLS sequence is arginine- and lysine-rich, its DNA/RNA aggregating behaviour could be hypothesized. Electrostatic forces indeed play an important role in the interactions of small molecules and proteins with nucleic acids, due to the polyanionic nature of the latter. Arginine and lysine groups in peptides and proteins are heavily involved in H-bonds in DNA/RNA–protein complexes. Most of the small molecule compounds targeting nucleic acids carry multiple positive charges as well.84,85 DNA photo-cleavage experiments were performed on a closed circular plasmid DNA (namely pcDNA3, see Fig. 3A). The DNA was incubated with various concentrations of Re-PLPG-NLS, and irradiated with UVA for 10 minutes. Re-PLPG-NLS cleaved the plasmids upon light irradiation in a concentration-dependent manner, converting the supercoiled DNA form (intact) to a circular relaxed/nicked form (damaged). Linearization of pcDNA3 was observed at a concentration as low as 10 μM, and complete conversion of the supercoiled to the nicked configuration occurred at a concentration of 50 μM (Fig. 3A). DNA incubated with the rhenium bioconjugate in the dark did not show a significant effect up to the highest concentration used in this study (50 μM). It was also observed that the plasmid treated with Re-PLPG-NLS displayed a smear band for high concentrations of Re-PLPG-NLS in the dark. This effect is due to NLS/DNA interactions, which produced the deformed band, as confirmed by DNA photo-cleavage experiments on NLS alone (see Fig. S24 in the ESI†).As RNA is one of the major components of nucleoli, we additionally investigated the effect of Re-PLPG-NLS on a 638 nucleotide long RNA sequence (D135-L14 ribozyme derived from the Sc.ai5γ group II intron of S. cerevisiae).86 Denaturing polyacrylamide gel electrophoresis (Fig. 3B) revealed a considerable decrease in intensity and shift of the RNA band due to Re-PLPG-NLS/RNA interactions and formation of non-migrating RNA agglomerates (Fig. S26B†) at high concentrations of Re-PLPG-NLS (100 μM). This effect occurred both in the absence and the presence of light, and no difference was observed for UVA-irradiated samples. Re-NH2 or Re-PLPG alone did not affect the RNA (Fig. S27 and S28†). Given the localization of Re-PLPG-NLS in RNA-rich nucleoli, it is possible that the strong dark toxicity of this compound is linked to its effect on RNA.
Morphological features induced by the targeted rhenium complex
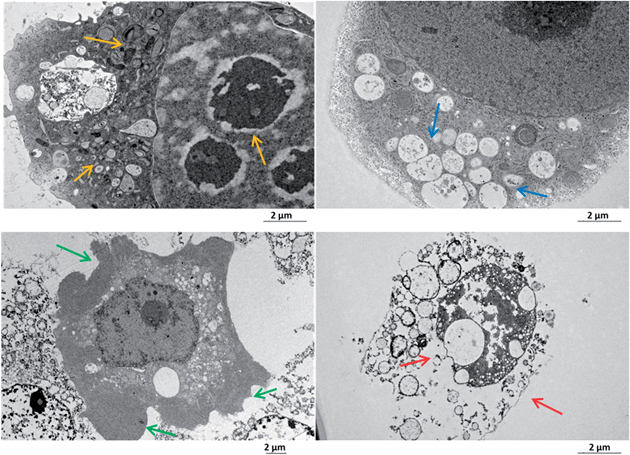
The deep cellular onset provoked by the target Re-PLPG-NLS is not just related to an isolated biochemical pathway or biological structure, but is also mirrored by macroscopic morphological features. Major alterations can be conveniently monitored by transmission electron microscopy (TEM), which is a particularly powerful technique to distinguish sub-cellular compartments, hence giving precious insights into the overall cell stress response.87–90Due to its traceability through confocal microscopy, its valuable anti-proliferative effects both in the dark and upon irradiation, and its strong interaction with DNA and RNA, Re-PLPG-NLS was chosen for this further biological characterization. TEM images of irradiated and non-irradiated HeLa cells incubated for 2 h with Re-PLPG-NLS were recorded. As shown in Fig. 4, the resulting images (selection from a total pool of 40–60 images per sample, see Fig. S29 in the ESI† for other examples) suggested that Re-PLPG-NLS induced the onset of severe cellular stress, with features ascribable to late apoptosis and necrosis. The sudden occurrence of membrane blebs, cytoplasm condensation, organelle packaging, extended vacuolation and nuclear pyknosis indicated an initial triggering of apoptosis. However, the extremely rapid uptake and activity of Re-PLPG-NLS (confirmed by ICP-MS measurements, see “Quantification of Re-PLPG-NLS uptake in whole cells and nucleoli” section) caused the balance to shift towards necrosis-related effects, such as loss of membrane integrity and nuclear fragmentation (see Fig. 4).
Moreover, coherent with the insights obtained from the cellular localization studies, major attention was paid to the specific effects triggered in nucleoli by Re-PLPG-NLS. Treatment with the rhenium conjugate influenced the nucleoli morphology. In good agreement with established data, the target sub-organelle presented a stress-altered morphology, characterized by indicators such as the formation of nuclear caps around the nucleolar body, separation of the fibrillar centre from the granular centre, unravelling of the fibrillar centre and migration of the whole nucleolar body to the peripheral part of the nucleus (Fig. S30 in the ESI†).91,92 All these features represent the main characteristics of nucleolar segregation, which could contribute to the initial apoptotic event, as well as to the overall cytotoxicity of Re-PLPG-NLS.
