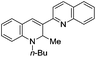
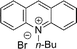
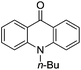
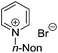
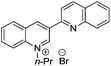
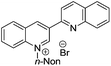
3-Substituted biquinolinium inhibitors of AraC family transcriptional activator VirF from S. flexneri obtained through in situ chemical ionization of 3,4-disubstituted dihydroquinolines†
Prashi Jaina,
Jiaqin Lib,
Patrick Porubskya,
Benjamin Neuenswanderc,
Susan M. Eganb,
Jeffrey Aubéacde and
Steven Rogers*d
aCenter for Chemical Methodologies and Library Development, The University of Kansas, 2034 Becker Drive, Lawrence, Kansas 66047, USA
bDepartment of Molecular Biosciences, University of Kansas, Lawrence, KS, USA
cSpecialized Chemistry Center, The University of Kansas, 2034 Becker Drive, Lawrence, Kansas 66047, USA
dUniversity of Kansas Center of Biomedical Research Excellence, Center for Cancer Experimental Therapeutics, 2034 Becker Drive, Lawrence, KS, USA. E-mail: sarogers@ku.edu
eDepartment of Medicinal Chemistry, University of Kansas, Lawrence, KS, USA
First published on 18th August 2014
Abstract
During a structure–activity relationship optimization campaign to develop an inhibitor of AraC family transcriptional activators, we discovered an unexpected transformation of a previously reported inhibitor that occurs under the assay conditions. Once placed in the assay media, the 3,4-disubstituted dihydroquinoline core of the active analogue rapidly undergoes a decomposition reaction to a quaternary 3-substituted biquinolinium. Further examination established an SAR for this chemotype while also demonstrating its resilience to irreversible binding of biologically relevant nucleophiles.
Introduction
The World Health Organization (WHO) recently published a warning that microbial resistance to antibiotics has become a “major threat to public health”.1 A dearth of new FDA approved antimicrobial medicines coupled with the rapid development of resistance to available antibiotics is creating a health care void that necessitates the development of new antimicrobial strategies to combat microbial infectious disease.2 One such strategy involves targeting non-growth essential virulence factors, which may avoid triggering resistance mechanism development by not placing selective pressure on microbes. As an example of this, the protein members of the AraC family of bacterial transcription activators activate expression of genes encoding virulence factors in various pathogenic bacteria such as Vibrio cholerae (ToxT),3 enterotoxigenic Escherichia coli (Rns/CfaD) (ETEC),4 Pseudomonas aeruginosa (ExsA)5 and Shigella flexneri (VirF),6 while other members of the family regulate bacterial stress responses or carbon metabolic operons.7 Furthermore, inhibition or deletion of AraC family activators has been demonstrated to ameliorate infections in vitro and in animal models.8Our study focuses on VirF, an AraC family transcriptional activator from Shigella flexneri, the causative agent of bacillary dysentery, or shigellosis.9 This disease affects over 165 million people worldwide while leading to approximately 1.1 million deaths annually.10 VirF is necessary for the expression of virulence genes associated with the first step of S. flexneri pathogenesis, which is rectal, and colonic epithelial cell invasion as well as the subsequent cell-to-cell dissemination.11 Using cell-based reporter gene assays with S. flexneri and E. coli, coupled with in vitro DNA-binding assays with purified VirF, we recently determined that SE-1 (Fig. 1) is an inhibitor of transcription activation and DNA binding by VirF.12 Furthermore, through examining mRNA concentrations using real-time quantitative reverse transcription-PCR (qRT-PCR), it was found that SE-1 reduced the expression of VirF-dependent virulence genes such as icsS, icsB, ipaB, and virB in S. flexneri. This initial study prompted us to seek analogues of SE-1 with greater potency that could serve as a lead VirF inhibitor.
