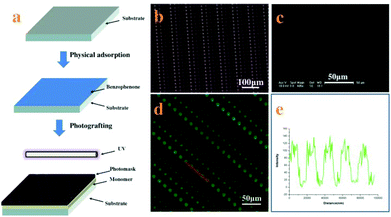
Precise patterning of the SEBS surface by UV lithography to evaluate the platelet function through single platelet adhesion†
Wei
Ye
a,
Qiang
Shi
*a,
Shing-Chung
Wong
b,
Jianwen
Hou
a,
Xiaodong
Xu
c and
Jinghua
Yin
*a
aPolymer Physics and Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, 130022, China. E-mail: shiqiang@ciac.ac.cn; yinjh@ciac.ac.cn
bDepartment of Mechanical Engineering, University of Akron, Akron, Ohio 44325-3903, USA
cPolymer Materials Research Center and Key Laboratory of Superlight Materials and Surface Technology, Ministry of Education, College of Materials Science and Chemical Engineering, Harbin Engineering University, Harbin 150001, China
First published on 30th April 2014
Abstract
Platelets have exhibited capabilities beyond clotting in recent years. Most of their functions are related to the nature of platelet adhesion. Establishing a facile method to understand the platelet adhesion and assess the platelet function through the mechanism and mechanics of adhesion is highly desired. Here, we report a generally applicable UV lithography technique with a photomask, which performs selective surface functionalization on large substrate areas, for creating stable, physical adhesive sites in the range of 12 μm to 3 μm. Our study demonstrated that the patterned surface facilitated probing of single platelet adhesion in a quantitative manner, and rendered platelets sensitive to adhesive proteins even at a low protein concentration. In addition, the platelet function in the presence of antiplatelet (anticancer) agents on platelets could be accurately estimated based on single platelet adhesion (SPA). This work paves a new way to understand and assess the blood platelet function. The SPA assay methodology has the potential to enable a rapid, accurate point-of-care platform suitable for evaluation of platelet function, detection of dysfunctional platelets, and assay of drug effects on platelets in cancer patients.
1. Introduction
Platelets have exhibited capabilities no one imagined they had in recent years.1 These small anucleate cells that circulate in the blood of mammals are not only critical effectors of hemostasis, blood clotting and wound repair, but also defenders against microbes2 and engineers which shape the vascular system in newborns. Yet for all their benefits, platelets can lodge in blood vessels, depriving tissue of oxygen and instigating a stroke and heart attack; in addition, they can foster cancer, rheumatoid arthritis, and other diseases.3 A platelet biochemical defect,4 poor platelet quality5 or the presence of antiplatelet agents6,7 can reduce or abolish platelet function. Understanding the mechanisms that drive platelet function and assessing the platelet function effectively are therefore important in both scientific and clinical areas. Platelets are highly adhesive in nature and almost all functions of blood platelets are related to platelet adhesive properties.7–13 It is our goal to quantitatively understand and evaluate platelet functions through single platelet adhesion on substrates.Micropatterning provides a powerful tool to create and model cues on soft materials, which define the microenvironment of platelets in spatially confined areas.8,9 A common approach is to create a passive surface and then introduce a pattern of proteins on the substrate which allows platelets to adhere to the pattern according to their natural preferences.14 Polymers, such as polyethylene glycol (PEG) and polymer of 2-methacryloyloxyethyl phosphorylcholine (MPC), and bioactive ligands, such as fibrinogen and some related synthetic peptides, are used extensively.14–19 However, evaluation of the platelet function on a micropatterned surface is not successful due to multiple steps for surface patterning and failure to probe adhesive behavior in a quantitative manner.8,15 In addition, the influences of microenvironmental geometry on the platelet function are often misinterpreted when patterned proteins are used to control platelet adhesion.17 Therefore, development of a facile method to pattern the surface, which can control single platelet adhesion with the guidance of physical properties and evaluate the platelet function in a quantitative manner, will facilitate establishing the adhesion–function relationship of platelets and find potential applications in clinical diagnostics of platelet-related disease and point-of-care testing.18–20
Here we present a simple method of surface patterning with conventional UV illumination and a photomask to create stable and physical adhesive sites with diameters from 12 μm to 3 μm over a large area. Our strategy is based on a controlled polymerization of MPC on the whole surface of a substrate and degradation of the obtained polymer of MPC in UV-exposed domains (adhesive sites) with UV irradiation in one step.15 The styrene-b-(ethylene-co-butylene)-b-styrene elastomer (SEBS) is used as a substrate due to its unique nanostructure, good biocompatibility and outstanding stability under physiological conditions.21 The single platelet adhesion on the patterned surface is studied in a quantitative manner and the platelet function in the presence of antiplatelet (anti-cancer) agents is accurately evaluated based on the single platelet adhesion assay. We demonstrate that single platelet adhesion on the patterned surface occurs in a controlled manner following the steps of (i) initiation, (ii) spreading and (iii) stabilization. The initiation is a rate-determining step in a confined environment. Patterned surfaces make platelets sensitive to adhesive proteins and enable assessment of the platelet function through single platelet adhesion. Our work paves a new way to understand and evaluate the platelet function both in research and clinical diagnostics. The single platelet adhesion assay methodology has the potential to enable a rapid, accurate point-of-care platform suitable for evaluation of the platelet function, detection of dysfunctional platelets and administration of antiplatelet agents.