Mechanism of cell death induced by Re-PLPG-NLS
The mobilization of phosphatidylserine from the inner to the outer layer of the cellular membrane promotes phagocytosis and is considered a clear marker of an ongoing early apoptotic event.93 Under severe stress, the loss of membrane integrity enables a large pool of solutes to penetrate the plasma membrane (which is normally impermeable to them), and represents a signal of the occurrence of necrosis. The combination of two staining methods allows evaluation of the cell death mechanism in large cell populations by flow cytometry, using the Annexin V/propidium iodide (PI) assay. The rapid triggering of cell death processes after short incubation treatment of HeLa cells with Re-PLPG-NLS investigated by TEM showed a combination of apoptotic and necrotic stimuli. Hence, it was of interest to study the contributions of these two pathways to the cytotoxicity of Re-PLPG-NLS. In agreement with the TEM data, Re-PLPG-NLS induced a drastic cellular effect after only 2 h incubation with HeLa cells, with less than 10% of cells still viable (see Fig. 5 and S31 in the ESI†). The majority of the cell population displayed membrane symmetry due to phosphatidylserine migration (Annexin V positive) and membrane permeabilisation, which allowed PI internalization (PI positive). The data support the thesis that Re-PLPG-NLS induces severe cellular stress after a short incubation time, to which cells react by activating apoptosis. However, the very dynamic onset of these events resulted in a pronounced late apoptotic profile (see Fig. 5). These effects could be observed for both probes in the dark and upon irradiation, and were irreversible since the cells did not show any recovery over a time frame of 24 h. Camptothecin was used as a positive control.Conclusions
In this article, we described the efficient caging of a Re(I) tricarbonyl N,N-bis(quinolinoyl) complex (Re-NH2) with a photo-linker (PLPG) to give Re-PLPG. The latter was further conjugated to two different peptides for potential organelle/cellular uptake (i.e. bombesin or NLS) to give Re-PLPG-NLS or Re-PLPG-Bombesin. Photolysis experiments showed that complete release of Re-NH2 could be achieved using a very low light dose (1.2 J cm−2). To the best of our knowledge, these are the first two examples of the selective release of an organometallic complex from a bioconjugate using light activation. A first biological screening on different cancer and non-tumorigenic cell lines demonstrated the very valuable toxicity of the target complexes, with IC50 values in the range of the established anti-cancer drug cisplatin. It is noteworthy that, while Re-PLPG-Bombesin showed a more marked 1O2-dependent cytotoxicity at low irradiation energy (2.58 J cm−2), with a certain preference for cancer cells over MRC-5, Re-PLPG-NLS showed a stronger (2-fold) anti-proliferative effect on malignant HeLa cells compared with Re-NLS. In addition, Re-PLPG-NLS affected non-cancerous MRC-5 cells less, compared with Re-NLS. Due to this interesting behaviour, Re-PLPG-NLS was chosen for in-depth biological characterization. Cellular biodistribution studies using ICP-MS indicated an outstanding cellular uptake, close to 50% of all available Re, with very rapid and strong accumulation in the nucleoli. The penetration into sub-cellular compartments (nucleus/nucleoli) allowed interaction between the Re complex and nucleic acids. Re-PLPG-NLS was able to photo-cleave DNA and created non-migrating RNA agglomerates. This molecular dysfunction contributed to the induction of late apoptotic and necrotic effects, mirrored by morphological features such as nucleolar segregation, pyknosis and extended vacuolation, which resulted in cell death.In conclusion, in this article, we present, for the first time, a strategy (organometallic complex – PLPG – targeting vector) to selectively release an organometallic complex upon light irradiation. Current efforts are aimed at increasing the ratio of dark vs. UV light toxicity and producing this photo-cytotoxic action in the PDT window.
Experimental section
Materials
Chemicals and solvents were of reagent grade or better, purchased from commercial suppliers and used without further purification unless otherwise specified. The plasmid pcDNA3 was obtained from Invitrogen.Instrumentation and methods
1H and 13C NMR measurements were carried out on Bruker 400 and 500 spectrometers and referenced to residual solvent peaks. UV spectra were recorded on a Cary 50 Scan Varian spectrophotometer. ESI-MS, LC-MS and UPLC-MS were performed using a Bruker Daltonics HCT 6000 mass spectrometer. LC-MS and UPLC-MS spectra were recorded on a Waters Acquity™ system equipped with a PDA detector and an auto-sampler. LC-MS was carried out using an Agilent Zorbax 300 SB-C18 analytical column (3.5 μm particle size, 300 Å pore size, 150 × 4.6 mm). The LC run (flow rate: 0.5 mL min−1) was performed with a linear gradient of A (double distilled water containing 0.1% v/v formic acid) and B (acetonitrile containing 0.1% v/v formic acid): t = 0 min, 5% B; t = 3 min, 5% B; t = 17 min, 100% B; t = 20 min, 100% B; t = 25 min, 5% B. UPLC-MS was performed on an Acquity UPLC BEH C18 column (2.1 × 50 mm, 1.7 μm). The UPLC run (flow rate: 0.6 mL min−1) was carried out with a linear gradient of A (double distilled water containing 0.1% v/v formic acid) and B (acetonitrile containing 0.1% v/v formic acid): t = 0 min, 5% B; t = 0.25 min, 5% B; t = 1.5 min, 100% B; t = 2.5 min, 100% B. High resolution ESI-MS spectra were recorded using a Bruker ESQUIRE-LC quadrupole ion trap instrument. Preparative HPLC purification was carried out on a Varian ProStar system and an Agilent Zorbax 300 SB-C18 prep column (5 μm particle size, 300 Å pore size, 150 × 21.1 mm. Flow rate: 20 mL min−1). The runs were performed with a linear gradient of A (double distilled water containing 0.1% v/v trifluoroacetic acid (TFA)) and B (acetonitrile containing 0.1% v/v TFA, Sigma-Aldrich HPLC grade). Preparative runs: t = 0 min, 5% B; t = 25 min, 100% B; t= 30 min, 100% B; t = 32 min, 5% B. Photolysis quantum yields were determined using an Edinburgh LP-920 setup equipped with a Continuum Surelite laser (355 nm). Cell culture irradiation was performed using a Rayonet RPR-200 photochemical reactor with 6 bulbs (14 W each) and maximum intensity output at 350 nm. Samples were irradiated in a fluorescence quartz cuvette (width 1 cm) placed in the centre of the reactor. The light intensity at that spot, measured with an X11 optometer (Gigahertz-Optik), was 42 W m−2. The temperature inside the reactor was 30 °C.Synthesis and characterization
NLS (Cys-Arg-DArg-Arg-Lys(CONH2)). The peptide was synthesised following a previously published procedure. The analytical data were in agreement with the literature report.1
Bombesin (Cys-βAla-βAla-Gln-Trp-Ala-Val-Gly-His-Cha-Nle-(CONH2)). The peptide was synthesised following a previously published procedure. The analytical data were in agreement with the literature report.1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 mixture of methanol–acetonitrile–water (45 mL). The reaction mixture was stirred overnight at room temperature. MeOH was removed using a rotary evaporator, while water and acetonitrile were removed by lyophilisation. The compound was purified by preparative HPLC to afford a yellow-white solid. Characterisation data: ESI-MS m/z 830.3 [M + H]2+, 553.9 [M + 2H]3+, 415.7 [M + 3H]4+. ESI HRMS calcd for [C68H92N22O14ReS]/z [M + H]2+ 830.32599, found 830.32526, calcd for [M + 2H]3+ 553.88660, found 553.88599, calcd for [M + 3H]4+ 415.66691, found 415.66631.
