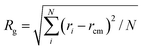Interaction of fullerene chains and a lipid membrane via computer simulations†
Wen-de Tian*a,
Kang Chenb and
Yu-qiang Ma*ab
aCenter for Soft Condensed Matter Physics and Interdisciplinary Research, Soochow University, Suzhou 215006, China. E-mail: tianwende@suda.edu.cn
bNational Laboratory of Solid State Microstructures and Department of Physics, Nanjing University, Nanjing 210093, China. E-mail: myqiang@nju.edu.cn
First published on 26th June 2014
Abstract
Coarse-grained molecular dynamics simulations were applied to investigate the interaction of fullerene polymers with a lipid membrane. It was found that the average center-of-mass distance between the polymer and membrane becomes larger with an increase of functionalization degree of the fullerene polymer. Free energy calculation shows the functionalization is energetically helpful to the adsorption of polymers on the membrane. For polymers with functionalized fullerenes, we found that the penetration of hydrophobic part into the lipid bilayer is realized by the thermally activated turnover of polymers.
Introduction
The rapid development of nanotechnology and synthesis methods have led to exciting innovations in many fields including the biomedical science and technology.1 Nanomaterials are of great interest for application in the areas of specific targeting, drug delivery, and enhanced bioimaging.2,3 An important example receiving considerable attention is the buckminsterfullerene (C60). Possible uses of C60 and its derivatives as X-ray contrast agents, antioxidant drugs for neurodegenerative diseases, and inhibitors of the allergic response have been suggested.4 However, chemical treatments are generally required for their potential biological applications in order to improve the solubility of C60. For instance, functionalizations with –OH and –NH2 have been used to increase its hydrophilicity.5 Recently, fullerenes were used as building blocks to construct novel materials such as the side-chain fullerene polymers, main-chain fullerene polymers, and star-shaped fullerene polymers.6 The introduction of fullerenes can enhance the properties of polymers such as photoconductivity. Meanwhile, the polymers can overcome the drawbacks of fullerenes (e.g., poor solubility) for large-scale applications. For example, amphiphilic fullerene polymers have been used for drug delivery via self-assembled supramolecular structures, which nearly can not realized by a single C60.7 Main-chain fullerene polymers are a type of polymer characterized by the presence of the fullerene spheres in the polymer backbone. Water-soluble poly(fullerocyclodextrin)s prepared by Geckeler and coworkers showed an excellent DNA-cleaving activity in photodynamic cancer therapy.8 For biomedical applications, it is of significantly importance to characterize the interactions of fullerene polymers with cellular components such as DNA, proteins, especially, the plasma membrane, by which cells respond to extracellular environment.Molecular dynamics (MD) simulations have played important roles in understanding the molecular mechanism of biosystems.9–11 All atom molecular dynamics (AAMD) simulations have been used to study the interaction of a C60 and its derivatives with lipid membranes.12 Bedrov et al.13 focused on the passive transport of C60 fullerene via atomistic simulations in the framework of CHARMM force field, and found that there is no free energy barrier for transport of the fullerene from the aqueous phase into the lipid core of membrane. Qiao et al.14 reported that the C60(OH)20 molecule is excluded by a dipalmitoylphosphatidylcholine (DPPC) bilayer but causes the area per neighboring lipid molecule reduced by 25%. The interactions of cationic amino functionalized C60 with a palmitoyl-oleyl-phosphatidyl-choline (POPC) membrane has also been studied by the CHARMM force field.15 However, it is difficult to study the interactions of fullerene polymers with membranes by atomistic models due to the limitation of system size and time scale.16 Therefore coarse-grained models have been applied to study the related systems.17 Wong-Ekkabut et al.18 simulated the translocation of fullerene clusters through a lipid membrane employing MARTINI force field, and suggested that high concentrations of fullerene are unlikely to cause mechanical damage to the membrane. Fundamental studies are still required to understand the interactions of fullerene–polymeric materials with biological membranes.19
In the present paper, we study the interactions of fullerene chains composed of eight C60s, some of which are surface-modified, with a DPPC membrane through coarse grained molecular dynamics simulations (CGMD) within the framework of the MARTINI force field.21,22 The surface properties of fullerenes play an important role in determining their cell uptake and translocation. It becomes more and more difficult for the polymer penetrating into the membrane with the increase of functionalization degree of the fullerene polymer. However, the PMF calculation shows that the adsorption of functionalized fullerene polymers on the membrane is energetically favorable. In addition, we also found that the thermal fluctuation may assist the turnover of functionalized fullerene polymer for penetrating into a membrane. The paper is organized as follows. We first introduce the CG model and give an overview of the simulation settings. Then we discuss on the results of structures, dynamics, and free energies of the interacting systems. Finally, a short summary is concluded.