2. Materials and methods
2.1. Chemicals and materials
SEBS copolymer with 29 wt% styrene (Kraton G 1652) was purchased from Shell Chemicals. Benzophenone (BP) was supplied by Peking Ruichen Chemical (China). 2-Methacryloyloxyethyl phosphorylcholine (MPC) was purchased from Nanjing Letianran Science and Technology Research Center, China. Fluorescein isothiocyanate-labeled bovine serum fibrinogen (BFg) and Fluoresce in isothiocyanate labeled phalloidine were purchased from Sigma-Aldrich. Phosphate-buffered saline (PBS 0.9% NaCl, 0.01 M phosphate buffer, pH 7.4) used for the platelet adhesion experiment was prepared freshly. ABT-737 was purchased from Selleck Chemicals. Other reagents were of AR grade and used without further purification.2.2. Preparation of SEBS films
SEBS was dissolved in xylene to form 15% (w/w) solutions and poured onto a clean glass. After the solvent evaporated, smooth SEBS films (0.2 mm thick) were obtained. The SEBS films were cut into strips of 2 cm × 1.5 cm and ultrasonically washed with deionized water and ethanol for 30 min, and then placed in a vacuum oven for 24 h to dry.2.3. Surface micro-patterning
The SEBS films were immersed in an ethanol solution of BP (1.5 wt%) for 30 min and dried at room temperature. Then the film was placed on the quartz plate (3 mm thick) and coated with 5 wt% aqueous solution of MPC; the pre-designed chrome photo-mask was placed on the SEBS surface directly. Then the sandwiched system (shown in Fig. 1a) was exposed to UV light (high-pressure mercury lamp, 400 W, main wavelength 380 nm) for 10 min. All the films were washed with deionized water and ethanol to remove the residual monomer, followed by drying in a vacuum oven for at least 24 h.2.4. Platelet adhesion assay
Fresh blood collected from a healthy rabbit was immediately mixed with 3.8 wt% sodium citrate solution at a dilution ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. (The experiments were carried out in accordance with the guidelines issued by the Ethical Committee of the Chinese Academy of Sciences.) Platelet rich plasma (PRP) was obtained from the supernatant after centrifugation of whole blood at 1000 rpm for 15 min. The SEBS films were immersed in PBS (pH 7.4) at 37 °C for 2 h to equilibrate the surfaces. Then PRP was deposited onto each sample and allowed to adhere for a given time at 37 °C. The non-adherent platelets were rinsed away with PBS several times. The samples were fixed with a freshly prepared solution of 2.5 wt% glutaraldehyde in PBS at 37 °C for at least 2 h. After rinsing with PBS, the adherent platelets were dehydrated in ascending ethanol–water mixtures (10, 30, 50, 70, 90 and 100%) for 30 min in each step, dried under vacuum and finally sputter-coated with gold. Platelet adhesion was characterized by field-emission scanning electron microscopy (SEM, Sirion-100, FEI, USA). Platelets were stained for F-actin with fluorescein isothiocyanate labeled phalloidine, and then characterized using a confocal laser scanning microscope (Zeiss LSM 710 confocal microscope).
1. (The experiments were carried out in accordance with the guidelines issued by the Ethical Committee of the Chinese Academy of Sciences.) Platelet rich plasma (PRP) was obtained from the supernatant after centrifugation of whole blood at 1000 rpm for 15 min. The SEBS films were immersed in PBS (pH 7.4) at 37 °C for 2 h to equilibrate the surfaces. Then PRP was deposited onto each sample and allowed to adhere for a given time at 37 °C. The non-adherent platelets were rinsed away with PBS several times. The samples were fixed with a freshly prepared solution of 2.5 wt% glutaraldehyde in PBS at 37 °C for at least 2 h. After rinsing with PBS, the adherent platelets were dehydrated in ascending ethanol–water mixtures (10, 30, 50, 70, 90 and 100%) for 30 min in each step, dried under vacuum and finally sputter-coated with gold. Platelet adhesion was characterized by field-emission scanning electron microscopy (SEM, Sirion-100, FEI, USA). Platelets were stained for F-actin with fluorescein isothiocyanate labeled phalloidine, and then characterized using a confocal laser scanning microscope (Zeiss LSM 710 confocal microscope).
