Fabrication of phospho-phytase/heteroatomic hierarchical Fe-ZSM-5 zeolite (HHFeZ) bio-conjugates for eco-sustainable utilization of phytate-phosphorus†
Wenzhong Zhanga,
Fang Xu*a and
Deju Wangb
aSchool of Environmental Science and Engineering, Shanghai Jiao Tong University, Shanghai 200240, China. E-mail: xufangzh@sjtu.edu.cn; Fax: +86-21-54741065; Tel: +86-21-34201548
bShanghai Research Institute of Petrochemical Technology, SINOPEC, Shanghai 201208, China
First published on 26th March 2014
Abstract
The originally fabricated phytase/HHFeZ bio-conjugates could appropriately enhance enzyme stabilization after heteroatomic hierarchical Fe-ZSM-5 zeolite (HHFeZ) was developed as a novel matrix for affinitive immobilization of Escherichia coli (E. coli) phytase. The immobilization strategy was primarily dependent on the affinitive coordination between heteroatomic iron atoms anchored into the HHFeZ frameworks with multiple sites of phosphorylation in phytase. In addition, the ordered multiporous structures of HHFeZ contributed to phytase immobilization and stabilization. The conditions for phytase immobilization were optimized under a solution pH of 4.5 in a batch mode, and the maximum phytase immobilization capacity was determined to be 2.01 mg mg−1. Compared to those of free phytase, the thermal and proteolytic resistance of phytase/HHFeZ bio-conjugates were effectively improved, and the enzymatic activity was maintained over a broad pH range. The phytase/HHFeZ bio-conjugates would be an innovative choice for eco-sustainable exploitation and utilization of phytate-phosphorus, especially with respect to eutrophication control.
1. Introduction
Approximately two-thirds of the total phosphorus (P) in feedstuffs occurs in the form of phytate (myo-inositol 1,2,3,4,5,6-hexaphosphate salts),1 which is biologically unavailable due to insufficient secretion of phytase (EC 3.1.3.8 or EC 3.1.3.26) in the gastrointestinal system of monogastric animals.2 In general, most of phytate-P is directly excreted, and the supplementary inorganic phosphate (e.g., dicalcium phosphate) is always excessive.3 Therefore, the correspondent P dispersion (both organic and inorganic) poses an environmental concern.4 Recently, microbial phytase has been exploited to the efficient utilization of phytate-P in control of subsequent P pollution control.5,6 However, the stabilization of biocatalyst is of most importance for its widespread application.7 Enzyme immobilization manipulation may be the most ideal methods for promoting the operational and storage stability of phytase. Currently, various matrices have been employed for phytase immobilization.5,8–11 It is essential to note that well-tailored nanomaterials may constitute unique host supports.12–14 Menezes-Blackburn et al. recently attempted to immobilize Escherichia coli (E. coli) phytase on allophanic synthetic compounds and montmorillonite nanoclays.15 However, phytases enzyme could be expressed from several species of bacteria, yeast and fungi. E. coli phytase had the highest specific activity of any tested phytases.16 Moreover, E. coli phytase was identified by us containing multiple sites of phosphorylation. Therefore, the E. coli phytase was the most suitable candidate for transitional metal ion driven affinitive immobilization process.17 Zeolites are a kind of non-toxic crystalline material in possession of regular inner channels and uniform open pores. In a previous study, hierarchical heteroatomic Fe-ZSM-5 zeolite (HHFeZ) was developed by our group for the selective immobilization of low-abundant phosphorylated proteins from complex sample.18 Herein, the novel concept continues for the oriented immobilization of E. coli phospho-phytase on HHFeZ as affinitive matrix. The phytase immobilization was primarily based on the specific coordination between heteroatomic iron atoms in the frameworks of HHFeZ and multiple sites of phosphorylation in phytase. The iron atoms as host sites homogeneously distributed into the microenvironment of HHFeZ effectively facilitate phytase immobilization and activity. At the same time, the hierarchical porosity of HHFeZ may contribute to phytase immobilization and stabilization.19,20 The ZSM-5 framework contains a configuration with linked tetrahedra consisting of eight five-membered rings. The channels in ZSM-5 have a specific diameter between the diameters of small-pore LTA-type and large-pore FAU-type channels.21 The originally fabricated phytase/HHFeZ bio-conjugates could allow for appropriate loading of phytase. Interestingly, the HHFeZ support also provides good accessibility for organic phytate molecules possessing six phosphate groups. Simultaneously, it is supposed that inorganic phosphate moieties generated through the catalytic reaction may be released from the HHFeZ support. The specific advantages of the phytase/HHFeZ bio-conjugates compared to free phytase have been extensively evaluated, which primarily rely on the enhanced enzyme stability against thermal/proteolysis denaturation, and reusability. The innovative phytase/HHFeZ bio-conjugates are of great potential in biotechnological application for eco-sustainable utilization of phytate-P in feedstuffs and in the control of relevant P pollution.2. Experimental