Model and methods
Five types of main-chain fullerene polymers (see Fig. 1) are considered to interact with a DPPC bilayer. Polymers consist of eight C60s and/or its derivatives. The C60 was modeled as a hydrophobic particle while its derivative as a hydrophilic particle. The eight fullerenes were connected via a bond without bond angle constraint which was modeled by a harmonic potential with K = 1250 kJ mol−1 nm−2 and R0 = 0.47 nm to represent a simple fullerene polymer chain. The relative toxicity of fullerene-based polymers is mostly attributed to the ratio of the hydrophilic/hydrophobic fullerenes, which can lead to cell membrane disruption and subsequent cellular death.20 We change the ratio of C60 on the polymers from 1 to 0 so as to explore how the degree of functionalization of C60–polymer might influence the interaction of the polymers with a lipid bilayer (Fig. 1). The CG simulations were on the basis of the MARTINI force field21 in which 4 to 1 mapping strategy was used. The force field and the topologies for C60 and DPPC lipid were downloaded from the website of Marrink group (http://md.chem.rug.nl/marrink/coarsegrain.html), where four bead types (Q0+, Qa−, Na, and C1) were used for DPPC and CNP bead type was used for C60. In order to model the C60's derivative with hydrophilicity, the bead type of C60 was replaced with P4. The detailed description of our model was given in the ESI.†In each unbiased simulation, one polymer molecule interacts with a pre-equilibrated bilayer of 512 DPPC lipids. The polymer was initially placed 4 nm away from the center of mass (c.o.m.) of the bilayer. The system was then solvated with CG waters and energy-minimized for up to 1000 steps using a steepest-descent algorithm to remove any steric conflicts. Typically, each system was simulated for 1.0 μs. The data of the last 400 ns was used for analysis.
The umbrella sampling method23 was used to calculate the potential of mean force (PMF). Twenty-seven independent windows (spaced by 0.2 nm) along the normal direction, z, of bilayer (z = 5.0 to 0.0 nm from the bilayer center) were allocated for sampling. The z coordinate of the c.o.m. of each polymer was restrained relative to the z coordinate of the c.o.m. of the bilayer with a harmonic biasing potential with a force constant of 1000 kJ mol−1 nm−2. For each window, 1.0 μs CGMD simulation was carried out. Data was analyzed with the WHAM method.24
A Berendsen thermostat/barostat25 was used to maintain a constant temperature of 310 K and a constant pressure of 1 atm with compressibility of 5.0 × 10−5 bar−1 in the NPzP‖T ensemble. The time step of integration was 0.025 ps. A cutoff of 1.2 nm was used for electrostatic interactions and van der Waals interactions. The Lennard-Jones potential was smoothly shifted to zero between 0.9 and 1.2 nm and the Coulomb potential was smoothly shifted to zero between 0 and 1.2 nm. The relative dielectric constant is 15 for explicit water screening.22 All simulations were implemented with GROMACS package of version 4.5 (ref. 26) and visualized by VMD.27
Results and discussion
In this section, we present the structural information such as number-density distribution, dynamics, and thermodynamics of the fullerene polymers interacting with a lipid membrane. Some analytical methods were given in ESI.†Structure
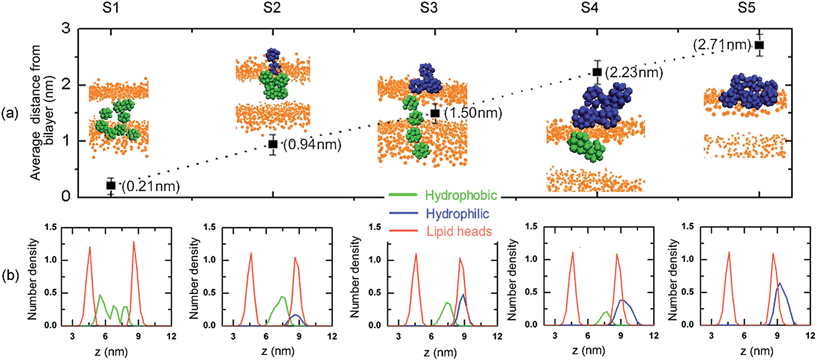