2 mixture of methanol–acetonitrile–water (45 mL). The reaction mixture was stirred overnight at room temperature. MeOH was removed using a rotary evaporator, while water and acetonitrile were removed by lyophilisation. The compound was purified by preparative HPLC to afford a yellow-white solid. Characterisation data: ESI-MS m/z 830.3 [M + H]2+, 553.9 [M + 2H]3+, 415.7 [M + 3H]4+. ESI HRMS calcd for [C68H92N22O14ReS]/z [M + H]2+ 830.32599, found 830.32526, calcd for [M + 2H]3+ 553.88660, found 553.88599, calcd for [M + 3H]4+ 415.66691, found 415.66631.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of acetonitrile–water (45 mL). The reaction mixture was stirred overnight at room temperature. The solvent was removed using a lyophiliser, and the crude compound was purified by HPLC to afford a yellow-white solid. Characterisation data: ESI-MS m/z 1075.3 [M + H]2+, 717.4 [M + 2H]3+, 538.4 [M + 3H]4+. ESI HRMS calcd for [C68H92N22O14ReS]/z [M + H]2+ 1075.42557, found 1075.42494, calcd for [M + 2H]3+ 717.28632, found 717.28571. Anal. found: C, 49.97; H, 4.90; N, 12.01. Calcd for [C90H109F9N19O22ReS]: C, 49.69; H, 4.94; N, 12.38.
1 mixture of acetonitrile–water (45 mL). The reaction mixture was stirred overnight at room temperature. The solvent was removed using a lyophiliser, and the crude compound was purified by HPLC to afford a yellow-white solid. Characterisation data: ESI-MS m/z 1075.3 [M + H]2+, 717.4 [M + 2H]3+, 538.4 [M + 3H]4+. ESI HRMS calcd for [C68H92N22O14ReS]/z [M + H]2+ 1075.42557, found 1075.42494, calcd for [M + 2H]3+ 717.28632, found 717.28571. Anal. found: C, 49.97; H, 4.90; N, 12.01. Calcd for [C90H109F9N19O22ReS]: C, 49.69; H, 4.94; N, 12.38.
Determination of photo-uncaging quantum yields
20 mM DMSO stock solutions of the caged compounds were diluted in phosphate buffer (pH = 7.4) to achieve an optical density at 355 nm (OD(λ = 355 nm)) of approximately 0.2. Aliquots of 100 μL were irradiated at 355 nm using an Edinburgh LP-920 setup equipped with a Continuum Surelite laser. The laser was slightly misaligned to reach a suitable irradiation power. After certain irradiation intervals (number of laser shots), the solutions were transferred to amber HPLC vials with 200 μL inlets. 20 μL of the solutions were injected into an LC-MS system. The percentage of remaining compound was plotted against the number of laser shots and fitted with a single exponential decay curve:C = C0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−kx) exp(−kx) |
The initial slope (less than 20% conversion) of the curve (Scompound) was determined by taking the tangent to the first (<20% decomposition) points of the exponential fit. It was then used to calculate the photolysis quantum yield by comparing it to the quantum yield of azobenzene photoisomerization. For that purpose, trans-azobenzene was dissolved in methanol to obtain an OD(λ = 355 nm) of approximately 0.2. It was then irradiated under the same conditions as the compounds for the measurements. UV/vis absorbance of the azobenzene solution at 355 nm was then monitored. The remaining amount of trans-azobenzene was then plotted against the number of laser shots, and the initial slope of the curve (Sreference) was then obtained. The photolysis quantum yields were then calculated using:
Photolysis in UV reactor
20 mM DMSO stock solutions of the compounds to be irradiated were diluted with 4 mL of PBS buffer (pH 7.4) to obtain absorbance of 0.2 at 350 nm. The solutions were then irradiated in fluorescence quartz cuvettes (width 1 cm). At each irradiation time, the solution compositions were analysed using LC-MS. The relative concentrations of the photolysed compounds were calculated from the areas of the corresponding LC-MS peaks. Concentration % plotted as a function of photolysis time (or irradiation dose) could be fitted with a single exponential (first order kinetics law):C = C0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−kx) exp(−kx) |
Cell culture
Human cervical carcinoma cells (HeLa) were cultured in DMEM (Gibco) with 5% fetal calf serum (FCS, Gibco), 100 U mL−1 penicillin and 100 μg mL−1 streptomycin at 37 °C and 5% CO2. Normal lung fibroblasts (MRC-5) were maintained in F-10 medium (Gibco) supplemented with 10% FCS (Gibco), 100 U mL−1 penicillin and 100 μg mL−1 streptomycin at 37 °C and 5% CO2. Prostate carcinoma cells (PC-3) were cultured in Ham's F-12K (Kaighn's) Medium (Gibco) supplemented with 10% FCS (Gibco), 100 U mL−1 penicillin and 100 μg mL−1 streptomycin at 37 °C and 5% CO2.Cytotoxicity studies