2.5. Protein adsorption
Fluoresce in isothiocyanate-labeled bovine serum fibrinogen (FITC-labeled BFg) was dissolved in PBS (pH 7.4) at a concentration of 0.1 mg ml−1. Then BFg was deposited onto each sample and incubated for 1.5 h at room temperature in a dark environment to allow fibrinogen to adsorb onto the virgin and patterned SEBS surface. Following protein incubation, the films were rinsed with PBS and deionized water to remove any weakly adsorbed protein. Fibrinogen, fibronectin and collagen (rat tail tendon collagen type I) were pre-adsorbed onto patterned SEBS films at a concentration of 0.1 mg ml−1. Washed rabbit platelets with the concentration of 2 × 106 per ml were seeded on the pre-adsorbed protein films at 37 °C for 15 min, 20 min, 30 min, 60 min and 90 min, respectively. After fixing with 2.5 wt% glutaraldehyde and dehydrating in ethanol-water mixtures with varied ethanol contents (10, 30, 50, 70, 90 and 100%), platelet was visualized by field-emission scanning electron microscopy (FESEM).2.6. Anti-cancer agents
Bcl-2 homology 3 (BH3) proteins play an important role in inducing apoptosis by inhibiting the function of Bcl-2 family proteins. In the experiment with the patterned SEBS surface, ABT-737 as a typical BH3 mimetic was added to the platelet rich plasma (PRP). ABT-737 solution (100 μg ml−1 in DMSO) was diluted to 1.5, 3, and 6 μg ml−1 in PBS. PRP was pretreated for 1 or 2 hours at 37 °C with ABT-737. The platelet adhesion assay was performed according to the procedure described above.2.7. Statistics
The analysis involved both counting of the adhered platelets (∼0.01 mm2 per SEM image) and the analysis of platelet morphology using the free software Image-J. The data from multiple separate experiments were analyzed and reported as the mean ± standard error (SE) of the mean. The data were analyzed by the one-way ANOVA method; the statistical significance was accepted when p < 0.05.3. Results and discussion
3.1. Fabrication of the patterned SEBS surface
Micropatterned surfaces of SEBS are fabricated by UV illumination with a photomask. The process to pattern the surface of SEBS is shown in Fig. 1a. The SEBS films first adsorb benzophenone (BP) physically, followed by coating with an MPC aqueous solution; then the photomask is put on the SEBS surface directly and irradiated with UV for 10 min. Based on grafting polymerization of MPC on the whole surface of SEBS and subsequent degradation of PMPC on UV-exposed domains through the photomask, platelet adhesive sites are created on these areas, while UV-unexposed domains are hydrophilic and resist platelet adhesion.15 The resulting micro-patterned surface is characterized by SEM. Fig. 1c shows that the structure of the photomask (Fig. 1b) is well duplicated on the SEBS surface. The array of adhesive sites is clearly observed and the diameters of adhesive sites are approximately 12, 9, 6, and 3 μm, respectively. As the adhesive sites are more hydrophobic than other places on the patterned surface, BFg can be easily adsorbed at the adhesive sites and is used to further characterize the structure of the SEBS surface.22Fig. 1d shows the fluorescence image of BFg adsorbed on the patterned surface; the array of blue spots indicates the perfect structure of adhesive sites. The line intensity of the fluorescence scan (Fig. 1f) confirms that the space between the adhesive sites is in a well-controlled manner.23 These results demonstrate that this method can create uniform and fine adhesive sites on a patterned structure.Obviously, the patterned surface fabricated in this way has many advantages for studying platelet adhesion and evaluating its function: (i) it provides an unique platform to probe single platelet adhesion induced by physical properties of the surface; (ii) the size of adhesive sites ranges from 12 μm to 3 μm, which enables quantitatively investigating the single platelet adhesive behavior, platelet–substrate and platelet–platelet interaction in a confined microenvironment; (iii) the platelet function can be accurately assessed based on a single platelet adhesion assay.
3.2. Single platelet adhesion on the patterned surface
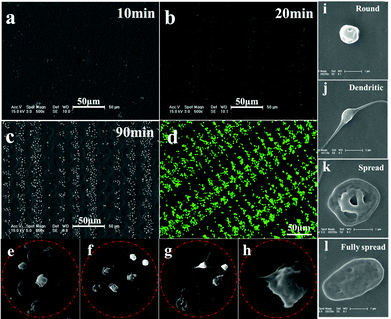
To probe adhesive behavior of single platelet on the patterned SEBS substrate, PRP is incubated on the patterned surface for a specific time at 37 °C, and then fixed with 2.5 wt% glutaraldehyde. The SEM images of platelet adhesion on the patterned surface of SEBS are shown in Fig. 2.The lack of hydrophilicity renders the platelets adherent and spread selectively on adhesive sites.15,18 After 10 min of incubation, only a few single platelet attach onto the 9 and 12 μm adhesive sites, while no adherent platelets are observed at 6 and 3 μm adhesive sites (Fig. 2a). After 20 min incubation, some adherent platelets appear at 6 and 3 μm adhesive sites (Fig. 2b). After 30 min incubation, more platelets adhere to the adhesive sites and an array of adherent platelets emerges (Fig. 2c). The array of platelets at 120 min is clearly observed by confocal laser scanning microscopy (Fig. 2d). Fig. 2e–2h show the morphology of adherent platelets at varied adhesive sites. The round shape is dominant at 3 μm sites; round shaped, dendritic and fully spread platelets are found at 6, 9 and 12 μm adhesive sites, respectively. The fully spread adherent platelets are highest at 12 μm adhesive sites. The morphology change is controlled by actin polymerization of platelets.13 The limited space may inhibit the polymerization of actin, resulting in a round shape at 3 μm adhesive sites. In contrast, relatively large room allows the polymerization of actin to form dendritic and fully spread platelets at 12 μm adhesive sites; as a result, fully spread platelets are dominant at 12 μm adhesive sites. The round shape of platelets appearing at the adhesive sites indicates that these platelets remain inactive, which is in agreement with the results obtained by Ruggeri et al.24 In addition, the size of adhesive sites determines the maximal number of adherent platelets. For example, 1 adherent platelet in 3 μm adhesive sites, 2–3 platelets in 6 μm sites, 4–5 platelets in 9 μm sites, and 5–6 platelets in 12 μm sites. The morphology and number of platelets at varied adhesive sites indicate the geometric confinement on platelet adhesion.