In this manuscript, we detail our activity optimization efforts of SE-1,12 which was originally identified through screening of a commercially obtained small molecule library, and show that SE-1 and related analogues undergo ionization in aqueous medium to afford a quaternary 3-substituted biquinolinium salt (Fig. 1). Although the decomposition product is a synthetic precursor to the 3,4-disubstituted dihydroquinoline core, its ionization in biological media was unexpected. Thus, we present evidence here that the biquinolinium salt is the causative agent responsible for the observed inhibitory activity toward bacterial AraC family activators, discuss structure–activity relationships (SARs) associated with this chemotype, and demonstrate that it is stable in the presence of biologically relevant nucleophiles.
Results and discussion
To gain preliminary SAR information to guide our synthetic endeavors, we procured and screened 24 commercially available analogues of SE-1, resulting in three compounds exhibiting notable activity in comparison to SE-1 in the VirF in vivo dose-response assay in E. coli (Fig. 2). The assays were performed as previously described.12 Briefly, the analogues were dissolved in 100% dimethyl sulfoxide (DMSO). Bacterial cultures were grown to an optical density at 600 nm (OD600) of ∼0.1 and then mixed with various concentrations of analogues (final concentration of DMSO, 6.2%), induced with 1 mM IPTG (final concentration) for 3 hours at 37 °C, lysed, and β-galactosidase activity was measured. Uninduced (no IPTG and no analogues) and uninhibited (1 mM IPTG and no analogues) controls were used to normalize β-galactosidase activity, as described previously.12 To eliminate from consideration any analogues that nonspecifically inhibited β-galactosidase activity or cell growth, we also tested these analogues on a control strain that carried a lacZ reporter fusion (hts-lacZ), with a synthetic promoter that was repressed by LacI and did not require VirF for activation, as described previously.12 We found that analogues E1, A2 and C4 inhibited expression of the VirF-activated virB-lacZ fusion to the same extent as SE-1, with an IC50 of 7 μM, 10 μM and 11 μM respectively. Other 4-substitutions were assessed such as an indole, a phenyl ring, a benzimidazole and a napthyl ring in which all analogues were found to be inactive (full list can be found in ESI†). Analogues without N-substitution were also found to not exhibit activator inhibition.To follow-up on our commercially accessible SAR, a series of 3,4-disubstituted dihydroquinolines were synthesized as shown in Scheme 1 that was adapted from protocols developed by Aksenov.13 We focused on the 4-methylnitro substitution instead of the malonic ester due to potential esterase liability and instead of the nitrile because E1 was found to be more active than C4. Briefly, various aryl boronic acids were coupled via Suzuki reaction with 3-bromoquinolines through the treatment with bis(triphenylphosphine)palladium dichloride, potassium carbonate in a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of dimethylformamide and water while heating to 150 °C for 30 minutes under microwave conditions. The resulting 3-substituted quinolines were subsequently N-substituted via electrophilic attack from various benzyl and alkyl halides, followed by a nucleophilic attack at the 4-position of the resulting quinolinium halide through the treatment with nitromethane under basic conditions in dioxane at room temperature, giving the desired 3,4-disubstituted dihydroquinolines. Both the 3,4-disubstituted dihydroquinolines and quinolinium halide synthetic intermediates were screened for inhibitory activity against VirF.
1 mixture of dimethylformamide and water while heating to 150 °C for 30 minutes under microwave conditions. The resulting 3-substituted quinolines were subsequently N-substituted via electrophilic attack from various benzyl and alkyl halides, followed by a nucleophilic attack at the 4-position of the resulting quinolinium halide through the treatment with nitromethane under basic conditions in dioxane at room temperature, giving the desired 3,4-disubstituted dihydroquinolines. Both the 3,4-disubstituted dihydroquinolines and quinolinium halide synthetic intermediates were screened for inhibitory activity against VirF.
