Microfluidics & nanotechnology: towards fully integrated analytical devices for the detection of cancer biomarkers
Abstract
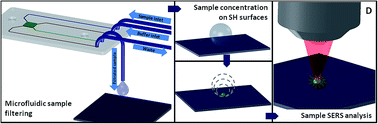
In this paper, we describe an innovative modular microfluidic platform allowing filtering, concentration and analysis of peptides from a complex mixture. The platform is composed of a microfluidic filtering device and a superhydrophobic surface integrating surface enhanced Raman scattering (SERS) sensors. The microfluidic device was used to filter specific peptides (MW 1553.73 D) derived from the BRCA1 protein, a tumor-suppressor molecule which plays a pivotal role in the development of breast cancers, from albumin (66.5 KD), the most represented protein in human plasma. The filtering process consisted of driving the complex mixture through a porous membrane having a cut-off of 12–14 kD by hydrodynamic flow. The filtered samples coming out of the microfluidic device were subsequently deposited on a superhydrophobic surface formed by micro pillars on top of which nanograins were fabricated. The nanograins coupled to a Raman spectroscopy instrument acted as a SERS sensor and allowed analysis of the filtered sample on top of the surface once it evaporated. By using the presented platform, we demonstrate being able to sort small peptides from bigger proteins and to detect them by using a label-free technique at a resolution down to 0.1 ng μL−1. The combination of microfluidics and nanotechnology to develop the presented microfluidic platform may give rise to a new generation of biosensors capable of detecting low concentration samples from complex mixtures without the need for any sample pretreatment or labelling. The developed devices could have future applications in the field of early diagnosis of severe illnesses, e.g. early cancer detection.

 Please wait while we load your content...
Please wait while we load your content...