The cytotoxicity of the rhenium complexes and their bioconjugates towards HeLa, MRC-5 and PC-3 cells in the presence and absence of UV irradiation was measured using a fluorometric cell viability assay using resazurin (Promocell GmbH). Cells were seeded in triplicate in 96-well plates at a density of 4 × 103 cells per well in 100 μL medium 24 h prior to treatment. To assess cytotoxicity, cells were treated with increasing concentrations of the compounds for 48 h, whereas, to assess phototoxicity, cells were treated with increasing concentrations of the compounds for 4 h only. After that, the medium was removed and replaced by fresh complete medium prior to 10 min UVA irradiation (2.58 J cm−2). Cells were then returned to the incubator for 48 h. After incubation, the medium was replaced by 100 μL complete medium containing resazurin (0.2 mg mL−1 final concentration). After 4 h of incubation at 37 °C (overnight incubation for PC-3 due to slower metabolism of this cell line), fluorescence of the highly red fluorescent resorufin product at 590 nm emission was quantified using a SpectraMax M5 microplate reader with a 540 nm excitation wavelength.In vitro fluorescence evaluation
Cellular localization of the luminescent rhenium complexes and bioconjugates was assessed using fluorescence microscopy. HeLa cells were grown in 2 mL medium on 18 mm Menzel-glaser coverslips at a density of 1 × 105 cells per mL and incubated with rhenium complexes at 50 μM for 2 h. Cells were fixed in 4% formaldehyde solution and mounted on slides for viewing by confocal microscopy on a CLSM Leica SP5 microscope. The rhenium complexes were excited at 405 nm, and emission above 420 nm was recorded.DNA photo-cleavage
The DNA photo-cleavage effect provoked by complex Re-PLP-NLS was investigated by electrophoresis. Supercoiled pcDNA3 plasmid (0.10 μg) was treated with increasing concentrations of the rhenium compounds in buffer (50 mM Tris–HCl, 18 mM NaCl, pH 7.2), incubated for 30 minutes at 25 °C and irradiated at 350 nm for 10 minutes (Rayomet Chamber Reactor, 2.58 J cm−2). A series of negative controls, in which the plasmid was treated with NLS or Re-PLP-NLS in the dark at different incubation temperatures and in the presence of BstXI restriction enzyme (to visualize the linearized form), were used for comparative purposes (see Fig. S18†). After irradiation, sample loading buffer (250 mg xylene cyanol in 33 mL of 150 mM Tris–HCl buffer, pH 7.6, 0.1 M EDTA, 20% Ficoll 400 in 100 mL of H2O) was added, and the probes were analyzed by electrophoresis in 0.8% agarose in 1× TBE at 70 V (Biorad Powerpack 1000, BioRad) for 2 h. The gel was pre-stained with 0.5 μg mL−1 ethidium bromide, photographed and examined using an AlphaDigiDoc 1000 CCD camera (Buchner Biotec AG) and AlphaImager software v1.3.0.7.RNA
Determination of the rhenium complex content in biological samples
Whole cells. To perform ICP-MS measurements, HeLa cells were seeded two days before treatment at a concentration of 4 × 105 cells per mL in a 75 cm2 cell culture flask until 80% confluence, and incubated with the target complexes, namely Re-NH2, Re-NLS or Re-PLPG-NLS, at 20 μM for 2 hours. The medium was removed, and the cells were washed with PBS and trypsinized. After re-suspension in PBS, the pellets were collected by centrifugation at 5500 rpm for 4.5 minutes. The pellets were re-dissolved in 500 μL of PBS, lysed by a freeze–thaw cycle and treated in an ultrasonic bath (Digitana AG) for 20 minutes. The lysates were lyophilized using an Alpha 2-4 LD plus (CHRIST).
Cell nucleoli. Nucleoli of HeLa cells were collected following an established procedure with appropriate modifications.81 Briefly, HeLa cells were seeded two days before treatment at a concentration of 4 × 105 cells per mL in a 75 cm2 cell culture flask until 80% confluence. Cells were treated with 20 μM Re-NH2, Re-NLS or Re-PLPG-NLS for 2 h, and the pellets were collected and washed 3 times with ice cold PBS. The pellets were incubated with lysis buffer (10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, pH 7.9) containing a protease inhibitors cocktail (1/500 v/v, batch number: P8340, Sigma-Aldrich) for 10 minutes, the membranes were disrupted and the cells were homogenized in a 7 mL Dounce Homogenizator with a tight pestle (type A, 20 strokes). The suspensions were centrifuged at 250g at 4 °C in a Centrifuge 5810R (Eppendorf) for 5 minutes, and the supernatant was removed. The pellets were re-dissolved in 3 mL of a sucrose solution (0.25 M sucrose, 10 mM MgCl2) and layered with 3 mL of a second hypertonic sucrose solution (0.35 M sucrose, 0.5 mM MgCl2). The suspensions were centrifuged at 1450g at 4 °C for 5 minutes. The supernatant was discarded, and the pellets were re-suspended in 3 mL of the second sucrose solution. The suspensions containing the nuclear fraction were sonicated 5–6 times in 10 s bursts (Bandelin Sonoplus GM70, 50% cycle, intervals of 10 s), layered with a third hypertonic sucrose solution (0.88 M sucrose, 0.5 mM MgCl2) and centrifuged at 3000g at 4 °C for 10 minutes. The pellets were re-suspended in the second sucrose solution (0.35 M sucrose, 0.5 mM MgCl2), centrifuged at 1450g at 4 °C for 5 minutes to obtain the pure nucleoli extracts (see Fig. S17†), and stored at −80 °C. All the steps of the isolation procedure were monitored under a phase contrast microscope on Menzel-gläser coverslips (Olympus IX81). The collected nucleoli solutions were lyophilized using an Alpha 2-4 LD plus (CHRIST).