 | ||
| Scheme 1 Reagents and conditions: (a) PdCl2(PPh3)2, K2CO3, DMF/H2O, MW, 150 °C; (b) R2Br, MW, 150 °C, 2 h (c) MeNO2, K2CO3, H2O, dioxane, rt, 4 h. | ||
Initially, seven 3,4-disubstituted dihydroquinolines, three quinolinium synthetic precursors and a new batch of SE-1 (1) were synthesized and screened for VirF activator inhibition (Table 1). Replacing the n-butyl group with a 4-chlorobenzyl (5) resulted in a significant loss of activity against VirF activator inhibition. Replacing the quinoline with other aryl groups such as 5-methyl pyridine (6), 3-chlorophenyl (7) resulted in analogues with very modest inhibitory activity, giving IC50 values >100 μM. Diminished activity was also observed when substituting the nitromethyl of the 1,4-dihydroquinoline with a phenyl (8) or methyl (11) as well in the same substitution of a 1,2-dihydroquinolines (entries 9 & 10). However, the resynthesized SE-1 (Table 1) repeated the activity observed from the HTS sample with an IC50 of 11 μM against VirF and >100 μM in the control strain.
As a first clue to the instability of these 1,4-dihydroquinoline cores, we observed that the quaternary amine quinolinium samples (Table 1, entries 2–4) exhibited the same, if not better activity against VirF while not affecting the control strain. This can be observed when comparing Table 1, entries 2 to SE-1 (1), entries 3 to 5 and entries 4 to E1. Furthermore, the 3,4-disubstituted dihydroquinolines (Table 1, SE-1 and entries 5–11) exhibited notable stability issues during their synthesis because they readily decomposed if purification attempts were made on normal phase chromatography with silica gel or reverse phase chromatography with C18-bonded silica gel. All 3,4-disubstituted dihydroquinolines had to be purified via recrystallization. We also noted that upon 1H NMR analysis of various dihydroquinoline samples in DMSO-d6 at room temperature two days after the samples had been prepared and analyzed, that a significant reversion to the quinolinium precursor was observed. These observations prompted us to examine (1) the latent instability of SE-1 and its analogues, (2) if the observed VirF inhibition activity was simply due to the quaternary species and (3) if non-quaternary adducts could be derived that demonstrated stability and VirF inhibitory activity.
To examine the aqueous stability of the 3,4-disubstituted dihydroquinoline analogues, samples were prepared at 10 μM in PBS at pH 7.4 (1% DMSO) and analyzed via auto-sampler by RP HPLC/UV/HRMS after the sample had been prepared and then every hour on the hour for 48 hours at room temperature. We found that SE-1 had degraded completely to the corresponding quaternary salt by the 0th time point, or about 30 minutes elapsed time including assay preparation. Given that the VirF inhibition assays typically run 8–24 hours, the stability analysis predicted that no detectable concentration of the 3,4-disubstituted dihydroquinoline species was present during the assay and that the activity was simply due to the quinolinium salt. Since most quaternary ammonium salts have limited pharmaceutical relevance, save topical and oral hygiene applications due to their hemolytic nature,14 we began exploring non-quaternary analogues of our most potent VirF activator inhibitors.
The 3,4-disubstituted dihydroquinoline core was substituted with an indole (Table 2, entries 12 and 13), 9-acridone (15) and 4-quinolone (16), resulting in no inhibitory activity towards VirF or in the control strain assay. Interestingly, we also screened acridinium (14), pyridinium (17) and triethylammonium (18) salt analogues that would behave similarly to other cationic quaternary ammonium chemical detergents such as cetylpyridinium chloride (CPC) to compare the activity profiles to our more active quinolinium compounds. The triethylammonium analogue exhibited no inhibitory activity in our assays whereas the acridinium and pyridinium exhibited good to modest, non-specific activity against both the VirF and control strain, indicating that the activity we see with our quinolinium compounds is not simply due to detergent effects. This prompted us to continue optimizing the activity of the quinolinium core.
