DNA-mediated phase transfer of DNA-functionalized quantum dots using reverse micelles†
Tomoharu Kato,
Yuhei Fujimoto,
Ayane Shimomura and
Tatsuo Maruyama*
Department of Chemical Science and Engineering, Graduate School of Engineering, Kobe University, Kobe 657-8501, Japan. E-mail: tmarutcm@crystal.kobe-u.ac.jp; Fax: +81-78-803-6070; Tel: +81-78-803-6070
First published on 28th October 2014
Abstract
We report the selective phase transfer of DNA-functionalized CdTe quantum dots (DNA-QDs) from an aqueous phase to an organic phase using reverse micelles and a DNA surfactant. The DNA surfactant recognized DNA tethered to QDs and transferred the DNA-QDs to an organic phase via the DNA hybridization. Extraction efficiency strongly depended on the nucleotide complementarity, the number of DNA oligonucleotides tethered to a QD and the DNA-QDs size.
Metal1,2 and semiconductor nanoparticles3–5 exhibit unique optical and electronic properties beyond their bulk analogs. They have great application potential in electronic devices, sensors, diagnostics, and biological visualization. Their unique properties are strongly influenced by the nanoparticle size and size distribution. To date, much effort has been made to synthesize nanoparticles with a narrow size distribution in colloidal solution. However, most of the approaches adopted for nanoparticle synthesis are based on nucleation and growth mechanisms that result in fluctuation and heterogeneity in solution, generating variable nanoparticle size distributions. Several research groups reported the size-selective separation of nanoparticles via precipitation,6,7 supercritical fluids,8,9 gas pressurization,10 high-performance liquid chromatography,11,12 and size-exclusion chromatography.13 However, Sweeney et al. pointed out the difficulty in the separation of water-soluble gold nanoparticles using these techniques.14 In the last decade, there have been a few reports on the size-selective separation of water-soluble nanoparticles using membrane separation and size-exclusion chromatography.15,16 These techniques, however, are limited to throughput and chemical composition owing to interactions between the nanoparticles and membrane matrices or column supports (e.g., membrane fouling or irreversible adsorption). Additionally, these approaches do not recognize the surface properties of nanoparticles or allow separation based on the surface chemistry of nanoparticles.
Liquid–liquid extraction (solvent extraction) is a powerful technique, which can be scaled up, for separation and purification of molecular substrates. Phase transfer of molecules is a key step in liquid–liquid extraction. Energetic efforts have been made using solubility control and molecular recognition to achieve high selectivity for separation in the extraction. To our knowledge, however, there are no reports on liquid–liquid extraction of solid nanoparticles based on size- or surface-selective separation without chemical transformation (e.g., ligand exchange). In the present study, we examined phase transfer of semiconductor nanoparticles in surface- and size-selective manners using reverse micelles and a DNA surfactant (Scheme 1). A reverse micelle, which is a nano-scaled water pool surrounded by surfactant molecules in an organic solvent, can be combined with liquid–liquid extraction to transfer water-soluble molecules from an aqueous solution into an organic solvent without chemical transformation.17 DNA duplex is maintained in the water pool of a reverse micelle.18 Our previous report demonstrated that reverse micelles could be used as nano-containers to extract DNA oligonucleotides and that high selectivity was achieved in the reverse micellar extraction using a DNA surfactant.19 Here, we adopted DNA-functionalized CdTe quantum dots (DNA-QDs), which have been widely used in the biological field, as target solid nanoparticles. These were successfully applied in the liquid–liquid extraction of DNA-QDs from an aqueous phase to an organic phase in both surface- and size-selective manners (Scheme 1).