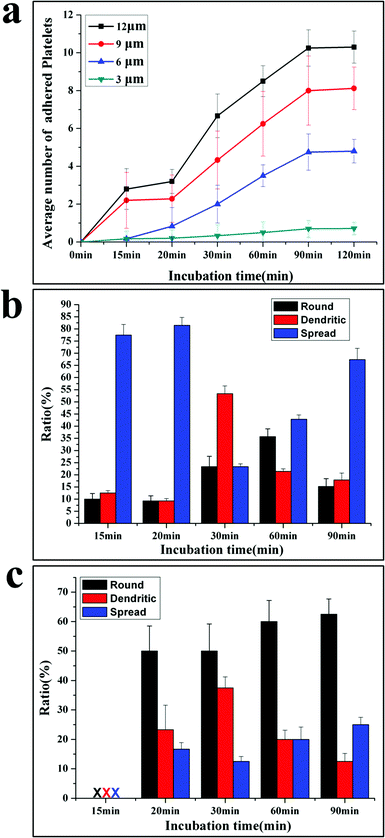
Based on the SEM picture, the time dependent platelet adhesion is plotted in Fig. 3a. The adhesive behaviours are different at varied adhesive sites. The average number (AN) of platelets at 3 μm sites increases linearly with time from 0.2 at 10 min to nearly 1 at 90 min. The AN of platelets adherent at 6, 9 and 12 μm sites first increases with the incubation time, then reaches the plateau at 90 min and does not increase any more, exhibiting three stages including the initiation, extension and stabilization (or termination) of adhesion, in agreement with the well-established mechanism of platelet adhesion and activation.10
Interestingly, acceleration occurs in the extension stage of platelets adherent at 6, 9 and 12 μm sites after 30 min incubation, showing that the platelet–platelet interactions speed up the adhesion process. After reaching the maximal number of adherent platelets, no more platelets can attach to the adhesive sites, indicating that the termination of adhesion is controlled by the size of adhesive sites.25 This is a very important function for normal platelets, since the extent of the platelet response to injury is subject to tight regulation.12 Appropriate platelet activation limits the extent of blood loss following vascular injury and promotes subsequent wound healing without causing vascular occlusion.26 This work shows that the geometric confinement may be one factor to control the termination of thrombus. The time evolution of morphology for adherent platelets at 12 μm and 3 μm adhesive sites is shown in Fig. 3b and 3c, respectively. At 12 μm adhesive sites, the attached platelets are in fully spread shape after 15–20 min incubation, and then some platelets extend with a dendritic shape, resulting in a high ratio of the dendritic shape. As newly adherent platelets become stable with a fully spread shape, the fully spread shape is dominant again at 90 min incubation (Fig. 3c).
At 3 μm adhesive sites, no platelets are detected after 15 min incubation; a few platelets with a round shape appear at 20 min. Then some adherent platelets begin to form dendritic, lowering the ratio of the round shape. But the round shape is dominant until 90 min incubation (Fig. 3b).
The morphology change shows the importance of stable adhesion of single platelet in initiating the adhesion.27 The platelets first attach at adhesive sites and need to stabilize their adhesion by increasing adhesive forces. For the platelets at 12 μm adhesive sites, they can form protrusions and then contract into a more compact structure (fully spread) to increase the attachment area, but at 3 μm adhesive sites platelets cannot extend and contract due to the limited space, they stabilize the adhesion only by increasing the attachment time.28 Thus, after 15 min incubation, platelets adhere at 12 μm adhesive sites with a fully spread shape, while after 20 min incubation, platelets appear at 3 μm adhesive sites with a round shape. The morphology change of adherent platelets at 3 and 12 μm adhesive sites confirms the three-stage adhesion under the confinement10 and the initiation of adhesion is the rate-determining step for the adhesion process.
The patterned surfaces facilitate probing of platelet adhesive behaviour in a quantitative manner. The data for adhesion induced by physical properties are listed in Table 1. Table 1 shows that the initial rate of adhesion (the slope of the adhesion curve before 15 min incubation), the rate of spread (average rate from 15 min to 90 min), the ratio of the fully spread shape and the maximal number of adherent platelets are proportional to the size of adhesive sites. The initial rate is slower than the spreading rate at each adhesive site, confirming that initiation is a rate-determining step in the confined environment. The data for adhesion at 3 μm and 12 μm adhesive sites are especially useful to shed light on the feature of adhesion. The 3 μm adhesive site is comparable to the size of single platelets, geometric confinements make it difficult for platelets to approach and attach onto this site, and only the most active platelets can reach and form stable adhesion at 3 μm adhesive sites through receptor–ligand reaction. The initial adhesive rate is a good indicator to assess the activity of platelets and the reactivity of platelets and the substrate.8 For the 12 μm adhesive site, the number of platelets initially attached on the adhesive site is more than other sites, and these adherent platelets can release a number of biologically active substances and extend into the dendritic shape to induce adjacent platelet adhesion, resulting in the acceleration of the adhesion process.7 Therefore, the rate of acceleration, the shape change and the maximal number of adherent platelets at 12 μm adhesive sites are indicators of platelet activation and platelet–platelet interaction.28 The data of adhesion at 3 and 12 μm adhesive sites provide the standard to evaluate the platelet function.