Various N-substituted and 3-aryl analogues were synthesized with aim to optimize the quinolinium core (Table 2, entries 19–24). As seen previously, N-substitution of n-butyl with methyl (4) gave decreased activity with a 90 μM IC50 value, whereas i-propyl (19), n-propyl (20), n-hexyl (21) and pent-4-en-1-yl (24) all gave similar activity to SE-1 with IC50 values of 12 μM, 16 μM, 11 μM and 11 μM respectively, while not inhibiting the control strain. Replacing the 3-quinoline substituent with a naphthalene (23) was also well tolerated and gave an IC50 value of 9 μM against VirF and >100 μM against the control strain, comparable activity to the parent compound (Table 1, entry 1). A modest increase in activity was attained while elongating the N-alkyl chain to n-nonyl (22); giving an IC50 value of 3 μM against VirF, but notable selectivity was lost because of control strain inhibition with an IC50 value of 25 μM. A planktonic bacterial growth curve analysis of both the VirF and control strains in the presence of Table 1, entry 6 demonstrated that this compound inhibited the growth of the bacteria, explaining the increase in activity against both strains. This toxic effect on bacterial growth was not seen with the other active compounds.
Given the inherent electrophillic nature of cationic species such quinoliniums, we assessed the stability of some of these analogues towards the thiol based nucleophiles dithiothreitol and glutathione to mimic interactions with thiol containing amino acid residues of proteins in a biological system. Briefly, compounds were dissolved at 10 μM in PBS at pH 7.4 (1% DMSO) and incubated at room temperature with either no thiol source as a negative control, 50 μM glutathione (GSH). The mixtures were sampled every hour for eight hours for 48 hours and analyzed by RP HPLC/UV/HRMS. The masses of potential adducts were searched for in the final samples to determine if any detectable adduct formed. Ethacrynic acid, a known Michael acceptor, was used as a positive control. In both of the quinolinium salts assayed (entries 2 and 3), no GSH adducts were observed, suggesting these compounds to be relatively stable despite their expected reactivity, giving this chemotype possible application toward developing biological probes while suggesting that these inhibitors may not be acting in a covalent manner.
Conclusions
Through analysis of SAR data and chemical stability behavior, we have discovered that our previously reported 3,4-disubstituted dihydroquinoline inhibitor of AraC transcription activators,12 readily fragments in assay media to a quinolinium core, which we ascribe as the active agent against this target. We note that SE-1 and analogues are available from several commercial vendors; and the present results suggest that the ionized precursors may well be responsible for any observed biological activity for this chemotype. Although we did not increase VirF activator inhibition potency of this class of compounds, we expounded the SAR and showed that the activity of the quinolinium analogues does depend on specific structure and was not due to generic detergent effects.Experimental section
All air- and moisture-sensitive reactions were carried out in flame- or oven-dried glassware under argon atmosphere using standard gastight syringes, cannula, and septa. Stirring was achieved with oven-dried magnetic stir bars. Flash column chromatography was performed with SiO2 from Sorbent Technology (30930M-25, Silica Gel 60 A, 40−63 μm) or by using an automated chromatography instrument with an appropriately sized column. Thin layer chromatography was performed on silica gel w/UV254 plates (1624126, sorbent technologies). 1H and 13C NMR spectra were recorded on instruments operating at 400 or 500 MHz and 100 or 126 MHz respectively. High-resolution mass spectrometry (HRMS) spectra were obtained on an ESI-TOF mass spectrometer. The analytical method utilized a Waters Aquity BEH C18 column (2.1 × 50 mm, 1.7 μm) eluting with a linear gradient of 95% water (modified to pH 9.8 through addition of NH4OH) to 100% CH3CN at 0.6 mL min−1 flow rate where purity was determined using UV peak area at 214 nm. Melting points were determined using an automated apparatus with digital imaging capability.General procedure for 4-substituted biquinolines
To a solution of nitromethane (50 μL, 1.0 mmol) in THF (1 mL), solution of K2CO3 (21.8 mg, 0.15 mmol) in water (0.2 mL) was added gradually with stirring and the mixture was stirred for 5 min. The 3-(2-quinolyl)quinolinium halide (1a–e) (0.75 mmol) was added and the mixture stirred for 4 h at 23 °C until a homogeneous solution was formed. The reaction was monitored by TLC/UPLC. The solution was poured into water (5 mL) and extracted with ethyl acetate (3 × 5 mL). The organic layers were separated, combined, washed with brine, dried over Na2SO4, and evaporated in vacuo.Series of quaternary quinolinium salts
Miscellaneous scaffolds
Antibacterial activity
Acknowledgements
Research reported in this publication was supported by two Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (8P30GM103495 and P30GM103326, SME) and the NIH University of Kansas Chemical Methodology and Library Development Center funded by the NIH Institute of General Medical Sciences (P50GM069663).References and notes
- Organization, W. H. Antimicrobial Resistance: Global Report on Surveillance 2014, 2014, pp. 1–257, http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1.