2.1 Materials, reagents and buffers
E. coli appA coded phytase (5000 U g−1) was purchased from Chifei Biochemical, China. Sodium phytate was obtained from Ruibio, Germany. White carbon black (Degussa, China), colloidal silica (30 wt%, Sigma, USA), and tetrapropylammonium hydroxide (Sigma, USA) were utilized for zeolite synthesis.Various buffering systems (all in 0.01 M) were prepared including glycine–HCl (pH 2.0, 2.5, 3.0, 3.5), acetate buffer solution (ABS, pH 4.0, 4.5, 5.0, 6.0, 6.5), and Tris–HCl (pH 7.0, 8.0, 9.0). In addition, a specific buffer solution was employed to determine the pH conditions required for phytase immobilization and the pH-activity dependence curve. Gastric fluid (GF, pH 1.2 pepsin–NaCl solution) was simulated according to The United States Pharmacopeia-National Formulary.22
2.2 Zeolite synthesis and characterization
HHFeZ, hierarchical Na-ZSM-5 zeolite (HNaZ), and zeolite Na-A were hydrothermally synthesized according to previously published protocols.18,23 The crystal structures of the materials were confirmed by X-ray powder diffraction (XRD; D/max-2200/PC, Rigaku, Japan). The zeolite morphology was investigated by field emission scanning electron microscopy (FE-SEM, Nova NanoSEM 230, FEI, USA). The surface area and porosity were examined using N2 adsorption–desorption isotherms (ASAP 2010M + C, Micromeritics, USA). The introduced iron composition was analyzed by X-ray fluorescence spectrometry (XRF; XRF-1800, Shimadzu, Japan), and the chemical states of the heteroatomic iron atoms in HHFeZ were determined by Mössbauer spectrometry (9.25 × 108 Bq 57Co/Pd radioactive source). The surface topologies of the HHFeZ and HHFeZ/phytase bio-conjugates were visualized via tapping-mode atomic force microscopy (AFM, NanoNavi E-Sweep, SII, Japan). The Fe3+ ions that were potentially released were measured by inductively coupled plasma optical emission spectrometry (iCAP6300, Thermo Fisher, USA) after 30 mg of HHFeZ was incubated in 10 mL of GF at 39 °C for specified periods.2.3 Characterization of E. coli phosphorylated phytase
After the raw phytase powder was thoroughly mixed with ABS at a pH level of 4.5 in an ultrasonic bath, the acquired phytase solution was filtered and stored at −20 °C. Then, 10 μg of phytase was loaded on an SDS-PAGE gel. The corresponding band (∼47 kDa) was excised and applied for trypsin digestion (Promega, USA). Next, the digested peptides were separated by nano-HPLC and introduced into an LTQ-Orbitrap mass spectrometer (ThermoFisher, USA). Both full-MS and MS/MS spectra were analyzed by MASCOT (Matrix Science, UK; v 2.3.02) and X! Tandem (The GPM; v 2010.12.01.1), which were configured to search the NCBI E. coli protein database (downloaded in Jan. 2013). Scaffold (Proteome Software, USA; v 4.2.1) was used to validate the MS/MS based identification. The detailed protocols were the same as those used by Zhang et al.182.4 Determination of phytase concentration and activity
The protein concentration of phytase was assayed using the Bradford method (Protein assay kit, Bio-Rad, USA). The phytase activity was determined by measuring the amount of inorganic phosphate released from sodium phytate according to a method reported by Menezes-Blackburn et al.152.5 Fabrication of phytase/HHFeZ bio-conjugates
Phytase was immobilized on HHFeZ in a batch mode process. First, 30 mg of HHFeZ was mixed with 1 mL of ABS (pH 4.5) under ultrasonic bath until a homogenous suspension was acquired. Then, 9 mL of a diluted phytase solution was dropped into the suspension under stirring and subsequently agitated on an orbital shaker at 200 min−1 overnight. Next, the acquired phytase/HHFeZ bio-conjugates were completely washed using ABS with centrifugation (1000g, 15 min) and re-suspension cycles. Finally, the fabricated phytase/HHFeZ bio-conjugates were air dried in a desiccator.The phytase immobilization capacity was optimized with respect to the effect of the solution pH (2.0–9.0). The immobilization kinetics were determined at certain time intervals (10–100 min). The immobilization isotherms were established using phytase dilutions (0.5–7.5 mg mL−1). The effect of the ionic strength on phytase immobilization was studied in the presence of sodium chloride.