The average distance between the c.o.m of fullerene polymer and the c.o.m of bilayer are shown in Fig. 2a. It is about 0.21 nm for S1, which means that the polymer consisting of pristine C60 fully penetrates into the lipid bilayer. The snapshot shows that the C60 chain swells its configuration as in a good solution. The result is consistent with the finding of Li et al.,28 who suggested that the bilayer was less able to accommodate the aggregated C60 pair. For the totally functionalized fullerene chain (S5), the polymer adheres to the lipid bilayer, where the distance is about 2.71 nm. It demonstrates that the polymer can not penetrate into the bilayer as displayed by the equilibrated configuration. The average distances are 0.94 nm, 1.50 nm, and 2.23 nm for the S2, S3, and S4, respectively. The snapshots reveal that the hydrophobic part of the fullerene polymers enters the bilayer and makes contact with the lipid tails. The structure resembles the lipid-anchored proteins which are anchored to one leaflet of cell membrane. The average distance increases with the increase of the ratio of functionalized C60, which implies that the functionalization is a pathway to control the penetration of fullerene polymers.Fig. 2b shows the corresponding density profiles of hydrophobic part and hydrophilic part of polymers as well as the lipid head groups of five systems. It can be seen that the distribution of fullerene in S1 is between the lipid head groups of two leaflets of bilayer, while the distribution of fullerene polymer in S5 is above the top leaflet of bilayer around the lipid head groups. For the other three systems, the hydrophilic and hydrophobic parts of the polymer distribute on the two sides of lipid head groups. And, the c.o.m distance between the two parts is about 1.5 nm. As expected, the simulations showed that more polar derivatives prefer to the water phase while less polar ones end up in the bilayer interior.29 The above results suggest that the more surface-modified the fullerene polymer is, the larger the average c.o.m distance is and the harder it is to penetrate into the membrane.
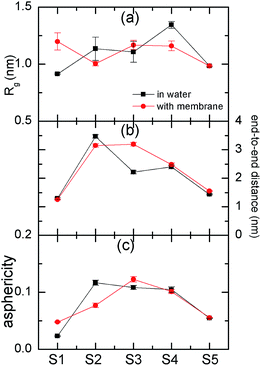
Further, the structures of fullerene polymers in water solution and after interacting with membrane were shown in Fig. 3. For S1 and S3, we find that the radius of gyration, Rg, in membrane is slightly larger than that in water, while the Rg for S2 and S4 in membrane is smaller than that in water. The profiles of end-to-end distance and asphericity as a function of functionalization degree seems like a horse-saddle. This maybe because the amphiphilic fullerene polymers for S2, S3, and S4 were split into two parts; one part is in water and the other part is in membrane. Here the asphericity, a, is equal to 1 − 3〈(IxIy +IyIz + IzIx)/(Ix + Iy + Iz)2〉, where Ix, Iy, and Iz are the three eigenvalues of the radius of gyration tensor. The Rg, the end-to-end distance, and the asphericity for the S5 seems not to be changed after interacting with membrane because the fully functionalized fullerene polymer does not penetrate into the membrane (i.e., it is still in water solution). In addition, we also calculated the order parameter of lipid tail, s = 0.5〈3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos2
cos2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ − 1〉, where θ is the angle between the C1A–C4A bond vector of acl chain and the bilayer normal, and the angular brackets indicate averaging over time and lipid molecules. The order parameter of lipids around the polymer was weakly disturbed for all cases (see Fig. S2 in ESI†). Moreover, we find the polymers can make the local thinning of membrane, especially, for S1–S3 (Fig. 4). For S5, the polymer slightly change the membrane thickness due to its non-penetration.