- (a) J. Davies and D. Davies, Origins and evolution of antibiotic resistance, Microbiol. Mol. Biol. Rev., 2010, 74(3), 417–433 CrossRef CAS PubMed; (b) K. Bush, P. Courvalin, G. Dantas, J. Davies, B. Eisenstein, P. Huovinen, G. A. Jacoby, R. Kishony, B. N. Kreiswirth and E. Kutter, Tackling antibiotic resistance, Nat. Rev. Microbiol., 2011, 9(12), 894–896 CrossRef CAS PubMed; (c) B. K. English and A. H. Gaur, The use and abuse of antibiotics and the development of antibiotic resistance. In Hot Topics in Infection and Immunity in Children VI, Springer, 2010, pp. 73–82 Search PubMed; (d) H. Madhavan and S. Murali, Mechanisms of Development of Antibiotic Resistance in Bacteria Among Clinical Specimens, J. Clin. Biomed. Sci., 2011, 1(2), 43 Search PubMed; (e) B. Spellberg, J. G. Bartlett and D. N. Gilbert, The future of antibiotics and resistance, N. Engl. J. Med., 2013, 368(4), 299–302 CrossRef CAS PubMed; (f) D. I. Andersson and D. Hughes, Persistence of antibiotic resistance in bacterial populations, FEMS Microbiol. Rev., 2011, 35(5), 901–911 CrossRef CAS PubMed; (g) D. M. Shlaes, D. Sahm, C. Opiela and B. Spellberg, The FDA Reboot of Antibiotic Development, Antimicrob. Agents Chemother., 2013, 57(10), 4605–4607 CrossRef CAS PubMed; (h) D. Jabes, The antibiotic R&D pipeline: an update, Curr. Opin. Microbiol., 2011, 14(5), 564–569 CrossRef PubMed.
- D. Higgins, E. Nazareno and V. DiRita, The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators, J. Bacteriol., 1992, 174(21), 6974–6980 CAS.
- O. G. Gómez-Duarte and J. B. Kaper, A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli, Infect. Immun., 1995, 63(5), 1767–1776 Search PubMed.
- A. K. Hovey and D. W. Frank, Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon, J. Bacteriol., 1995, 177(15), 4427–4436 CAS.
- S. M. Egan, Growing repertoire of AraC/XylS activators, J. Bacteriol., 2002, 184(20), 5529–5532 CrossRef CAS.
- J. Yang, M. Tauschek and R. M. Robins-Browne, Control of bacterial virulence by AraC-like regulators that respond to chemical signals, Trends Microbiol., 2011, 19(3), 128–135 CrossRef CAS PubMed.
- L. K. Garrity-Ryan, O. K. Kim, J.-M. Balada-Llasat, V. J. Bartlett, A. K. Verma, M. L. Fisher, C. Castillo, W. Songsungthong, S. K. Tanaka and S. B. Levy, Small molecule inhibitors of LcrF, a Yersinia pseudotuberculosis transcription factor, attenuate virulence and limit infection in a murine pneumonia model, Infect. Immun., 2010, 78(11), 4683–4690 CrossRef CAS PubMed.