For comparison, HNaZ and Na-A based phytase bio-conjugates were studied in parallel.
2.6 Enzymatic stability of phytase/HHFeZ bio-conjugates
In comparison to that of free phytase, the enzymatic stability of phytase/HHFeZ bio-conjugates was evaluated by the detection of the residual phytase activities under specified conditions.To determine the enzyme thermal stability, free phytase and phytase/HHFeZ bio-conjugates were separately incubated in an 80 °C water bath for 0.1–10 min. The proteolytic resistance was investigated by exposing the bio-conjugates to GF at 39 °C for 2 and 24 h. The pH-activity dependence (2.0–9.0) was determined at 39 °C.
The apparent Michaelis–Menten constants (Km) and maximum reaction velocities (Vmax) of the free phytase and phytase/HHFeZ bio-conjugates were assayed according to the methods reported by Çelem et al.9
To evaluate their reusability, the bio-conjugates were recovered from the reaction media by centrifugation (1000g, 5 min) and subjected to repeated reactions.
All of the experiments were performed in triplicate.
3. Result and discussion
3.1 Characterization of HHFeZ and phytase
The MFI-type structure of HHFeZ was confirmed based on the XRD patterns obtained (Fig. S1 in ESI†). The SEM micrographs in Fig. 1A indicated that the spherical crystals of HHFeZ (10–20 μm in diameter) were constructed from hierarchical porous aggregates. Iron accounted for 6.27 wt% of HHFeZ according to the XRF analysis (Table S1†). Therefore, the Mössbauer spectroscopy was utilized to determine the iron speciation in HHFeZ (Fig. 1B). The spectra could be split into two lines of doublets where one doublet (tetrahedrally coordinated Fe3+, Fe3+Tet) exhibited an isomer shift (IS) of 0.296 mm s−1 and a quadrupole splitting (QS) of 0.835 mm s−1 and the other doublet (octahedrally coordinated Fe3+, Fe3+Oct) exhibited an IS of 0.390 mm s−1 and a QS of 1.101 mm s−1.24 These data indicated that the heteroatomic iron atoms in the framework positions of HHFeZ constituted the majority (Fe3+Tet/Fe3+Oct = 7.05). In addition, the framework iron atoms remained appropriately stable because only 2.53% and 4.22% of the iron was released into GF after 2 and 24 h of incubation, respectively. The BET surface area of HHFeZ was determined to be 267.7 m2 g−1. The hierarchical structure was clarified, and an adsorption average pore diameter of 4.1 nm was calculated using the BJH method (Fig. 2). | ||
| Fig. 2 N2 adsorption–desorption isotherms at 77 K (A) and pore size distribution calculated from the desorption branch using BJH method (B) of HHFeZ. | ||
E. coli appA-coded phytase was identified by mass spectrometry to possess two serine phosphorylation sites within the NAQMC[pS]LAGFTQIVNEAR and AGG[pS]IADFTGHR peptides. The database search results are shown in Table 1. Detailed MS/MS identification information is available in Fig. S2 and Tables S2 and S3.†
| Sequence | Mascot ion score | Mascot identity score | Mascot delta ion score | X! Tandem –log(e) score | Delta, Da | Delta, ppm |
|---|---|---|---|---|---|---|
| (R)AGG[pS]IADFTGHR(Q) | 34.1 | 25.8 | 28.6 | 1.42 | −0.00065 | −0.51 |
| (R)NAQGMC[pS]LAGFTQIVNEAR(I) | 49.3 | 29.4 | 27.0 | 3.48 | 000014 | 0.65 |
3.2 Oriented fabrication of phytase/HHFeZ bio-conjugates
As shown in Scheme 1, the fabrication of the phytase/HHFeZ bio-conjugates was primarily dependent on the affinitive coordination between iron atoms in the framework of HHFeZ and phosphate groups in the phospho-phytase. In addition, the electrostatic, hydrogen bonding, and van der Waals (vdW) forces may contribute to the immobilization of the phytase protein.25 In our studies, the phytase/HHFeZ bio-conjugates were orientedly fabricated by batch immobilization.The conditions for immobilization of phytase were optimized by varying the solution pH from 2.0 to 9.0. However, according to Fig. 3A, the most appropriate loading capacity for phytase was achieved when the solution pH was 3.0. As indicated in Fig. 3B, the phytase/HHFeZ bio-conjugates reserved the most enzymatic activity when the immobilization process was conducted at a solution pH of 4.5, which coincided with the optimal efficacy of the E. coli phytase.