Transmission electron microscopy
HeLa cells were seeded in 6-well plates containing carbon-coated sapphire discs and glass cover slips at a concentration of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL and cultured at 37 °C/5% CO2 for one day (>80% confluence on the discs). The medium was then removed and replaced with medium containing 20 μM Re-PLG-NLS. The cells were further incubated for 2 hours and irradiated at 350 nm for 10 minutes (2.58 J cm−2) or left non-irradiated (control). Subsequently, the cells were processed by chemical fixation and high-pressure freezing, as follows.
000 cells per mL and cultured at 37 °C/5% CO2 for one day (>80% confluence on the discs). The medium was then removed and replaced with medium containing 20 μM Re-PLG-NLS. The cells were further incubated for 2 hours and irradiated at 350 nm for 10 minutes (2.58 J cm−2) or left non-irradiated (control). Subsequently, the cells were processed by chemical fixation and high-pressure freezing, as follows.
Ultrathin 50 nm sections of all specimens were contrasted with uranyl acetate and lead citrate, and analysed with a Tecnai G2 Spirit or CM 100 transmission electron microscope (FEI, Eindhoven, The Netherlands) using an ORIUS 1000 CCD camera (Gatan, Munich, Germany).
Annexin V/PI assay
The induction of apoptosis and necrosis in HeLa cells treated with Re-PLPG-NLS was evaluated by means of the well-established Annexin V/PI assay, using flow cytometry (BD Pharmingen, Bioscience) according to the manufacturer’s instructions with minor changes (BD Pharmingen, Mat. no. 556419). Briefly, cells were seeded in a Petri dish at a concentration of 2 × 105 cells mL−1 and cultured at 37 °C/6% CO2 for 24 h. The medium was replaced with fresh medium containing 20 μM Re-PLPG-NLS. The cells were incubated for 2 h, and then the medium was replaced with fresh medium and the Petri dish was irradiated at 350 nm for 10 minutes (2.58 J cm−2). To observe the direct effects after irradiation, the cells were immediately collected for FACS analysis, whereas, for the recovery experiments, the medium was replaced and the cells were further incubated for 24 h at 37 °C/6% CO2. The cells were then trypsinized and pelleted, washed twice with ice cold PBS, centrifuged at 250g for 5 minutes and re-suspended in binding buffer (10 mM HEPES, 140 mM NaCl, and 2.5 mM CaCl2, pH 7.4) at a concentration of 1 × 106 cells per mL. 100 μL of the obtained suspension was transferred into an FACS culture tube (1 × 105 cells), and 5 μL of FITC Annexin V complex solution and 5 μL of a 50 μg mL−1 propidium iodide (PI) solution were added. Samples were incubated for 15 minutes at room temperature (25 °C) in the dark, 400 μL of binding buffer was added, and the probes were analyzed using a CynAn ADP9 flow cytometer with the FITC (for Annexin V-FITC, excitation = 488 nm, emission = 515–545 nm) and PE-Texas Red channels (for PI, excitation = 488 nm, emission = 564–606 nm). The data were analyzed using Summit v4.3 software. Camptothecin, used at 5 μM for 2 or 24 h, and untreated cells in the dark or under irradiation were used as positive controls.Acknowledgements
This work was supported by the Swiss National Science Foundation (Professorship no. PP00P2_133568 and Research Grants no. 200021_129910 and no. 200020_146776 to G.G.), the University of Zurich (G.G. and S.F.), the Stiftung für Wissenschaftliche Forschung of the University of Zurich (G.G. and S.F.), the Novartis Jubilee Foundation (G.G. and R.R.), the Stiftung zur Krebsbekämpfung (S.F.), the Huggenberger-Bischoff Stiftung (S.F.), the European Research Council (ERC Starting Grant to R.K.O.S.), the COST Action CM1105 (R.K.O.S. and G.G.), the State Secretariat for Education, Research and Innovation (R.K.O.S.) and the University of Zurich Priority Program (S.F.).Notes and references
- A. Leonidova, V. Pierroz, R. Rubbiani, J. Heier, S. Ferrari and G. Gasser, Dalton Trans., 2014, 43, 4287–4294 RSC.
- A. F. Taub, Dermatol. Clin., 2007, 25, 101–109 CrossRef CAS PubMed.
- D. M. A. Vera, M. H. Haynes, A. R. Ball, T. Dai, C. Astrakas, M. J. Kelso, M. R. Hamblin and G. P. Tegos, Photochem. Photobiol., 2012, 88, 499–511 CrossRef CAS PubMed.
- D. Vecchio, T. Dai, L. Huang, L. Fantetti, G. Roncucci and M. R. Hamblin, J. Biophotonics, 2013, 6, 733–742 CrossRef CAS PubMed.
- J.-H. Park, M.-Y. Ahn, Y.-C. Kim, S.-A. Kim, Y.-H. Moon, S.-G. Ahn and J.-H. Yoon, Biol. Pharm. Bull., 2012, 35, 509–514 CAS.
- F. Gad, T. Zahra, K. P. Francis, T. Hasan and M. R. Hamblin, Photochem. Photobiol. Sci., 2004, 3, 451–458 CAS.
- X. Ragas, T. Dat, G. P. Tegos, M. Agut, S. Nonell and M. R. Hamblin, Lasers Surg. Med., 2010, 42, 384–390 CrossRef PubMed.