| Adhesive sites | Adhesion rate number/min | The ratio of fully spread platelets/% | Maximum number of platelets | |
|---|---|---|---|---|
| V 0 (initial rate) | V s (rate of spreading) | |||
| 3 μm | 0.02 | 0.01 | 25 | 0.7 ± 0.4 |
| 6 μm | 0.04 | 0.12 | 30 | 4.8 ± 1 |
| 9 μm | 0.12 | 0.20 | 48 | 8.0 ± 1.8 |
| 12 μm | 0.16 | 0.37 | 70 | 10.2 ± 1 |
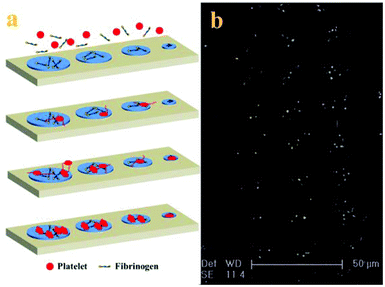
With the adhesive behaviour and morphology change of adherent platelets, the mechanism of platelet adhesion and activation on the patterned surface is tentatively proposed (shown in Fig. 4a). Because PRP usually contains about 2–3 mg ml−1 fibrinogen,29 when PRP comes into contact with the patterned surface, the fibrinogen first adsorbs onto the adhesive sites to mediate the platelet adhesion by binding with the receptor on the platelet membrane and then arrests the platelets at the adhesive sites.30 Platelets preferentially attach to large adhesive sites, but need a long time to adhere to small adhesive sites due to the confinement. The adherent platelets become stable and release a number of biologically active substances upon activation such as α-granules which contained vWF, coagulation factor V, and fibronectin to induce adjacent platelet adhesion, resulting in the acceleration of the adhesion process.7 At the same time, some platelets begin to form pseudopodia to probe and sense the geometry of their microenvironment, inducing adjacent platelet adhesion to the adhesive sites.31 Intracellular signalling downstream of agonist receptors activates integrin αIIbβ3 (GPIIb/IIIa), making cohesive interactions between platelets possible. The platelets not only use the protrusions to enable more physical connections with other platelets at the adhesive sites but also use the actin–myosin interactions to pull the adherent platelets into a more compact structure (fully spread).32 The number and morphology of adherent platelets at the final stage depend on the surface properties and the size of adhesive sites. The mechanism of platelet adhesion in the confined environment is confirmed by the morphology of adherent platelets on the patterned surface after 90 min incubation (Fig. 4b). It has been proposed that in the process platelets not only provide a surface that can facilitate leukocyte immigration into surrounding tissue but also serve as a source of inflammatory mediators and molecules that promote wound healing.1 Although platelet adhesion to the injured sites is thought to be spatial and temporal,10 platelet adhesion to the adhesive sites tends to happen orderly in a series of events. This difference may be due to the low concentration of platelets used in the experiment28 and confinement on the patterned surface.25,27 The controlled platelet adhesion facilitates the study of the characteristics of adhesive behaviours to evaluate the platelet function.
3.3. Sensitivity of single platelet adhesion to adhesive proteins
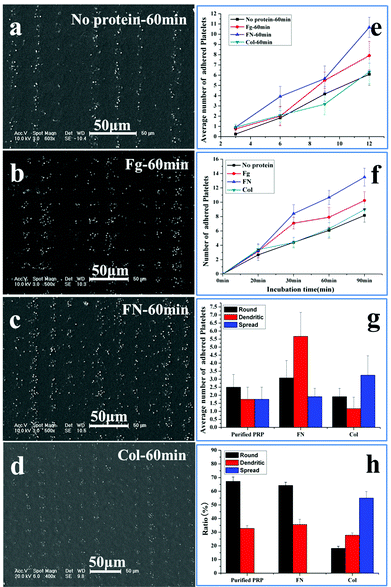
The sensitivity of platelet adhesive behaviour to adhesive proteins is a key standard to evaluate the accuracy of the patterned surface in assessing the platelet function.33–36 Fibrinogen, fibronectin and collagen are pre-adsorbed at adhesive sites to induce platelet adhesion. Fibrinogen is the major plasma protein, which is made up of three globular units connected by two rods. Its γ chain C-terminal dodecapeptide is the most important site in mediating platelet adhesion and aggregation. Fibronectin is present in plasma, the subendothelium of the vessel wall, and the α granules of platelets. It has similar binding sites to fibrinogen to the glycoprotein (GP) IIb–IIIa receptor (αIIbβ3) on the membrane of platelets.34 Collagens are among the major constituents that determine the thrombogenicity of the vessel wall. The interaction of collagen with platelet membrane receptors GP Ia–IIa and GPVI is an important step in their adhesion to the subendothelium.35 All the three proteins contribute to the platelet adhesion and aggregation on the substrate, but fibrinogen and fibronectin tend to support both platelet–surface and platelet–platelet interactions,6,34 while collagen shows the tendency to stabilize the adhesion.36Fig. 5 shows the SEM images of adhesion of the washed PRP on the patterned surface with adhesive proteins’ pre-adsorption after 60 min incubation. To decrease the effect of fibrinogen in plasma on adhesion, the PRP is washed with PBS; as a result, the concentration of fibrinogen is decreased by 92.8 ± 0.3% (ESI†). The density of adsorbed proteins on the adhesive site is measured by the bicinchoninic acid (BCA) protein assay. The amount of fibrinogen, fibronectin, and collagen adsorbed are 6.83 ± 0.30 μg cm−2, 4.38 ± 0.18 μg cm−2, and 1.09 ± 0.02 μg cm−2, respectively, similar to the results obtained by Latour et al.30 Compared with the controlled surface without protein adsorption (Fig. 5a), more platelets adhere on the surface with fibrinogen (Fig. 5b) and fibronectin pre-adsorption (Fig. 5c), but with a slight increase in the number of adherent platelets on the surface with collagen pre-adsorption (Fig. 5d). Furthermore, the size of adherent platelets on the surfaces with fibrinogen and fibronectin adsorption is much larger than that on the controlled surface and the surface with collagen adsorption, indicating that the adherent platelets tend to aggregate in the presence of fibrinogen and fibronectin.34
