Interleukin-8 gene silencing on pancreatic cancer cells using biodegradable polymer nanoplexes
Guimiao
Lin†
ab,
Chengbin
Yang†
a,
Rui
Hu
a,
Chih-Kuang
Chen
c,
Wing-Cheung
Law
d,
Tommy
Anderson
a,
Butian
Zhang
a,
Quoc Toan
Nguyen
e,
Hui Ting
Toh
e,
Ho Sup
Yoon
e,
Chong
Cheng‡
c and
Ken-Tye
Yong
*a
aSchool of Electrical and Electronic Engineering, Nanyang Technological University, Singapore 639798, Singapore. E-mail: ktyong@ntu.edu.sg
bThe Engineering Lab of Synthetic Biology and the Key Lab of Biomedical Engineering, School of Medicine, Shenzhen University, Shenzhen 518060, China
cDepartment of Chemical and Biological Engineering, University at Buffalo, The State University of New York, Buffalo, New York 14260, USA
dDepartment of Industrial and Systems Engineering, The Hong Kong Polytechnic University, Hung Hom, Kowloon, Hong Kong, P.R. China
eDivision of Structural Biology & Biochemistry, School of Biological Sciences, Nanyang Technological University, Singapore 639798, Singapore
First published on 3rd March 2014
Abstract
Pancreatic cancer is one of the deadliest cancers throughout the world with rarely efficient therapies currently available. Gene therapy on pancreatic cancer through small interfering RNA (siRNA)-based RNA interference (RNAi) has shown great potential and attracted much attention. However, due to the fragile nature of nucleic acid, the application of RNAi as a safe and efficient carrier faces great challenges. In this contribution, a self-assembly regime, which is based on well-defined cationic polylactides (CPLAs) with tertiary amine groups, has been used to encapsulate and protect siRNAs from fast degradation. CPLA is a safe and degradable formulation that allowed us to deliver siRNAs targeting the proangiogenic chemokine interleukin-8 (IL-8) to pancreatic cancer cells for gene therapy. Stable IL-8 siRNA–CPLA nanoplexes were successfully formed by electrostatic force and high gene transfection efficiencies were shown on two pancreatic cancer cell lines. We did not observe any cytotoxicity from these CPLAs over a large concentration range via cell viability evaluations. More importantly, the silencing of IL-8 gene expression significantly attenuated the proliferation of pancreatic cancer cells. Our preliminary results support the future development of gene therapy that might provide an effective and safe treatment approach towards pancreatic cancer.
1. Introduction
Pancreatic cancer ranks as the fourth leading cause of cancer related death in the United States, with yearly incidents of about 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cases.1,2 Every year it causes more than 200
000 cases.1,2 Every year it causes more than 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 patients to die throughout the world.3,4 Traditional therapies such as surgery, radiation and chemotherapy play an important role in the treatment of patients with pancreatic cancer.5,6 Yet unfortunately, even with combined modality therapy, the survival rates for pancreatic cancer patients are still very low.7 With the development of molecular techniques, new therapeutic strategies have been proposed, among which gene therapy has caught great attention. Several gene therapy-based clinical trials using virus vectors for transfection have shown quite promising results. More recently, the discovery of RNA interference (RNAi) has made small interfering RNAs (siRNAs) another rising topic of focus. siRNAs are double-stranded RNA molecules with a typical length of around 21 base pairs. In the RNAi process, they mediate the post-transcriptional gene silencing of a target gene and regulate its expression. During the past decade, RNAi-based therapeutic strategies have been extensively explored for cancer treatments.8–12 The success of RNAi gene therapy is complex and relies on two key factors, namely, therapeutic efficacy and the delivery efficiency.
000 patients to die throughout the world.3,4 Traditional therapies such as surgery, radiation and chemotherapy play an important role in the treatment of patients with pancreatic cancer.5,6 Yet unfortunately, even with combined modality therapy, the survival rates for pancreatic cancer patients are still very low.7 With the development of molecular techniques, new therapeutic strategies have been proposed, among which gene therapy has caught great attention. Several gene therapy-based clinical trials using virus vectors for transfection have shown quite promising results. More recently, the discovery of RNA interference (RNAi) has made small interfering RNAs (siRNAs) another rising topic of focus. siRNAs are double-stranded RNA molecules with a typical length of around 21 base pairs. In the RNAi process, they mediate the post-transcriptional gene silencing of a target gene and regulate its expression. During the past decade, RNAi-based therapeutic strategies have been extensively explored for cancer treatments.8–12 The success of RNAi gene therapy is complex and relies on two key factors, namely, therapeutic efficacy and the delivery efficiency.