- A. V. Jennison and N. K. Verma, Shigella flexneri infection: pathogenesis and vaccine development, FEMS Microbiol. Rev., 2004, 28(1), 43–58 CrossRef CAS PubMed.
- S. K. Niyogi, Shigellosis, J. Microbiol., 2005, 43(2), 133–143 Search PubMed.
- J. Mounier, T. Vasselon, R. Hellio, M. Lesourd and P. Sansonetti, Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole, Infect. Immun., 1992, 60(1), 237–248 CAS.
- (a) V. Koppolu, I. Osaka, J. M. Skredenske, B. Kettle, P. S. Hefty, J. Li and S. M. Egan, Small-Molecule Inhibitor of the Shigella flexneri Master Virulence Regulator VirF, Infect. Immun., 2013, 81(11), 4220–4231 CrossRef CAS PubMed; (b) J. M. Skredenske, V. Koppolu, A. Kolin, J. Deng, B. Kettle, B. Taylor and S. M. Egan, Identification of a Small-Molecule Inhibitor of Bacterial AraC Family Activators, J. Biomol. Screening, 2013, 18(5), 588–598 CrossRef PubMed.
- (a) A. Aksenov, O. Nadein, I. Borovlev and Y. I. Smushkevich, Investigations in the region of 2,3′-biquinolyl 5. Investigation of the reaction of stabilized C-nucleophiles with 1-alkyl-3-(2-quinolyl)-quinolinium halides, Chem. Heterocycl. Compd., 1998, 34(9), 1045–1049 CrossRef CAS; (b) A. Aksenov, Investigations on 2,3′-Biquinolyls. 15. Regioselectivity of the Addition of Nitromethane Anion to 1′-Alkyl-3′-(2-quinolyl) quinolinium Halides, Chem. Heterocycl. Compd., 2003, 39(6), 756–759 CrossRef CAS.
- (a) P. Renton-Harper, M. Addy, J. Moran, F. Doherty and R. Newcombe, A comparison of chlorhexidine, cetylpyridinium chloride, triclosan, and C31G mouthrinse products for plaque inhibition, J. Periodontol., 1996, 67(5), 486–489 CrossRef CAS PubMed; (b) N. Bodor and P. Buchwald, Soft drug design: general principles and recent applications, Med. Res. Rev., 2000, 20(1), 58–101 CrossRef CAS; (c) N. Bodor, J. J. Kaminski and S. Selk, Soft drugs. 1. Labile quaternary ammonium salts as soft antimicrobials, J. Med. Chem., 1980, 23(5), 469–474 CrossRef CAS.
- A. V. Aksenov and O. N. Nadein, Investigation of 2,3′-Biquinolyl. 10. The Regioselectivity of the Reaction of 2,3′-Biquinolyl and 1′-Alkyl-3-(2-quinolyl)quinolinium Halides with Halo Derivatives in the Presence of Metallic Lithium and Magnesium, Chem. Heterocycl. Compd., 2000, 36(11), 1314–1318 CrossRef CAS.
- M. Brasse, J. A. Ellman and R. G. Bergman, A facile, metal-and solvent-free, autoxidative coupling of quinolines with indoles and pyrroles, Chem. Commun., 2011, 47(17), 5019–5021 RSC.
- D. Garella, A. Barge, D. Upadhyaya, Z. Rodriguez, G. Palmisano and G. Cravotto, Fast, Solvent-Free, Microwave-Promoted Friedländer Annulation with a Reusable Solid Catalyst, Synth. Commun., 2009, 40(1), 120–128 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental protocols and structural characterization of target compounds. See DOI: 10.1039/c4ra08384a |
| This journal is © The Royal Society of Chemistry 2014 |