The number of washed platelets at varied adhesive sites is shown in Fig. 5e; at each site, protein adsorption increases the number of adherent platelets, except for collagen at the 9 μm site. This indicates that collagen has a slight effect on the platelet spreading.36 The time dependent occupation of adherent platelets at the 12 μm site is shown in Fig. 5f. The initial rate of adhesion with protein adsorption is similar (∼0.2 min−1), which is double of that without protein adsorption (0.1 min−1). The spreading rate without protein adsorption is shown to be about 0.2 min−1, but the rates are 0.1, 0.3 and 0.5 min−1 for collagen, fibrinogen and fibronectin, respectively. The obvious acceleration induced by fibrinogen and fibronectin at the spreading stage is due to the increased platelet–platelet interactions.28 Compared with the maximal number of adherent platelets without proteins (8.1), the number increases in the order of fibronectin (13.5), fibrinogen (10.3) and collagen (8.3). Fig. 5f shows that collagen has a slight effect on the platelet spreading while fibronectin and fibrinogen have a similar effect on the platelet adhesion and spreading.34 In the following experiments, only fibronectin is used for comparison.
The morphological changes at 12 μm adhesive sites after 60 min incubation are shown in Fig. 5g, without protein adsorption, the round shaped platelets are slightly higher in number than dendritic and fully spread shaped platelets; when the adhesive sites are adsorbed with fibronectin, the dendritic shape becomes much higher in number than the round and fully spread shape; however, when these sites are adsorbed with collagen, the fully spread shape becomes the highest in number. As round, dendritic and fully spread shapes change in order, the high ratio of dendritic shape indicates that fibronectin can support more interactions between platelets to form the linkers, while the high ratio of fully spread shape suggests that collagen tends to contract the platelets into a compact structure to stabilize the adhesion.36
The role of collagen in platelet adhesion is further confirmed by the morphology of platelets at 3 μm adhesive sites (Fig. 5h). No fully spread platelets are detected on the sites with fibronectin and without proteins due to the confinement. In contrast, the sites with collagen possess a high ratio of fully spread platelets, confirming the ability of collagen in stabilizing adhesion in spite of limited space. Our work shows that the response of platelets to adhesive proteins of low concentration (0.1 mg ml−1) can be clearly detected. The dominant roles of adhesive proteins in platelet adhesion can thus be distinguished based on the adhesive behaviour on the patterned surface, demonstrating that evaluation of the platelet function with adhesive behaviour is practical and accurate.
3.4. Evaluation of the platelet function based on single platelet adhesion
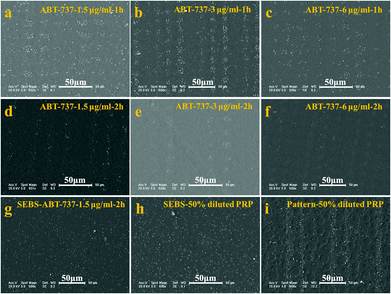
The antiplatelet and anticancer agents that are widely used to prevent cardiovascular events and tumours will inevitably affect the platelet function.11,37 As a consequence, evaluation of the platelet function is clinically important to test the toxicity of new agents towards platelets. BH3 mimetics are a new class of proapoptotic anticancer agents that have shown considerable promise in preclinical animal models and early-stage human trials.11 These agents act by inhibiting the prosurvival function of one or more Bcl-2-related proteins. It is well established that these agents can inhibit Bcl-xL induced rapid platelet death, resulting in thrombocytopenia. However, their impact on the function of residual circulating platelets remains an issue of debate.38–40 Some researchers reported that these agents, which perturb the normal function of the mitochondria, led to phosphatidylserine (PS) exposure and a procoagulant phenotype,38 indicating that BH3 mimetics may promote blood coagulation and thrombin generation in vivo.39 In contrast, other work showed that BH3 mimetics induced a transient thrombocytopathy that undermined the hemostatic function of platelets.37,40 To clarify the effect of BH3 mimetics on the function of platelets, the adhesive behaviour of platelets treated with ABT-737 on the patterned surface is studied.The SEM images of platelets adherent on the patterned surface are shown in Fig. 6. As shown in Fig. 6a–6f, not only the number of adherent platelets decreases with the increasing treatment time and the increasing dose of ABT-737, but also the pattern of platelets becomes undetectable with high dose and long treatment (e.g. 3 μg ml−1, 2 h and 6 μg ml−1, 2 h). These results confirm that ABT-737 reduces the platelet concentration in a dose- and time-dependent manner,11,37 but the effect of ABT-737 on the platelet function remains unclear because the decrease of platelet concentration leads to the similar incompetence of the platelet pattern.