Interleukin-8 (IL-8), alternatively known as CXCL8, was originally identified as an 8 kDa proinflammatory CXC chemokine which is secreted by multiple cell types, including neutrophils, monocytes, and endothelial cells.13 It has since been demonstrated that IL-8 is produced in response to multiple stimuli and associated with recruiting neutrophils, basophils, and T cells during immune system activation. IL-8 exerts its biological effects by binding to two cell surface G protein-coupled receptors, CXCR1 and CXCR2, which are produced by macrophages and other cell types such as epithelial and endothelial cells.14,15 However, IL-8 was also found to be overexpressed in many tumors,16 such as ovarian epithelial tumors and17 lung cancer,18 and exert profound effects on the tumor microenvironment as a proangiogenic cytokine.16,19 Tumor-derived IL-8 activates endothelial cells in the tumor vasculature to accelerate angiogenesis and promote the proliferation, invasion, migration and survival of cancer cells through autocrine signalling pathways.16 The extensive effects of IL-8 activity on tumor progression and development make it an ideal therapeutic target in pancreatic cancer.16,20 Targeting of IL-8 signalling may have great implications in the halting of tumor progression and assist in enhancing the sensitivity of tumors to chemotherapy or radiotherapy.
In practical applications, since naked siRNAs are extremely unstable and negatively charged, it is difficult for them to penetrate the cell membrane efficiently without a proper carrier.21 Previously, viral vectors have been demonstrated as promising gene delivery vehicles because of their high transfection efficiency.22 However, the application of viral vectors was severely impeded because of several bottlenecks, among which biosafety issues and immune response were of the most concern.22–26 As a result, non-viral carriers for siRNAs have emerged and been broadly explored.27,28 Unlike viral vectors, these non-viral alternatives have great advantages over viral vectors, such as increased biosafety, ease of design and synthesis, flexibility in chemical modifications and improved biocompatibility.29–33 In this study, highly biocompatible and degradable CPLAs were used for the delivery of siRNAs to pancreatic cancer cells targeting the proangiogenic chemokine IL-8 for gene therapy. Two different pancreatic cancer cell lines, Panc-1 and MiaPaCa-2, were used in the experiments for parallel comparison. The gene transfection efficiency was measured and the gene expression was examined. In addition, we evaluated the effect of IL-8 gene silencing on the proliferation of both cell lines. We found that the delivery of siRNAs by CPLAs was efficient and the gene expression of IL-8 was successfully suppressed. More importantly, the down-regulated IL-8 gene in turn attenuated the proliferation of the pancreatic cancer cells, indicating a new, effective and safe way towards pancreatic cancer therapy.
2. Materials and methods
2.1 Synthesis of CPLAs with 26 mol% amine group relative to PLA backbone repeat units
Materials: 4-dimethylaminopyridine (DMAP; 99+%) and L-lactide (L-LA) (98%) were purchased from Sigma-Aldrich, and 2,2′-dimethoxy-2-phenylacetophenone (DMPA; 98%) was purchased from Acros Organics. Dichloromethane (DCM; HPLC grade), acetone (HPLC grade), and benzyl alcohol (BnOH; HPLC grade) were purchased from Fisher Chemical. 2-(Diethylamino)ethanethiol hydrochloride (DEAET, 98+%) was purchased from Amfinecom Inc. All other chemicals were used without further purification.CPLA was prepared according to the method reported previously.33 In brief, in a 10 mL flask, allyl-functionalized PLA, DEAET, and photoinitiator DMPA were dissolved in CDCl3 (5 mL), resulting in a particular molar ratio. Then, the thiolene reaction was induced by UV irradiation (λmax = 365 nm) for 30 min to yield CPLA with 26 mol% tertiary amine cationic groups relative to backbone repeat units.