θ − 1〉, where θ is the angle between the C1A–C4A bond vector of acl chain and the bilayer normal, and the angular brackets indicate averaging over time and lipid molecules. The order parameter of lipids around the polymer was weakly disturbed for all cases (see Fig. S2 in ESI†). Moreover, we find the polymers can make the local thinning of membrane, especially, for S1–S3 (Fig. 4). For S5, the polymer slightly change the membrane thickness due to its non-penetration.
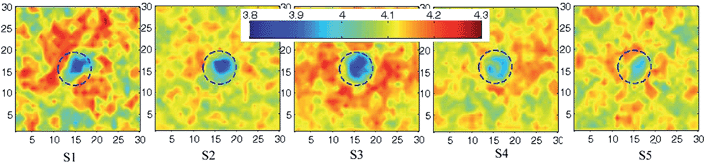 | ||
| Fig. 4 Thickness fluctuations across the bilayer in the x–y plane (grid resolution: 30 × 30) after the membrane interacting with the fullerene polymer for the five cases. The legend in units of nanometer shows the bilayer thickness values mapped to a rainbow color gradient. Grid-based membrane analysis tool30 was used for averaging the coordinates of last 100 ns of our simulations. | ||
Dynamics
The dynamics of how the fullerene polymer penetrates into the membrane was given in Fig. 5 for S1 and S3. It can be seen that the C60 polymer diffuses randomly in the water solution at the beginning. It reaches the down leaflet of bilayer at about 400 ns. After a while, the polymer enters the membrane and stays between the two leaflet of lipid bilayer (Fig. 5a). The time evolution of the z-distance between polymer center and bilayer center also manifests the above process (the inset of Fig. 5b). It has been suggested that the binding of apolar molecules to lipid membranes is often enthalpy-driven, with an unfavorable entropic contribution.31 When the polymer contacts with the bilayer, the potential energy between the polymer and lipid heads deceases firstly, accompanied with the slight increase of that between the polymer and water molecules (Fig. 5b). Then, the potential energy between the polymer and lipid tail groups decreases sharply. The potential energy decrease between the polymer and lipids is more than the energy increase between the polymer and water. To evaluate the penetration time of C60s, simulations were performed additionally ten times for each system. The average penetration time for the system is about 137 ns as the polymer was placed 3.4 nm about the center of mass of membrane (see Table S2 in ESI†). However, the polymer does not adsorb on the membrane over half of ten simulations of 1.0 μs due to the existence of energy barrier. Longer simulations are needed for capturing more penetration phenomena.In addition, we capture a typical penetration process for S3 (Fig. 5c). The polymer waits a long time, about 1 μs, to insert into the bilayer. Under the influence of thermal fluctuation, the hydrophobic part of polymer begins to contact with the bilayer, and then turn over into the interior of bilayer. During this process, the hydrophilic part of polymer acts as a cupule attached on the bilayer that ensures the polymer not to diffuse away. The time evolutions of c.o.m distance between the fullerene polymer and lipid bilayer, and the order parameter of the polymer relative to the bilayer norm show that the process of turnover of the polymer is very short, about 25 ns (Fig. 5d). This behavior is similar to the thermally activated barrier hopping process. The conclusion is consistent with the finding of Qiao et al.,14 who suggest a single pristine C60 molecule can “jump” into the DPPC bilayer. The polymer can rapidly penetrate into the membrane as long as it finds a suitable way with the help of thermal motion. Similar behaviors are also found in the S2 and S4. This may imply that the turnover behavior for penetration may be a general phenomenon for the amphiphilic polymers when interacting with cell membranes. Additionally we found that the polymer of S5 just adsorb on the membrane without penetration for the total simulations.