- H. Deppe, T. Mucke, S. Wagenpfeil, M. Kesting and A. Sculean, Quintessence International, 2013, 44, 609–618 Search PubMed.
- G. Bozzini, P. Colin, N. Betrouni, C. A. Maurage, X. Leroy, S. Simonin, C. Martin-Schmitt, A. Villers and S. Mordon, Photodiagn. Photodyn. Ther., 2013, 10, 296–303 CrossRef CAS PubMed.
- M. J. Brown, Tumor Hypoxia in Cancer Therapy, in Methods in Enzymology, ed. H. Sies and B. Brüne, Academic Press, 2007, vol. 435, pp. 295–321 Search PubMed.
- G. Mayer and A. Heckel, Angew. Chem., Int. Ed., 2006, 45, 4900–4921 CrossRef CAS PubMed.
- M. Goeldner and R. Crivens, Dynamic Studies in Biology, Wiley-VCH, 2005 Search PubMed.
- C. P. McCoy, C. Rooney, C. R. Edwards, D. S. Jones and S. P. Gorman, J. Am. Chem. Soc., 2007, 129, 9572–9573 CrossRef CAS PubMed.
- P. D. Kehayova, C. D. Woodrell, P. J. Dostal, P. P. Chandra and A. Jain, Photochem. Photobiol. Sci., 2002, 1, 774–779 CAS.
- Y. Wei, Y. Yan, D. Pei and B. Gong, Bioorg. Med. Chem. Lett., 1998, 8, 2419–2422 CrossRef CAS.
- M. Noguchi, M. Skwarczynski, H. Prakash, S. Hirota, T. Kimura, Y. Hayashi and Y. Kiso, Bioorg. Med. Chem., 2008, 16, 5389–5397 CrossRef CAS PubMed.
- W. Lin, D. Peng, B. Wang, L. Long, C. Guo and J. Yuan, Eur. J. Org. Chem., 2008, 2008, 793–796 CrossRef.
- S. Ibsen, E. Zahavy, W. Wrasdilo, M. Berns, M. Chan and S. Esener, Pharm. Res., 2010, 27, 1848–1860 CrossRef CAS PubMed.
- N. Ueberschaar, H.-M. Dahse, T. Bretschneider and C. Hertweck, Angew. Chem., Int. Ed., 2013, 52, 6185–6189 CrossRef CAS PubMed.
- M. Bio, P. Rajaputra, G. Nkepang, S. G. Awuah, A. M. L. Hossion and Y. You, J. Med. Chem., 2013, 56, 3936–3942 CrossRef CAS PubMed.
- T. Joshi, V. Pierroz, C. Mari, L. Gemperle, S. Ferrari and G. Gasser, Angew. Chem., Int. Ed., 2014, 53, 2960–2963 CrossRef CAS PubMed.
- N. J. Farrer, L. Salassa and P. J. Sadler, Dalton Trans., 2009, 10690–10701 RSC.
- D. Maeda, H. Shimakoshi, M. Abe and Y. Hisaeda, Dalton Trans., 2009, 140–145 RSC.
- A. F. Westendorf, J. A. Woods, K. Korpis, N. J. Farrer, L. Salassa, K. S. Robinson, V. Appleyard, K. Murray, R. Grünert, A. M. Thompson, P. J. Sadler and P. J. Bednarski, Mol. Cancer Ther., 2012, 11, 1894–1904 CrossRef CAS PubMed.
- C. R. Maldonado, N. Gomez-Blanco, M. Jauregui-Osoro, V. G. Brunton, L. Yate and J. C. Mareque-Rivas, Chem. Commun., 2013, 49, 3985–3987 RSC.
- J. S. Butler and P. J. Sadler, Curr. Opin. Chem. Biol., 2013, 17, 175–188 CrossRef CAS PubMed.
- J. Mlcouskova, J. Stepankova and V. Brabec, J. Biol. Inorg. Chem., 2012, 17, 891–898 CrossRef CAS PubMed.
- Y. Zhao, G. M. Roberts, S. E. Greenough, N. J. Farrer, M. J. Paterson, W. H. Powell, V. G. Stavros and P. J. Sadler, Angew. Chem., Int. Ed., 2012, 51, 11263–11266 CrossRef CAS PubMed.
- M. Roy, T. Bhowmick, R. Santhanagopal, S. Ramakumar and A. R. Chakravarty, Dalton Trans., 2009, 4671–4682 RSC.
- J. Talib, D. G. Harman, C. T. Dillon, J. Aldrich-Wright, J. L. Beck and S. F. Ralph, Dalton Trans., 2009, 504–513 RSC.
- A. K. Patra, T. Bhowmick, S. Roy, S. Ramakumar and A. R. Chakravarty, Inorg. Chem., 2009, 48, 2932–2943 CrossRef CAS PubMed.
- T. K. Goswami, M. Roy, M. Nethaji and A. R. Chakravarty, Organometallics, 2009, 28, 1992–1994 CrossRef CAS.
- F. Barragan, P. Lopez-Senin, L. Salassa, S. Betanzos-Lara, A. Habtemariam, V. Moreno, P. J. Sadler and V. Marchan, J. Am. Chem. Soc., 2011, 133, 14098–14108 CrossRef CAS PubMed.
- B. S. Howerton, D. K. Heidary and E. C. Glazer, J. Am. Chem. Soc., 2012, 134, 8324–8327 CrossRef CAS PubMed.
- E. Wachter, D. K. Heidary, B. S. Howerton, S. Parkin and E. C. Glazer, Chem. Commun., 2012, 48, 9649–9651 RSC.