According to the number of adherent platelets on the blank surface without a pattern, the concentration of live platelets is estimated to be about 50% concentration of PRP after PRP is treated with 1.5 μg ml−1 ABT-737 for 2 h (ESI, Fig. S5†). The adhesion of treated platelets (1.5 μg ml−1, 2 h) and diluted PRP (50%) is thus performed on a blank film and patterned surface, respectively, to clarify the effect of ABT-737 on the platelet function.
Slight differences are observed in morphology and adherent number between adhesion on the blank surfaces with treated PRP (Fig. 6g) and with dilute PRP (Fig. 6h). In contrast, the obvious distinctions in the platelet pattern on the patterned surfaces treated with PRP (Fig. 6d) and with diluted PRP (Fig. 6i) are detected, and the pattern of adherent platelets of diluted PRP is much clearer than the pattern of treated platelets, showing the advantage of the patterned surface in detection.15 The average numbers of adherent platelets treated with ABT-737 at 3 and 12 μm are about 1/2 and 1/3 of the number of diluted platelets adherent at 3 and 12 μm sites, respectively, confirming that ABT-737 induces not only decreased platelet number (thrombocytopenia) but also reduced adhesive ability.37 Based on the single platelet adhesion on the patterned surface, our research successfully assesses the platelet function in the presence of antiplatelet agents, which provides a new avenue for accurate detection of dysfunctional platelets and drug effects in cancer patients.41
4. Conclusions
In this work, we have developed a facile and stable UV lithography technique combined with a photomask to create physical adhesive sites in the range of 12 μm to 3 μm on the patterned surface of SEBS. With the simplicity and effectiveness of the patterned surface, single platelet adhesion was controlled and quantitatively probed. The patterned surface rendered platelets sensitive to adhesive proteins and enabled assessment of the platelet function through single platelet adhesion. Our work paves a new way to understand and evaluate the platelet function both in research and clinical diagnostics. The methodology has the potential to enable a rapid, accurate point-of-care platform suitable for evaluation of platelet function, detection of dysfunctional platelets and administration of antiplatelet agents.Acknowledgements
The authors acknowledge the financial support from the National Natural Science Foundation of China (Project no. 51273199, 21274150 and 51103030).Notes and references
- M. Leslie, Science, 2010, 328, 562–564 CrossRef CAS PubMed.
- C. R. Engwerda and M. F. Good, Science, 2012, 338, 1304–1305 CrossRef PubMed.
- G. A. Zimmerman and A. S. Weyrich, Science, 2010, 327, 528–529 CrossRef CAS PubMed.
- S. Vaiyapuri, L. A. Moraes, T. Sage, M. S. Ali, K. R. Lewis, M. P. Mahaut-Smith, E. Oviedo-Orta, A. M. Simon and J. M. Gibbins, Nat. Commun., 2013, 4 Search PubMed.
- K. M. Hoffmeister, E. C. Josefsson, N. A. Isaac, H. Clausen, J. H. Hartwig and T. P. Stossel, Science, 2003, 301, 1531–1534 CrossRef CAS PubMed.
- K. D. Mason, M. R. Carpinelli, J. I. Fletcher, J. E. Collinge, A. A. Hilton, S. Ellis, P. N. Kelly, P. G. Ekert, D. Metcalf and A. W. Roberts, Cell, 2007, 128, 1173–1186 CrossRef CAS PubMed.
- A. Lopez-Alonso, B. Jose, M. Somers, K. Egan, D. P. Foley, A. J. Ricco, S. Ramström, L. Basabe-Desmonts and D. Kenny, Anal. Chem., 2013, 85, 6497–6504 CrossRef CAS PubMed.
- Z. M. Ruggeri and G. L. Mendolicchio, Circ. Res., 2007, 100, 1673–1685 CrossRef CAS PubMed.
- T. Ekblad, L. Faxälv, O. Andersson, N. Wallmark, A. Larsson, T. L. Lindahl and B. Liedberg, Adv. Funct. Mater., 2010, 20, 2396–2403 CrossRef CAS.
- L. Brass, Am. Soc. Hematol. Educ. Program, 2010, 2010, 387–396 CrossRef PubMed.
- B. Savage, E. Saldívar and Z. M. Ruggeri, Cell, 1996, 84, 289–297 CrossRef CAS.
- L. Brass, in Platelets, ed. A. Michelson, Academic press, London, 2007, pp. 367–398 Search PubMed.
- Z. Li, E. S. Kim and E. L. Bearer, Blood, 2002, 99, 4466–4474 CrossRef CAS PubMed.
- L. Basabe-Desmonts, S. Ramstrom, G. Meade, S. O'neill, A. Riaz, L. Lee, A. Ricco and D. Kenny, Langmuir, 2010, 26, 14700–14706 CrossRef CAS PubMed.