The immobilization capacity of phytase increased with the contact time and attained a maximum value after 80 min, as shown in Fig. 4A. The immobilization kinetics data were fitted to pseudo-first-order (eqn (1)) and pseudo-second-order (eqn (2)) models:
 | (1) |
 | (2) |
The linearized pseudo first-order and second-order models are shown in Fig. 4B and C. The associated coefficients (Table 2) indicate that the immobilization followed second-order kinetics.
| Zeolite matrices | HHFeZ | HNaZ | Na-A | |
|---|---|---|---|---|
| a Particle diffusion did not occur. | ||||
| qe (exp.)/g g−1 | 1.70 ± 0.06 | 1.12 ± 0.05 | 0.887 ± 0.06 | |
| Pseudo-first-order kinetics | ||||
| qe (cal.)/g g−1 | 1.74 ± 0.24 | 0.756 ± 0.051 | 0.794 ± 0.025 | |
| k1/min−1 | 0.0790 ± 0.0069 | 0.0428 ± 0.0033 | 0.0392 ± 0.0016 | |
| R2 | 0.915 | 0.933 | 0.981 | |
| Pseudo-second-order kinetics | ||||
| qe (cal.)/g g−1 | 1.79 ± 0.01 | 1.15 ± 0.04 | 0.972 ± 0.054 | |
| k2/g g−1 min−1 | 0.118 ± 0.002 | 0.171 ± 0.04 | 0.0853 ± 0.0076 | |
| R2 | 0.999 | 0.996 | 0.993 | |
| Intra-particle diffusion kinetics | ||||
| Film diffusion | kp/min−1/2 | 0.425 ± 0.011 | 0.313 ± 0.002 | 0.126 ± 0.002 |
| C/g g−1 | Fixed at 0 | |||
| R2 | 0.997 | 0.999 | 0.999 | |
| Particle diffusion | kp/min−1/2 | 0.0953 ± 0.0074 | 0.0500 ± 0.0014 | N.A.a |
| C/g g−1 | 0.928 ± 0.047 | 0.642 ± 0.009 | N.A.a | |
| R2 | 0.965 | 0.996 | N.A.a | |
| Surface adsorption | kp/min−1/2 | 0.00104 ± 0.0001 | 0.0166 ± 0.0013 | 0.0508 ± 0.0021 |
| C/g g−1 | 1.69 ± 0.01 | 0.954 ± 0.012 | 0.391 ± 0.017 | |
| R2 | 0.961 | 0.989 | 0.988 | |
The isotherms of phytase immobilization (Fig. 5A) were described by the Langmuir (eqn (3)), Freundlich (eqn (4)), and Tempkin (eqn (5)) models:26
 | (3) |
 | (4) |
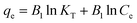 | (5) |
The linearized isotherm models are shown in Fig. 5B–D, which obeyed the Langmuir model (Table 3), suggesting that the immobilization of phytase was a monolayer process. In addition, the maximum loading capacity of phytase was 2.01 mg mg−1 for HHFeZ, higher than the loading capacities of HNaZ and Na-A, 1.18 mg mg−1 and 1.04 mg mg−1, respectively.