2.2 Cell culture
Human pancreatic cancer cells, Panc-1 (CRL-1469, American Type Culture Collection) and MiaPaCa-2 (CRL-1420), were maintained in culture with Dulbecco's Modified Eagle's Medium (DMEM, Hyclone), supplemented with 10% fetal bovine serum (FBS, Hyclone), 100 μg mL−1 penicillin (Gibco) and 100 μg mL−1 streptomycin (Gibco). Cells were cultured at 37 °C in a humidified atmosphere with 5% CO2.2.3 RNA extraction and real-time quantitative polymerase chain reaction (Q-PCR)
Total RNA was extracted from Panc-1 cells using TRIzol reagent (Invitrogen) and the amount was quantified by a spectrophotometer (Nano-Drop ND-1000). Total RNA (2 μg) was reverse transcribed to cDNA using the reverse transcriptase kit from Promega according to the manufacturer's instructions. Relative expression of IL-8 mRNA was assessed using the SYBR green master mix from Promega to perform Q-PCR using a real-time PCR instrument (Applied Biosystems 7500) that detects and plots the increase in fluorescence versus PCR cycle number to produce a continuous measure of PCR amplification. To provide a precise quantification of initial target in each PCR reaction, the amplification plot was examined at a point during the early log phase of product accumulation. This was accomplished by assigning a fluorescence threshold above background and determining the time point at which each sample's amplification plot reached the threshold (defined as the threshold cycle number or CT). Differences in the threshold cycle number were used to quantify the relative amount of PCR target contained within each tube. Relative mRNA expression was quantified and expressed as transcript accumulation index (TAI = 2 − delta·delta CT), calculated using the comparative CT method. All data were controlled for quantity of RNA input by performing measurements on an endogenous reference gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH).2.4 ELISA assay
The level of IL-8 in cell culture supernatants was determined using standard sandwich ELISA Kits (EH2IL8, Thermo Scientific) following the manufacturer's instructions.2.5 Transfection
The day before transfection, cells were seeded onto 6-well plates in DMEM medium without antibiotics to give 30–50% confluence at the time of transfection. A 20 μL CPLA (1 mg ml−1) dispersion was mixed with 20 μL of 10 μM IL-8 siRNAFAM (sense: 5′-FAM-GGAUUUUCCUAGAUAUUGCdTdT-3; antisense: 5-GCAAUAUCUAGGAAAAUCCdTdT-3) with a gentle vortex and left undisturbed for 20 minutes. Before transfection, the culture medium was replaced with 960 μL of OPTI-MEM (Invitrogen), the above mentioned CPLAs–siRNAFAM mixture was then added to the medium and the cells were continuously cultured. Four hours later, 500 μL DMEM medium with 30% FBS was added to the medium. Free siRNAFAM was also used in another parallel experiment at the same dosage level. A commercial transfection reagent Oligofectamine (Invitrogen) coupled siRNAFAM was used as a positive control. Gene expression was monitored at 48 hours post-transfection. For transfection efficiency examination, fluorescence imaging and flow cytometry analysis were performed at 4 hours post-treatment.2.6 Fluorescence imaging
In vitro fluorescence microscopy images were obtained using a fluorescence microscope (Eclipse-Ti, Nikon). The cells were washed and fixed with 4% formaldehyde before imaging, and the nuclei were stained with DAPI (Sigma). To image the cells, filter sets for DAPI (excitation at 405 nm and emission was collected with a band pass filter 450/50 nm) and FITC (excited with 488 nm laser and emission was collected with a band pass filter 525/50 nm) were applied for DAPI and FAM (with excitation/emission maximum at 492/518 nm) signals, respectively.2.7 Flow cytometry
For the flow cytometry experiments, the cells were washed twice with phosphate-buffered saline (PBS) and harvested by trypsinization. The FAM served as the luminescent marker (filter set for FITC was applied) to determine the transfection efficiency quantitatively. The samples were analyzed using a FACSCalibur flow cytometer (Becton Dickinson, Mississauga, CA, USA).2.8 Gene expression analysis
48 hours after transfection, total RNA was extracted from Panc-1 cells using TRIzol reagent (Invitrogen) and the amount was quantified by a micro-spectrophotometer (Nano-Drop ND-1000). Total RNA (2 μg) was reverse transcribed to cDNA using the reverse transcriptase kit from Promega according to the manufacturer's instructions. Semi-quantitative PCR was used to determine the IL-8 relative mRNA expression level normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), one of the most commonly used housekeeping genes adopted in comparisons of gene expression. The PCR products were electrophoresed on 2% ethidium bromide-stained agarose gel and observed under a UV transilluminator (Bio-Rad). Primers used were as before (Table 1).| Gene name | Primers |