- W. Ye, Q. Shi, S. C. Wong, J. Hou, H. Shi and J. Yin, Macromol. Biosci., 2013, 13, 676–681 CrossRef CAS PubMed.
- C. Choi, I. Hwang, Y.-L. Cho, S. Y. Han, D. H. Jo, D. Jung, D. W. Moon, E. J. Kim, C. S. Jeon and J. H. Kim, ACS Appl. Mater. Interfaces, 2013, 5, 697–702 CAS.
- A. Kita, Y. Sakurai, D. R. Myers, R. Rounsevell, J. N. Huang, T. J. Seok, K. Yu, M. C. Wu, D. A. Fletcher and W. A. Lam, PLoS One, 2011, 6, e26437 CAS.
- X. Zhu, D. Jańczewski, S. S. C. Lee, S. L. Teo and G. J. Vancso, ACS Appl. Mater. Interfaces, 2013, 5, 5961–5968 CAS.
- L. E. Corum, C. D. Eichinger, T. W. Hsiao and V. Hlady, Langmuir, 2011, 27, 8316–8322 CrossRef CAS PubMed.
- M. J. Price, S. Endemann, R. R. Gollapudi, R. Valencia, C. T. Stinis, J. P. Levisay, A. Ernst, N. S. Sawhney, R. A. Schatz and P. S. Teirstein, Eur. Heart J., 2008, 29, 992–1000 CrossRef PubMed.
- J. Hou, Q. Shi, P. Stagnaro, W. Ye, J. Jin, L. Conzatti and J. Yin, Colloids Surf., B, 2013, 111, 333–341 CrossRef CAS PubMed.
- T. Konno, H. Hasuda, K. Ishihara and Y. Ito, Biomaterials, 2005, 26, 1381–1388 CrossRef CAS PubMed.
- M. R. Lorenz, V. Holzapfel, A. Musyanovych, K. Nothelfer, P. Walther, H. Frank, K. Landfester, H. Schrezenmeier and V. Mailänder, Biomaterials, 2006, 27, 2820–2828 CrossRef CAS PubMed.
- Z. M. Ruggeri, J. N. Orje, R. Habermann, A. B. Federici and A. J. Reininger, Blood, 2006, 108, 1903–1910 CrossRef CAS PubMed.
- C. S. Chen, M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, Science, 1997, 276, 1425–1428 CrossRef CAS.
- J. Zhao, L. Song, Q. Shi, S. Luan and J. Yin, ACS Appl. Mater. Interfaces, 2013, 5, 5260–5268 CAS.
- C. Yan, J. Sun and J. Ding, Biomaterials, 2011, 32, 3931–3938 CrossRef CAS PubMed.
- M. J. Maxwell, E. Westein, W. S. Nesbitt, S. Giuliano, S. M. Dopheide and S. P. Jackson, Blood, 2007, 109, 566–576 CrossRef CAS PubMed.
- C. H. Jo, J. E. Kim, K. S. Yoon and S. Shin, Am. J. Sports Med., 2012, 40, 1035–1045 CrossRef PubMed.
- B. Sivaraman and R. A. Latour, Biomaterials, 2010, 31, 832–839 CrossRef CAS PubMed.
- P. K. Mattila and P. Lappalainen, Nat. Rev. Mol. Cell Biol., 2008, 9, 446–454 CrossRef CAS PubMed.
- W. A. Lam, O. Chaudhuri, A. Crow, K. D. Webster, A. Kita, J. Huang and D. A. Fletcher, Nat. Mater., 2010, 10, 61–66 CrossRef PubMed.
- S. Beumer, M. IJsseldijk, P. G. de Groot and J. J. Sixma, Blood, 1994, 84, 3724–3733 CAS.
- T. Zaidi, L. McIntire, D. Farrell and P. Thiagarajan, Blood, 1996, 88, 2967–2972 CAS.
- E. Saelman, H. K. Nieuwenhuis, K. M. Hese, P. G. de Groot, H. Heijnen, E. Sage, S. Williams, L. McKeown, H. Gralnick and J. Sixma, Blood, 1994, 83, 1244–1250 CAS.
- B. Nieswandt and S. P. Watson, Blood, 2003, 102, 449–461 CrossRef CAS PubMed.
- S. M. Schoenwaelder, K. E. Jarman, E. E. Gardiner, J. Qiao, M. J. White, E. C. Josefsson, I. Alwis, A. Ono, A. Willcox and R. K. Andrews, Blood, 2011, 118, 1663–1674 CrossRef CAS PubMed.
- G. Dale, J. Thromb. Haemost., 2005, 3, 2185–2192 CrossRef CAS PubMed.
- S. M. Schoenwaelder, Y. Yuan, E. C. Josefsson, M. J. White, Y. Yao, K. D. Mason, L. A. O'Reilly, K. J. Henley, A. Ono and S. Hsiao, Blood, 2009, 114, 663–666 CrossRef CAS PubMed.
- S. M. Schoenwaelder and S. P. Jackson, Blood, 2012, 119, 1320–1321 CrossRef CAS PubMed.
- L. J. Gay and B. Felding-Habermann, Nat. Rev. Cancer, 2011, 11, 123–134 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4bm00072b |
| This journal is © The Royal Society of Chemistry 2014 |