| Zeolite matrices | HHFeZ | HNaZ | Na-A |
|---|---|---|---|
| Langmuir model | |||
| Qm/mg mg−1 | 2.01 ± 0.04 | 1.18 ± 0.02 | 1.04 ± 0.01 |
| KL/mL mg−1 | 3.96 ± 0.15 | 2.87 ± 0.23 | 1.13 ± 0.04 |
| R2 | 0.992 | 0.998 | 0.992 |
| Freundlich model | |||
| KF | 1.69 ± 0.09 | 0.725 ± 0.079 | 0.466 ± 0.005 |
| n | 1.87 ± 0.02 | 2.29 ± 0.01 | 1.90 ± 0.01 |
| R2 | 0.919 | 0.958 | 0.963 |
| Tempkin model | |||
| B1/mg mg−1 | 0.414 ± 0.029 | 0.842 ± 0.015 | 0.565 ± 0.012 |
| KT | 50.7 ± 15.4 | 31.7 ± 7.3 | 10.9 ± 1.44 |
| R2 | 0.967 | 0.974 | 0.983 |
The enzyme, substrate, and product all possess phosphate groups when E. coli phytase catalyzes phytate to form phosphate and inositol. After the phospho-phytase is immobilized on HHFeZ, the organic phytate molecules can easily diffuse and be transported inside the hydrophobic ZSM-5 type zeolite.27 Therefore, the organic phytate, which is a substrate that contains six phosphate groups, also tends to have an affinity for HHFeZ. According to our studies, the produced inorganic orthophosphate ions could be subtly released into the reaction media solution. Therefore, the fabricated phytase/HHFeZ bio-conjugates can serve as an enzyme bioreactor for phytate-P bioavailability.
3.3 HHFeZ as a suitable host for phospho-phytase immobilization
The surface morphologies of the HHFeZ and phytase/HHFeZ bio-conjugates were visualized using tapping mode AFM, as shown in Fig. 6. The HHFeZ surface exhibited jagged crystal-like morphology with root-mean square (RMS) roughnesses of 123.0 and 101.6 nm over areas measuring 2 × 2 and 1 × 2 μm2, respectively. However, the relevant RMS roughnesses decreased to 12.7 and 4.3 nm, respectively, after the immobilization of phytase. Based further on the height profiles of the selected directions, the phytase was confirmed to be orientedly immobilized on the surface of HHFeZ.28Menezes-Blackburn et al. determined that the iron coating of allophanic materials facilitated the immobilization and stabilization of phytase.15 In this study, the oriented immobilization of phospho-phytase on HHFeZ was primarily dependent on iron atom active sites that were homogenously introduced into the zeolite. The force of the coordination between iron and the phosphate group was stronger than that of physical adsorption but weaker than that of covalent bonding. In addition, electrostatic forces may have affected the immobilization, as shown in Fig. 3A. When the solution pH was lower than the isoelectric point (pI = 7.5) of phytase,16 electrostatic attractive forces between the negative zeolite framework and positive phytase promoted the immobilization. The effect of the competition of H+ was limited when the solution pH was less than 3.0. When the solution pH > pI, the negatively charged phytase may have been repelled by the zeolite framework. As shown in Fig. S3,† the immobilization capacity of phytase on HHFeZ was barely affected in the presence of 0.1 and 0.5 M NaCl. However, the amount of phytase immobilized on zeolite Na-A decreased as the concentration of aqueous Na+ increased. The zeolite Na-A possessed only micropores (pore diameter < 1 nm),23 which may have prevented the entry of phytase. The N2 adsorption–desorption isotherms (Fig. 2) indicated that the micro- and mesopore areas of HHFeZ were 220.4 and 43.6 m2 g−1, respectively. Therefore, the relevant studies confirmed that the hierarchical, rigid, and uniform multiporous structures of HHFeZ promoted the access and diffusion of phytase. Correspondently, the hydrogen bonding and vdW forces between the internal zeolite surface and the biomolecules facilitated the immobilization process.
