Theoretical analyses of the fluorescence lifetimes of the d-amino acid oxidase–benzoate complex dimer from porcine kidney: molecular dynamics simulation and photoinduced electron transfer†
Abstract
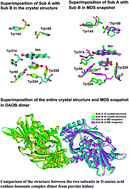
The mechanism of photoinduced electron transfer (ET) from benzoate (Bz) and aromatic amino acids to the excited isoalloxazine (Iso*) in the D-amino acid oxidase–benzoate complex (DAOB) dimer from porcine kidney was studied using molecular dynamics simulation (MDS) and an electron transfer theory, and compared with that in the DAOB monomer. The DAOB dimer displayed two fluorescent lifetime components of 0.85 ps and 4.8 ps, as reported. The ET parameters contained in the Kakitani and Mataga (KM) model were determined so as to reproduce these lifetimes with MDS atomic coordinates. The Bz–isoalloxazine (Iso) distances were 0.66 nm in subunit A (Sub A), 0.68 nm in subunit B (Sub B) and 0.61 nm in the monomer. The fluorescent lifetimes of 4.8 ps and 0.85 ps were found to originate from Sub A and Sub B, respectively. In Sub A, Tyr228 was the fastest ET donor followed by Bz and Tyr55, while Bz was followed by Tyr228 and Tyr314 in Sub B. The ET rate from Bz was fastest in Sub B, followed by that in Sub A and the DAOB monomer. The static dielectric constants obtained near Iso were 2.4–2.6 in the DAOB dimer and monomer and 5.8–5.9 in holo D-amino oxidase (DAAO). The different dielectric constants could account for the experimental fluorescence peak observed for DAOB (524 nm) and DAAO (530 nm). Logarithmic ET rates decreased linearly with the donor–acceptor distance expressed by both center to center distance (Rc) and edge to edge distance (Re) in Sub A and Sub B of DAOB dimer and monomer, which reveals that the conventional Dutton rule holds in the ET processes in DAOB. The logarithmic ET rates were decomposed into the electronic coupling (EC), square root (SQ) and exponential (GTRAM) terms. It was found that both the EC term and the GTRAM term also decreased linearly with Rc. The sum of the slopes in the EC and GTRAM vs. Rc plots coincided with the slopes in the logarithmic ET rate vs. Rc functions, suggesting that the GTRAM term makes a significant contribution to the linear relations between logarithmic ET rate and Rc.

 Please wait while we load your content...
Please wait while we load your content...