Potential of mean force
The potential of mean force was calculated to obtain the free energy profiles (as a function of the position of the fullerene polymer along the z axis normal to the bilayer) from bulk water to the core of the bilayer (Fig. 6a). The PMF profile of S1 displays a weak barrier of 1–2 KBT near 3.75 nm due to the hydrophobic interactions as the polymer approaches the membrane. This barrier pushes the polymer to diffuse away as shown in the subsection of dynamics. If the barrier is overcome by thermal activation, the free energy decreases till the center of bilayer, and the polymer is pulled into the bilayer core. For S2, the PMF profiles decrease as the polymers approach the membrane from 5.0 nm to 3.0 nm. The functionalization of two C60s on the polymer seems to have ability to eliminate the barrier of pure C60 polymer for penetration. For S3–S5, there is a minimum in the PMF at the water/membrane interface. That's why the partly functionalized fullerene polymer prefers adsorbing on the membrane. The position of minimal energy is about 2` nm from the c.o.m of bilayer in S5, which is similar to that of the unbiased simulation. | ||
| Fig. 6 (a) Potentials of mean force (PMFs) as a function of position along the bilayer normal for five systems. (b) Well depth (blue/square; left-hand scale) of the energy minima (derived from the PMFs in Fig. 4a) for various systems. Also shown is the value of the PMFs at the center of the bilayer (red/circles, right-hand scale). (c) Distance between the energy minima (derived from the PMFs in Fig. 4a) and the c.o.m of lipid bilayer. Error bars are shown. | ||
The energy minima is −110 KBT, −75 KBT, −50 KBT, −40 KBT, and −20 KBT for the S1, S2, S3, S4, and S5, respectively (Fig. 6b). This means that the adsorption or penetration of polymers is strongly energetically favorable. However, the free energies in the center of bilayer are about −60 KBT, −25 KBT, 30 KBT, and 105 KBT for S2, S3, S4, and S5, respectively (Fig. 6c). Compared with the energy minima, it suggests that the polymers can hardly penetrate across the bilayer. The positions of energy minima for five systems is shown in Fig. S3.† It can be found that the distance is corresponding to the value of average distance in Fig. 2a, which is in accordance with that of our unbiased simulations.
Conclusion
In conclusion, the interactions of main-chain fullerene polymers with a DPPC membrane were firstly investigated by coarse-grained molecular dynamics simulations. We found that the C60 polymer chain can fully partition into the lipid bilayer. The more surface-modified the fullerene polymer is, the farther away the center-of-mass position of polymer is from the bilayer, implying the harder for the polymer to penetrate into the membrane. In addition, The PMF calculations indicate that the free energy is in favor of the adsorption of functionalized fullerene polymers on the membrane. The adsorption provides enough time for the partly functionalized polymers to find a path to penetrate into the membrane via the polymer turnover with the help of thermal fluctuation. Furthermore, we found that the passive penetration of fullerene polymers through the membrane is difficult although the adsorption is energetically favorable. It should be pointed out, however, that a recent paper32 suggested it is an artifact barrier of the CG model as a single C60 approaches the POPC membrane. For DPPC membrane, whether the barrier really exists or not should be checked via atomistic simulations although we found there has a small barrier in S1 system within the framework of CG model. Fullerene polymers, the fundamental unit of which is fullerene, combine the properties of polymer and fullerene, a type of nanoparticles. It is interesting to study their physicochemical properties when interacting with biomembrane. Our findings is valuable in designing new materials for bio-applications.Acknowledgements
The work was supported by the National Natural Science Foundation of China (nos 91027040, 21104056, 1174180, and 10974080.) and the National Basic Research Program of China (973 program of no. 2012CB821500).References
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543 CrossRef CAS PubMed.
- W. Tian and Y. Ma, Chem. Soc. Rev., 2013, 42, 705 RSC.
- K. Yang and Y. Ma, Nat. Nanotechnol., 2010, 5, 579 CrossRef CAS PubMed.
- R. Partha and J. L. Conyers, Int. J. Nanomed., 2009, 4, 261 CrossRef CAS.
- T. Ros and M. Prato, Chem. Commun., 1999, 663 RSC.