The effect of hierarchical structure on the immobilization of phytase is consistent with the immobilization kinetics observed (Fig. 4A). An intra-particle diffusion kinetics model (eqn (6)) was explored to fully elucidate the relevant processes:29
 | (6) |
In general, three consecutive steps occur (i) film diffusion (from solution to the external surface of the zeolite), (ii) particle diffusion (inside the pore-channel system), and (iii) surface adsorption. As indicated in Fig. 4D, three stages of excellent linearity for the kinetics of phytase immobilization on both HHFeZ and HNaZ were observed. The hierarchical structure provided these zeolites with multiporous cavities that allowed the biomolecule to diffuse and accommodate. Based on the diffusion rate constants reported in Table 3, the film and particle diffusion processes in the HHFeZ crystals occurred more rapidly than in the HNaZ samples.
3.4 Enhanced enzyme stability of phytase/HHFeZ bio-conjugates
The immobilization of phytase is essential to enhancing the enzyme's resistance to elevated temperatures and proteolysis.30 Typically, the enzyme has to endure 30 s to 2 min of exposure to temperatures of 70–95 °C during the feed pelleting process.31 However, free phytase was nearly inactive after 2 min of thermo-exposure. According to Fig. 7A, the phytase/HHFeZ bio-conjugates retained ca. 70% of their activity after exposure at 80 °C for 2 min. The phytase/HHFeZ bio-conjugates exhibited an 18-fold improvement in the enzyme half-life (t1/2 = 3.505 ± 0.063 min) compared to that of free phytase form (t1/2 = 0.188 ± 0.002 min).It was also noted that free phytase itself might be digested by proteases (pepsin or trypsin) in GF, leading to inactivation. As shown in Fig. 7B, the phytase/HHFeZ bio-conjugates preserved ca. 90% and 70% of the original activity of phytase after 2 and 24 h, respectively, of exposure in GF. However, after the same incubation time, the free phytase was completely inactivated. The enzyme can be effective over a broad pH range of 1.0 to 9.0, which occurs in the gastrointestinal systems of most poultry and livestock. The activity-pH dependence was effectively improved due to the immobilization of phytase on HHFeZ (Fig. 7C). The phytase/HHFeZ bio-conjugates exhibited appropriate activity within a pH range of 2–6.
The enzyme kinetics followed the Michaelis–Menten equation. According to the Lineweaver–Burk plots, the free phytase exhibited a Km value of 0.146 mM and a Vmax value of 1.21 U mg−1, and phytase/HHFeZ exhibited a Km value of 0.192 mM and a Vmax value of 1.68 U mg−1. The 1.3-fold higher Km value for the phytase/HHFeZ bio-conjugates was most likely due to increased diffusion limits.9
However, reusability must be considered when assessing the specific stability of the immobilization strategy. The activity of phytase immobilized on HHFeZ decreased by only ca. 10% after recycling five times (Fig. 7D). After 10 successive batch reactions, ca. 70% of the enzyme activity was retained. Therefore, in contrast to free phytase, the phytase/HHFeZ bio-conjugates may act as an enzymatic bioreactor for the sequential digestion of phytate into inorganic orthophosphate and inositol. Further studies using animal models are being performed.
3.5 HHFeZ as a suitable host for phospho-phytase stabilization
The immobilization of phytase may disrupt its active sites due to the possible formation of stable complexes and disruption of its substrate recognition ability. Lim et al. previously determined the peptide region for phytate binding and catalytic reaction in E. coli phytase.16 However, any structural changes in this peptide region leads to loss of enzyme activity. The two identified phospho-Ser residues in phytase responsible for the affinitive coordination with HHFeZ were not near the active region. Therefore, the activity of the fabricated HHFeZ/phytase bio-conjugates was delicately maintained.Thermal denaturation of the enzyme is primarily due to the destabilization of ionic and hydrophobic interactions. The loss of hydrogen bonds, vdW forces and ionic interactions may induce structural misfolding resulting in a decrease in enzymatic activity.32 Recently, it was reported that the phytase immobilized on probiotic Bacillus polyfermenticus spores exhibited a 6.5-fold improvement in t1/2 at 80 °C.10 The 18-fold longer t1/2 indicated that HHFeZ was a promising support for phytase immobilization. The abundant internal surface area of HHFeZ preserves the active structure of phytase due to interaction with the complex hydrogen bond/vdW force networks.21 Fei et al. stated that E. coli appA phytase begins to collapse from the C-terminal during thermo-exposure.33 Interestingly, phospho-Ser392 is located in the CS β-strand between a GM bend and an LAGFTQIVNEA α-helix close to the C-terminal. The enhanced thermostability of phytase may be due to the reinforcement of the C-terminal structure after oriented immobilization.