 | ||
| Scheme 1 Schematic illustration of phase transfer of DNA-QDs using reverse micelles and a DNA surfactant. | ||
Water-soluble CdTe QDs were synthesized according to the literature.20 The CdTe QDs were capped with mercaptopropionic acid. CdTe QDs were mixed with 5′-thiolated DNA oligonucleotide (12-mer) to prepare DNA-functionalized QDs (DNA-QDs).21 The details are given in the ESI† file. The DNA–surfactant (DNA surfactant 1) was synthesized according to our previous reports.19,22 An oleoyl group was introduced at the N-terminus of a single-stranded 5′-aminated DNA oligonucleotide (12-mer). The nucleotide sequences of the DNA surfactant and DNA–CdTe QDs are listed in Table S1.†
An organic phase (1 mL) was contacted with an aqueous phase (1 mL) for reverse micellar extraction of DNA-QDs. The aqueous phase (Tris–HCl buffer (pH 8, 25 mM), KCl (300 mM)) contained DNA-QDs (75 nM), and DNA surfactant 1 (300 nM). The organic phase (2,2,4-trimethyl pentane) contained dilauroyl phosphatidylcholine (DLPC, 15 mM) and 1-hexanol (4.5 vol%). DLPC was used as a zwitterion surfactant, and 1-hexanol was employed as a co-surfactant for the formation of reverse micelles. After the two phases were gently stirred at room temperature for 3 h, Kirk–Fisher analyses revealed that 1.5 wt% water was present in the organic phase, suggesting the formation of reverse micelles in the organic phase. Dynamic light scattering measurements showed that the diameter of the reverse micelles was ∼8 nm. This result was similar to that of the previous report.19
The fluorescence of the DNA-QDs in each phase was measured to determine the percentage extraction of the DNA-QDs. After contacting the two phases for 3 h, the fluorescence of the DNA-QDs in the aqueous phase decreased by 20% and the correspondent fluorescence was observed in the organic phase (Fig. 1a). We found that ∼20% of the DNA-QDs was extracted from the aqueous phase to the organic phase. It should be noted that the fluorescence intensity and spectrum of the DNA-QDs in the reverse micelles were comparable to those in the aqueous solution. In the absence of the DNA surfactant 1, DNA-QDs were not extracted.
We hypothesized that DNA surfactant 1, which was complementary to the nucleotide sequence of the DNA-QDs, facilitated extraction of the DNA-QDs via DNA hybridization (Scheme 1). To validate the hypothesis, we examined the liquid–liquid extraction of the DNA-QDs by varying the concentration of DNA surfactant 1 (Fig. 1b). The percentage extraction of DNA-QDs increased with increase in the DNA–surfactant 1 concentration, thus supporting our hypothesis. The use of DNA–surfactant 1 at 800 nM, which was twice the concentration of DNA tethered to the QDs (DNA/QD = 4; QD concentration, 100 nM QD), gave the highest percentage extraction (22%). The melting temperature (Tm) of 12-mer DNA (used for the DNA–surfactant 1) was 36.0 °C under the present buffer conditions, and was sufficiently high for the DNA hybridization.
One of the featured characteristics of DNA is the specificity in the duplex formation with its complementary strand. We then synthesized DNA–surfactant 2, featuring a different nucleotide sequence that was not complementary to the DNA tethered to the QDs (Table S1†). The use of the mismatched DNA surfactant did not lead to extraction of the DNA-QDs. These results suggested that the DNA surfactant recognized the DNA strand tethered to the QDs and enabled extraction of the DNA-QDs.
We then investigated the effect of extraction time on the percentage extraction of the DNA-QDs (Fig. S1†). As the extraction time increased, the percentage extraction increased. After 1 h, the percentage extraction reached a plateau (at 20%). Extraction of the DNA-QDs to an organic phase was relatively long because of the gentle stirring used and the low diffusion velocity of the QDs.