- C. Wang, Z. Guo, S. Fu, W. Wu and D. Zhu, Prog. Polym. Sci., 2004, 29, 1079 CrossRef CAS PubMed.
- R. Partha, M. Lackey, A. Hirsch, S. Casscells and J. L. Conyers, J. Nanobiotechnol., 2007, 5, 6 CrossRef PubMed.
- S. Samal, B. J. Choi and K. E. Geckeler, Chem. Commun., 2000, 1373 RSC.
- W. Tian and Y. Ma, Soft Matter, 2012, 8, 2627 RSC.
- W. Tian and Y. Ma, Soft Matter, 2012, 8, 6378 RSC.
- A. Jusufi, R. H. DeVane, W. Shinoda and M. L. Klein, Soft Matter, 2011, 7, 1139 RSC.
- P. Ke and M. Lamm, Phys. Chem. Chem. Phys., 2011, 13, 7273 RSC.
- D. Bedrov, G. D. Smith and H. Davandeand, J. Phys. Chem. B, 2008, 112, 2078 CrossRef CAS PubMed.
- R. Qiao, A. P. Roberts, A. S. Mount, S. J. Klaineand and P. Ke, Nano Lett., 2007, 7, 614 CrossRef CAS PubMed.
- S. Kraszewski, M. Tarek and C. Ramseyer, ACS Nano, 2011, 5, 8571 CrossRef CAS PubMed.
- C. Ren, W. Tian, I. Szleifer and Y. Ma, Macromolecules, 2011, 44, 1719 CrossRef CAS.
- L. Monticelli, E. Salonen, P. C. Ke and I. Vattulainen, Soft Matter, 2009, 5, 4433 RSC.
- J. Wong-Ekkabut, S. Baoukina, W. Triampo, I. M. Tang, D. P. Tieleman and L. Monticelli, Nat. Nanotechnol., 2008, 3, 363 CrossRef CAS PubMed.
- M. Schulz, A. Olubummo and W. H. Binder, Soft Matter, 2012, 8, 4849 RSC.
- C. M. Sayes, J. D. Fortner, W. Guo, D. Lyon, A. M. Boyd, K. D. Ausman, Y. J. Tao, B. Sitharaman, L. J. Wilson, J. B. Hughes, J. L. West and V. L. Colvin, Nano Lett., 2004, 4, 1881–1887 CrossRef CAS.
- S. J. Marrink, A. H. de Vries and A. E. Mark, J. Phys. Chem. B, 2004, 108, 750 CrossRef CAS.
- S. J. Marrink, H. J. Risselada, S. Yefimov, D. P. Tieleman and A. H. de Vries, J. Phys. Chem. B, 2007, 111, 7812 CrossRef CAS PubMed.
- G. M. Torrie and J. P. Valleau, J. Comput. Chem., 1992, 13, 187 CrossRef.
- S. Kumar, D. Bouzida, R. H. Swendsen, P. A. Kollman and J. M. Rosenberg, J. Comput. Chem., 1992, 13, 1011 CrossRef CAS.
- H. J. C. Berendsen, J. P. M. Postma, W. F. Van Gunsteren, A. Dinola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684 CrossRef CAS PubMed.
- D. Van Der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark and H. J. C. Berendsen, J. Comput. Chem., 2005, 26, 1701 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33 CrossRef CAS.
- L. W. Li, H. Davande, D. Bedrov and G. D. Smith, J. Phys. Chem. B, 2007, 111, 4067 CrossRef CAS PubMed.
- R. S. G. D'Rozario, C. L. Wee, E. J. Wallace and M. S. P. Sansom, Nanotechnology, 2009, 20, 115102 CrossRef PubMed.
- W. J. Allen, J. A. Lemkul and D. R. Bevan, J. Comput. Chem., 2009, 30, 1952–1958 CrossRef CAS PubMed.
- J. Seelig, Biochim. Biophys. Acta, 1997, 1331, 103 CrossRef CAS.
- L. Monticelli, J. Chem. Theory Comput., 2012, 8, 1370–1378 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra04593a |
| This journal is © The Royal Society of Chemistry 2014 |