It is important to note that the average kinetic diameter of pepsin (3 nm) is smaller than that of the zeolite pores.34 Therefore, approaching pepsin molecules can diffuse into the channels of HHFeZ. Phytase-pepsin interactions might be limited by pore blocking and conformational restrictions after immobilization.35 Therefore, the phytase/HHFeZ bio-conjugates perform well with respect to proteolytic resistance.
Upon reuse, the homogeneous distribution of iron atoms inside the HHFeZ framework provided sufficient strength for phytase binding. Simultaneously, the interaction was suitable for allowing phytase to be sufficiently flexible to maintain its enzyme catalytic activity. In addition, the activity of phytase was barely affected by the dehydration–rehydration process, which indicated that HHFeZ is a suitable support for phytase immobilization. The reusability of phytase/HHFeZ bio-conjugates was comparable to that of the phytase covalently immobilized on Sepabead EC-EP,9 in which ca. 80% of the activity was retained after 10 cycles.
4. Conclusions
Phytase/HHFeZ bio-conjugates were successfully fabricated through the oriented immobilization of E. coli phospho-phytase on HHFeZ. Heteroatomic iron atoms synthetically introduced into the framework of HHFeZ provided attractive sites for the selective affinity recognition of phospho-phytase. In addition, the abundant porous structures of HHFeZ provide an appropriate micro-environment for phytase stabilization. The phytase/HHFeZ bio-conjugates exhibited enhanced enzymatic stability. The reusability was satisfactory after 10 cycles of batch operation. The phytase/HHFeZ bio-conjugates should be further explored for the eco-sustainable utilization of phytate-P and amelioration of P pollution.Acknowledgements
This work was supported by the Ministry of Science and Technology, China (China-Slovakia 5-8 and 6-13), the National Ocean Public Beneficial Research Foundation (201005026), and the Shanghai Science and Technology Committee (12142200500). The authors wish to thank Dr Shujun Chang for recording the Mössbauer spectra. ICP-OES analysis was conducted at the Instrumental Analysis Center, Shanghai Jiao Tong University. The LC-MS/MS experiments and PTM identifications were performed at the Institutes of Biomedical Sciences, Fudan University.References
- A. Viveros, C. Centeno, A. Brenes, R. Canales and A. Lozano, J. Agric. Food Chem., 2000, 48, 4009 CrossRef CAS PubMed.
- K. Nahm, Crit. Rev. Environ. Sci. Technol., 2009, 39, 521 CrossRef CAS.
- E. Kebreab, A. V. Hansen and A. B. Strathe, Curr. Opin. Biotechnol., 2012, 23, 872 CrossRef CAS PubMed.
- B. Singh, D. Singh and K. Sharma, in Biotechnology for Environmental Management and Resource Recovery, Springer, 2013, pp. 239–260 Search PubMed.
- R. Greiner, U. Konietzny, D. M. Blackburn and M. A. Jorquera, Bioresour. Technol., 2013, 142, 375 CrossRef CAS PubMed.
- D. Menezes-Blackburn, M. A. Jorquera, L. Gianfreda, R. Greiner and M. de la Luz Mora, Biol. Fertil. Soils, 2013 DOI:10.1007/s00374-013-0872-9.
- J. B. Garrett, K. A. Kretz, E. O'Donoghue, J. Kerovuo, W. Kim, N. R. Barton, G. P. Hazlewood, J. M. Short, D. E. Robertson and K. A. Gray, Appl. Environ. Microbiol., 2004, 70, 3041 CrossRef CAS.
- M. Calabi-Floody, G. Velásquez, L. Gianfreda, S. Saggar, N. Bolan, C. Rumpel and M. L. Mora, Chemosphere, 2012, 89, 648 CrossRef CAS PubMed.
- E. B. Çelem and S. Önal, J. Mol. Catal. B: Enzym., 2009, 61, 150 CrossRef PubMed.