The diameter of the prepared reverse micelles was 8 nm. The length of 12-mer DNA oligonucleotide on the QDs was ∼4 nm. Although both a micelle and a DNA strand are flexible, DNA functionalization would increase the physical size of the DNA-QDs and influence the extraction efficiency. The combined effect of the number of DNA oligonucleotides tethered to a QD and size of the reverse micelles is expected to play a key role in the extraction. We then investigated the effect of molar ratio of the tethered DNA/QD on extraction (Fig. 2). The molar ratio of tethered DNA/QD was controlled by varying the concentration of 5′-thiolated DNA during the DNA-QDs synthesis. With increasing amounts of DNA tethered to a QD, the percent extraction of DNA-QDs increased. When the molar ratio of the tethered DNA/QD exceeded 4, the percent extraction decreased. These results suggested that the number of the DNA oligonucleotides tethered to a QD was essential for the extraction of DNA-QDs. The increase in the hydrophobic tail (derived from DNA surfactant 1) enabled phase transfer of a water-soluble QD to the organic phase. However, excessive amounts of DNA oligonucleotides on a QD resulted in lower extraction probably because of the resulting larger size of the DNA-QDs and electrostatic repulsion between the excess DNA oligonucleotides tethered to the QDs and DNA surfactant 1.23
Fig. 3 shows the effect of particle diameter of CdTe QDs on extraction. We synthesized CdTe QDs with a range of 3–5.5 nm in diameter by controlling the reflux time during QD synthesis. The DNA/QD ratio was set at 4. As observed, larger CdTe QDs resulted in lower percentage extraction. CdTe QDs (5.5 nm in diameter) were barely extracted probably because the size of the resulting DNA-QDs would be too large for encapsulation in the reverse micelles.
These results imply that the physical size of the nanoparticles was important for the extraction yield in the reverse micellar extraction. Although we also investigated the various extraction conditions (co-surfactant concentration, salt concentration, buffer type and temperature), the percentage extraction was still ∼20%. The increase of the size of reverse micelles might improve the percentage extraction. The appropriate selection of a surfactant for reverse micelles, alternate to DLPC, would produce reverse micelles large enough for the extraction of the QDs. We are currently studying the conditions for preparing large reverse micelles.
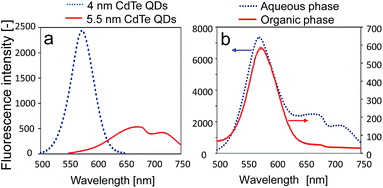
We finally investigated the size-dependent extraction of the DNA-QDs from a mixture of DNA-QDs with different diameters. DNA-QDs with two different diameters (4 and 5.5 nm) displayed maxima fluorescence at 580 and 675 nm, respectively, in an aqueous solution (Fig. 4a). These two types of DNA-QDs (75 nM) were added to an aqueous phase containing DNA surfactant 1 (300 nM), followed by the addition of an organic phase containing DLPC and 1-hexanol. Following gentle stirring for 3 h, the fluorescence of the CdTe QDs was measured. Only the fluorescence at 580 nm was observed in the organic phase (Fig. 4b). The percentage extraction values were 8.0% (QD, 4 nm) and 1.6% (QD, 5.5 nm). Thus, DNA-QDs with QDs of size 4 nm were preferentially extracted over DNA-QDs with QDs of size 5.5 nm, which were barely extracted. Thus, the present system affords size-selective extraction of DNA-QDs from a mixture of DNA-QDs to an organic phase.
Conclusions
We employed reverse micelles coupled with a DNA surfactant and succeeded in the DNA-mediated phase transfer of DNA-functionalized CdTe QDs from an aqueous phase to an organic phase for the first time. The present study reveals that precise molecular recognition of DNA can be used in liquid–liquid extraction for the selective separation of nano-scaled solid materials (QDs) and that the reverse micellar system can discriminate between differently sized QDs during extraction. The present approach using reverse micelles might be extended to the separation of nanometer-sized metal particles and rods. Due to the size limitation of reverse micelles, the available size of nanomaterials will be a few nanometers. Liquid–liquid extraction using an extracting agent has potential for the separation of solid nanomaterials in addition to its widespread use in the separation of molecular substrates.Acknowledgements
We thank Prof. M. Mizuhata, Prof. Y. Ueda and Prof. Takeuchi for the technical help and advice. This study was partly supported by The Tounen General Foundation and by the Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), Ministry of Education, Culture, Sports, Science and Technology (Japan).Notes and references
- M. C. Daniel and D. Astruc, Chem. Rev., 2004, 104, 293–346 CrossRef CAS PubMed.