- M. Frasconi, Z. Liu, J. Lei, Y. Wu, E. Strekalova, D. Malin, M. W. Ambrogio, X. Chen, Y. Y. Botros, V. L. Cryns, J.-P. Sauvage and J. F. Stoddart, J. Am. Chem. Soc., 2013, 135, 11603–11613 CrossRef CAS PubMed.
- A. Hussain, S. Gadadhar, T. K. Goswami, A. A. Karande and A. R. Chakravarty, Dalton Trans., 2012, 41, 885–895 RSC.
- A. Kastl, A. Wilbuer, A. L. Merkel, L. Feng, P. Di Fazio, M. Ocker and E. Meggers, Chem. Commun., 2012, 48, 1863–1865 RSC.
- Q.-X. Zhou, W.-H. Lei, Y.-J. Hou, Y.-J. Chen, C. Li, B.-W. Zhang and X.-S. Wang, Dalton Trans., 2013, 42, 2786–2791 RSC.
- L. Zayat, C. Calero, P. Albores, L. Baraldo and R. Etchenique, J. Am. Chem. Soc., 2003, 125, 882–883 CrossRef CAS PubMed.
- L. Zayat, M. Salierno and R. Etchenique, Inorg. Chem., 2006, 45, 1728–1731 CrossRef CAS PubMed.
- M. Salierno, C. Fameli and R. Etchenique, Eur. J. Inorg. Chem., 2008, 2008, 1125–1128 CrossRef.
- L. Zayat, M. G. Noval, J. Campi, C. I. Calero, D. J. Calvo and R. Etchenique, ChemBioChem, 2007, 8, 2035–2038 CrossRef CAS PubMed.
- T. Respondek, R. N. Garner, M. K. Herroon, I. Podgorski, C. Turro and J. J. Kodanko, J. Am. Chem. Soc., 2011, 133, 17164–17167 CrossRef CAS PubMed.
- K. L. Ciesienski, L. M. Hyman, D. T. Yang, K. L. Haas, M. G. Dickens, R. J. Holbrook and K. J. Franz, Eur. J. Inorg. Chem., 2010, 2010, 2224–2228 CrossRef.
- K. L. Ciesienski, K. L. Haas, M. G. Dickens, Y. T. Tesema and K. J. Franz, J. Am. Chem. Soc., 2008, 130, 12246–12247 CrossRef CAS PubMed.
- A. A. Kumbhar, A. T. Franks, R. J. Butcher and K. J. Franz, Chem. Commun., 2013, 49, 2460–2462 RSC.
- G. C. R. Ellis-Davies, Chem. Rev., 2008, 108, 1603–1613 CrossRef CAS PubMed.
- C. Gwizdala, D. P. Kennedy and S. C. Burdette, Chem. Commun., 2009, 6967–6969 RSC.
- H. M. D. Bandara, D. P. Kennedy, E. Akin, C. D. Incarvito and S. C. Burdette, Inorg. Chem., 2009, 48, 8445–8455 CrossRef CAS PubMed.
- C. Gwizdala, S. C. Gwizdala and C. Burdette, Curr. Opin. Chem. Biol., 2013, 17, 137–142 CrossRef CAS PubMed.
- H. W. Mbatia and S. C. Burdette, Biochemistry, 2012, 51, 7212–7224 CrossRef CAS PubMed.
- G. Gasser, I. Ott and N. Metzler-Nolte, J. Med. Chem., 2011, 54, 3–25 CrossRef CAS PubMed , and references therein.
- C. G. Hartinger, N. Metzler-Nolte and P. J. Dyson, Organometallics, 2012, 31, 5677–5685 CrossRef CAS , and references therein.
- P. C. A. Bruijincx and P. J. Sadler, Curr. Opin. Chem. Biol., 2008, 12, 197–206 CrossRef PubMed , and references therein.
- G. Gasser and N. Metzler-Nolte, Curr. Opin. Chem. Biol., 2012, 16, 84–91 CrossRef CAS PubMed , and references therein.
- G. Jaouen and N. Metzler-Nolte, Medicinal organometallic chemistry, Springer-Verlag, 2010 Search PubMed.
- I. Kitanovic, S. Can, H. Alborzinia, A. Kitanovic, V. Pierroz, A. Leonidova, A. Pinto, B. Spingler, S. Ferrari, R. Molteni, A. Steffen, N. Metzler-Nolte, S. Wolfl and G. Gasser, Chem.–Eur. J., 2014, 20, 2496–2507 CrossRef CAS PubMed.
- N. Viola-Villegas, A. E. Rabideau, M. Bartholoma, J. Zubieta and R. P. Doyle, J. Med. Chem., 2009, 52, 5253–5261 CrossRef CAS PubMed.
- X. Tang and I. J. Dmochowski, Nat. Protoc., 2007, 1, 3041–3048 CrossRef PubMed.
- C. P. Holmes, J. Org. Chem., 1997, 62, 2370–2380 CrossRef CAS PubMed.
- C.-y. Chang, B. Niblack, B. Walker and H. Bayley, Chem. Biol., 1995, 2, 391–400 CrossRef CAS.
- L. Niu, R. Wieboldt, D. Ramesh, B. K. Carpenter and G. P. Hess, Biochemistry, 1996, 35, 8136–8142 CrossRef CAS PubMed.
- M. J. Brubaker, D. H. Dyer, B. Stoddard and D. E. Koshland, Biochemistry, 1996, 35, 2854–2864 CrossRef CAS PubMed.
- E. Garcìa Garayoa, D. Rüegg, P. Bläuenstein, M. Zwimpfer, I. U. Khan, V. Maes, A. Blanc, A. G. Beck-Sickinger, D. A. Tourwé and P. A. Schubiger, Nucl. Med. Biol., 2007, 34, 17–28 CrossRef PubMed.
- E. Garcia Garayoa, C. Schweinsberg, V. Maes, D. Rüegg, A. Blanc, P. Bläuenstein, D. A. Tourwé, A. G. Beck-Sickinger and P. A. Schubiger, Eur. J. Nucl. Med. Mol. Imaging, 2007, 51, 42–50 CAS.