- E.-A. Cho, E.-J. Kim and J.-G. Pan, Enzyme Microb. Technol., 2011, 49, 66 CrossRef CAS PubMed.
- M. V. Ushasree, P. Gunasekaran and A. Pandey, Appl. Biochem. Biotechnol., 2012, 167, 981 CrossRef CAS PubMed.
- B. Malvi and S. S. Gupta, Chem. Commun., 2012, 48, 7853 RSC.
- E. T. Hwang, B. Lee, M. Zhang, S. Jun, J. Shim, J. Lee, J. Kim and M. B. Gu, Green Chem., 2012, 14, 1884 RSC.
- Y. Masuda, S. Kugimiya, Y. Kawachi and K. Kato, RSC Adv., 2014, 4, 3573 RSC.
- D. Menezes-Blackburn, M. Jorquera, L. Gianfreda, M. Rao, R. Greiner, E. Garrido and M. de la Luz Mora, Bioresour. Technol., 2011, 102, 9360 CrossRef CAS PubMed.
- D. Lim, S. Golovan, C. W. Forsberg and Z. Jia, Nat. Struct. Biol., 2000, 7, 108 CrossRef CAS PubMed.
- I. L. Batalha, C. R. Lowe and A. C. Roque, Trends Biotechnol., 2012, 30, 100 CrossRef CAS PubMed.
- W. Zhang, D. Wang, H. Sun, J. Yao, F. Xu and P. Yang, Anal. Methods, 2012, 4, 2644 RSC.
- C. Bernal, L. Sierra and M. Mesa, J. Mol. Catal. B: Enzym., 2012, 84, 166 CrossRef CAS PubMed.
- B. Luangon, A. Siyasukh, P. Winayanuwattikun, W. Tanthapanichakoon and N. Tonanon, J. Mol. Catal. B: Enzym., 2012, 75, 80 CrossRef CAS PubMed.
- G. Kokotailo, S. Lawton and D. Olson, Nature, 1978, 272, 437 CrossRef CAS.
- The United States Pharmacopeia-National Formulary, United States Pharmacopeial Convention, Rockville, MD, 2011 Search PubMed.
- R. A. Rakoczy and Y. Traa, Microporous Mesoporous Mater., 2003, 60, 69 CrossRef CAS.
- P. Fejes, J. B. Nagy, K. Lázár and J. Halász, Appl. Catal., A, 2000, 190, 117 CrossRef CAS.
- Y. Wang and F. Caruso, Chem. Mater., 2005, 17, 953 CrossRef CAS.
- K. Y. Foo and B. H. Hameed, Chem. Eng. J., 2010, 156, 2 CrossRef CAS PubMed.
- H. Nakamoto and H. Takahashi, Zeolites, 1982, 2, 67 CrossRef CAS.
- M. Mahmoudi and V. Serpooshan, J. Phys. Chem. C, 2011, 115, 18275 CAS.
- F.-C. Wu, R.-L. Tseng and R.-S. Juang, Chem. Eng. J., 2009, 153, 1 CrossRef CAS PubMed.
- S. Mitchell and J. Pérez-Ramírez, Catal. Today, 2011, 168, 28 CrossRef CAS PubMed.
- K. Lundblad, S. Issa, J. Hancock, K. Behnke, L. McKinney, S. Alavi, E. Prestløkken, J. Fledderus and M. Sørensen, Anim. Feed Sci. Technol., 2011, 169, 208 CrossRef PubMed.
- V. Sarath Babu, M. Kumar, N. Karanth and M. Thakur, Biosens. Bioelectron., 2004, 19, 1337 CrossRef CAS PubMed.
- B. Fei, Y. Cao, H. Xu, X. Li, T. Song, Z. Fei, D. Qiao and Y. Cao, Curr. Microbiol., 2013, 66, 374 CrossRef CAS PubMed.
- J. Tang, I. I. Slowing, Y. Huang, B. G. Trewyn, J. Hu, H. Liu and V. S.-Y. Lin, J. Colloid Interface Sci., 2011, 360, 488 CrossRef CAS PubMed.
- M. A. Rao, A. Violante and L. Gianfreda, Soil Biol. Biochem., 2000, 32, 1007 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra01385a |
| This journal is © The Royal Society of Chemistry 2014 |