- A. C. Templeton, M. P. Wuelfing and R. W. Murray, Acc. Chem. Res., 2000, 33, 27–36 CrossRef CAS PubMed.
- A. P. Alivisatos, P. F. Barbara, A. W. Castleman, J. Chang, D. A. Dixon, M. L. Klein, G. L. McLendon, J. S. Miller, M. A. Ratner, P. J. Rossky, S. I. Stupp and M. E. Thompson, Adv. Mater., 1998, 10, 1297–1336 CrossRef.
- S. A. Empedocles and M. G. Bawendi, Science, 1997, 278, 2114–2117 CrossRef CAS.
- A. P. Alivisatos, Science, 1996, 271, 933–937 CAS.
- C. P. Collier, T. Vossmeyer and J. R. Heath, Annu. Rev. Phys. Chem., 1998, 49, 371–404 CrossRef CAS PubMed.
- C. B. Murray, C. R. Kagan and M. G. Bawendi, Annu. Rev. Mater. Sci., 2000, 30, 545–610 CrossRef CAS.
- P. S. Shah, J. D. Holmes, K. P. Johnston and B. A. Korgel, J. Phys. Chem. B, 2002, 106, 2545–2551 CrossRef CAS.
- N. Z. Clarke, C. Waters, K. A. Johnson, J. Satherley and D. J. Schiffrin, Langmuir, 2001, 17, 6048–6050 CrossRef CAS.
- M. C. McLeod, M. Anand, C. L. Kitchens and C. B. Roberts, Nano Lett., 2005, 5, 461–465 CrossRef CAS PubMed.
- V. L. Jimenez, M. C. Leopold, C. Mazzitelli, J. W. Jorgenson and R. W. Murray, Anal. Chem., 2003, 75, 199–206 CrossRef CAS.
- J. P. Wilcoxon, J. E. Martin and P. Provencio, Langmuir, 2000, 16, 9912–9920 CrossRef CAS.
- A. M. Al-Somali, K. M. Krueger, J. C. Falkner and V. L. Colvin, Anal. Chem., 2004, 76, 5903–5910 CrossRef CAS PubMed.
- S. F. Sweeney, G. H. Woehrle and J. E. Hutchison, J. Am. Chem. Soc., 2006, 128, 3190–3197 CrossRef CAS PubMed.
- A. Akthakul, A. I. Hochbaum, F. Stellacci and A. M. Mayes, Adv. Mater., 2005, 17, 532–535 CrossRef CAS.
- L. Alamo-Nole, S. Bailon-Ruiz, O. Perales-Perez and F. R. Roman, Anal. Methods, 2012, 4, 3127–3132 RSC.
- K. E. Goklen and T. A. Hatton, Biotechnol. Prog., 1985, 1, 69–74 CrossRef CAS PubMed.
- T. Maruyama, L. C. Park, T. Shinohara and M. Goto, Biomacromolecules, 2004, 5, 49–53 CrossRef CAS PubMed.
- T. Maruyama, T. Hosogi and M. Goto, Chem. Commun., 2007, 4450–4452 RSC.
- S. H. Xu, C. L. Wang, H. Zhang, Z. Y. Wang, B. Yang and Y. P. Cui, Nanotechnology, 2011, 22, 315703 CrossRef PubMed.
- F. Li, H. Q. Zhang, B. Dever, X. F. Li and X. C. Le, Bioconjugate Chem., 2013, 24, 1790–1797 CrossRef CAS PubMed.
- T. Maruyama, H. Yamamura, M. Hiraki, Y. Kemori, H. Takata and M. Goto, Colloids Surf., B, 2008, 66, 119–124 CrossRef CAS PubMed.
- L. M. Demers, C. A. Mirkin, R. C. Mucic, R. A. Reynolds, R. L. Letsinger, R. Elghanian and G. Viswanadham, Anal. Chem., 2000, 72, 5535–5541 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: syntheses of QDs and DNA–surfactants, DNA sequences and reverse micellar extraction of DNA-QDs. See DOI: 10.1039/c4ra09683e |
| This journal is © The Royal Society of Chemistry 2014 |