- J. Olejnik, S. Sonar, E. Krzymañska-Olejnik and K. J. Rothschild, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 7590–7594 CrossRef CAS.
- G. Gauglitz and S. J. Hubig, J. Photochem., 1985, 30, 121–125 CrossRef CAS.
- R. T. Jensen, J. F. Battey, E. R. Spindel and R. V. Benya, Pharmacol. Rev., 2008, 60, 1–60 CrossRef CAS PubMed , and references therein.
- J. Berlanda, T. Kiesslich, V. Engelhardt, B. Krammer and K. Plaetzer, J. Photochem. Photobiol., B, 2010, 100, 173–180 CrossRef CAS PubMed.
- F. S. Mackay, J. A. Woods, P. Heringová, J. Kašpárková, A. M. Pizarro, S. A. Moggach, S. Parsons, V. Brabec and P. J. Sadler, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 20743–20748 CrossRef CAS PubMed.
- N. J. Farrer, J. A. Woods, L. Salassa, Y. Zhao, K. S. Robinson, G. Clarkson, F. S. Mackay and P. J. Sadler, Angew. Chem., Int. Ed., 2010, 49, 8905–8908 CrossRef CAS PubMed.
- B. Peña, A. David, C. Pavani, M. S. Baptista, J.-P. Pellois, C. Turro and K. R. Dunbar, Organometallics, 2014, 33, 1100–1103 CrossRef.
- R. Lincoln, L. Kohler, S. Monro, H. Yin, M. Stephenson, R. Zong, A. Chouai, C. Dorsey, R. Hennigar, R. P. Thummel and S. A. McFarland, J. Am. Chem. Soc., 2013, 135, 17161–17175 CrossRef CAS PubMed.
- H. Yin, M. Stephenson, J. Gibson, E. Sampson, G. Shi, T. Sainuddin, S. Monro and S. A. McFarland, Inorg. Chem., 2014, 53, 4548–4559 CrossRef CAS PubMed.
- D. A. Lutterman, P. K. L. Fu and C. Turro, J. Am. Chem. Soc., 2006, 128, 738–739 CrossRef CAS PubMed.
- A. A. Holder, D. F. Zigler, M. T. Tarrago-Trani, B. Storrie and K. J. Brewer, Inorg. Chem., 2007, 46, 4760–4762 CrossRef CAS PubMed.
- M. A. Sgambellone, A. David, R. N. Garner, K. R. Dunbar and C. Turro, J. Am. Chem. Soc., 2013, 135, 11274–11282 CrossRef CAS PubMed.
- G. Gasser, A. Pinto, S. Neumann, A. M. Sosniak, M. Seitz, K. Merz, R. Heumann and N. Metzler-Nolte, Dalton Trans., 2012, 41, 2304–2313 RSC.
- F.-M. Boisvert, J. van Koningsbruggen, A. Navascués and A. Lamond, Nat. Rev. Mol. Cell Biol., 2007, 8, 574–585 CrossRef CAS PubMed , and references therein.
- A. J. Pickard and U. Bierbach, ChemMedChem, 2013, 8, 1441 CrossRef CAS PubMed.
- G. Gasser, S. Neumann, I. Ott, M. Seitz, R. Heumann and N. Metzler-Nolte, Eur. J. Inorg. Chem., 2011, 36, 5471–5478 CrossRef.
- M. Muramatsu, K. Smetana and H. Busch, Cancer Res., 1963, 25, 693–697 Search PubMed.
- T. Hermann, Angew. Chem., Int. Ed., 2000, 39, 1890–1905 CrossRef.
- S. Phongtongpasuk, S. Paulus, J. Schnabl, R. K. O. Sigel, B. Spingler, M. J. Hannon and E. Freisinger, Angew. Chem., Int. Ed., 2013, 52, 11513–11516 CrossRef CAS PubMed.
- M. Steiner, K. S. Karunatilaka, R. K. O. Sigel and D. Rueda, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 13853–13858 CrossRef CAS PubMed.
- M. G. Sun, J. Williams, C. Munoz-Pinedo, G. A. Perkins, J. M. Brown, M. H. Ellisman, D. R. Green and T. G. Frey, Nat. Cell Biol., 2007, 9, 1057–1065 CrossRef CAS PubMed.
- M. R. Gill, D. Cecchini, M. G. Walker, R. S. Mulla, G. Battaglia, W. Smythe and J. A. Thomas, Chem. Sci., 2013, 4, 4512–4519 RSC.
- J. M. Hearn, I. Romero-Canelon, B. Qamar, Z. Liu, I. Hands-Portman and P. J. Sadler, ACS Chem. Biol., 2013, 8, 1335–1343 CrossRef CAS PubMed.
- D. V. Krysko, T. V. Berghe, K. D'Herde and P. Vandenabeele, Methods, 2008, 44, 205–221 CrossRef CAS PubMed.
- V. Sirri, S. Urcuqui-Inchima, P. Roussel and D. Hernandez-Verdun, Histochem. Cell Biol., 2008, 129, 13–31 CrossRef CAS PubMed.
- W. J. Welch and J. P. Suhan, J. Cell Biol., 1985, 101, 1198–1211 CrossRef CAS.
- M. Engeland, L. J. W. Nieland, F. C. S. Ramaekers, B. Schutte and C. P. M. Reutelingsperger, Cytometry, 1998, 31, 1–9 CrossRef.
- S. Gallo, M. Furler and R. K. O. Sigel, Chimia, 2005, 59, 812–816 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Characterization of compounds (NMR, MS and LC-MS spectra), additional data on photolysis, DNA and RNA gel electrophoresis, TEM images and FACS. See DOI: 10.1039/c3sc53550a |
| ‡ These authors have contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2014 |