|---|---|
| IL-8 | 5′-CTTCTAGGACAAGAGCCAGGAAGAAACCAC-3′ |
| 5′-GTCCAGACAGAGCTGTCTTCCATCAGAAAG-3′ | |
| CXCR1 | 5′-GAGCCCCGAATCTGACATTA-3′ |
| 5′-GCAGACACTGCAACACACCT-3′ | |
| CXCR2 | 5′-ATTCTGGGCATCCTTCACAG-3′ |
| 5′-TGCACTTAGGCAGGAGGTCT-3′ | |
| GAPDH | 5′-ACCACAGTCCATGCCATCAC-3′ |
| 5′-TCCACCACCCTGTTGCTGTA-3′ |
2.9 Cell viability
Cell viability was measured by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma) assay. Cells were seeded in a 96-well plate at a density of 5 × 103 cells per well and incubated with different concentrations of CPLAs for 24 or 48 hours. 20 μL of 5 mg ml−1 MTS in PBS was added and the cells were incubated for 4 hours at 37 °C with 5% CO2. 150 μL of 100% dimethylsulfoxide (DMSO, Sigma) was then added to solubilize the precipitate with 5 minutes gentle shaking. Absorbance was measured with a microplate reader (Bio-Rad) at a wavelength of 490 nm. The cell viability was obtained by normalizing the absorbance of the sample well against that from the control well and expressed as a percentage, assigning the viability of non-treated cells as 100%.2.10 Statistical analysis
All data were presented as means ± SD. The results were compared by analysis of variance (ANOVA). All statistical calculations were performed with the SPSS 11.0 software package. A p value less than 0.05 was regarded as a statistically significant difference.3. Results and discussion
Increased expression of IL-8 and/or its receptors has been identified in cancer cells.16 The tumor-derived IL-8 will then increase the NFκB transcription and the anti-apoptotic protein expression. As a result, profound effects on the tumor microenvironment will be initiated to maintain the viability of tumor cells.19 Here we have conducted in vitro experiments to examine the role of IL-8 and IL-8 receptors on the aggressive phenotype of two pancreatic cancer cell lines, Panc-1 and MiaPaCa-2. As shown in Fig. 1A, the RT-PCR derived gene expression of IL-8 and its receptors, CXCR1 and CXCR2, can be detected in both the Panc-1 and MiaPaCa-2 cell lines. More importantly, the results show that the IL-8 expression is significantly higher in MiaPaCa-2 than in Panc-1 cells, as characterized in both mRNA and protein levels (Fig. 1B and 1C). It is worth mentioning that the doubling time of MiaPaCa-2 cells in culture is about 40 hours, which is much less than that of Panc-1 cells (∼52 hours). This suggests a possible relationship between the IL-8 levels and the aggressiveness of the cancer cells. The targeted silencing of the IL-8 gene might be an effective way to halt the proliferation of pancreatic cancer cells.Our results showed that the IL-8 gene is highly expressed in pancreatic cancer cells and MiaPaCa-2 cells possess higher aggressive potential. One may be interested to know whether the RNAi mediated silencing of IL-8 in pancreatic cancer cells could eventually lead to the abrogation of proliferation. Prior to testing it using CPLA, a commercially available gold standard, Oligofectamine, was used as the transfection agent. Panc-1 and MiaPaCa-2 cells were treated with Oligofectamine–IL-8 siRNA nanoplexes (Oligo–IL-8) and the transfection results were compared with the control scrambled siRNA (Oligo–SC). Fig. 2A shows the gene expression levels of IL-8 in Panc-1 and MiaPaCa-2 cells at 48 hours post-treatment. As compared with those untreated or treated with the control scrambled siRNAs, the gene expressions of IL-8 in the samples treated with Oligo–IL-8 were remarkably inhibited (Fig. 2B). Fig. 2C shows that the cells treated with IL-8 siRNA manifested an obvious decrease in cell proliferation (p < 0.01) as determined by MTT assays while no significant effects were observed in the negative control, clearly showing the anti-cancer efficacy of IL-8 siRNA towards pancreatic carcinoma.
Oligofectamine has been widely used as a carrier system for siRNAs delivery in vitro and in vivo.34 Although it has shown promising results, a high dose of Oligofectamine is known to be cytotoxic.35 Furthermore, it may initiate an immune response and change the expression of non-target genes that are involved in critical cellular processes.34,36 For example, reports have shown that Oligofectamine can induce the overexpression of apoptosis related genes, such as the heat shock protein 70, caspase 8 isoform c and Bcl-2-related protein AL (Bcl-2 AL), and thus result in an increased tendency for early cell apoptosis.34,35 These drawbacks of Oligofectamine have greatly limited its further clinical applications. In contrast, degradable materials have shown superior capability of dealing with toxicity and clearance issues. CPLAs are soluble in water and positively charged. They can form nanoplexes with nucleic acids by electrostatic absorption. In addition, under physiochemical conditions, CPLAs can be degraded into oligomers (fragments of CPLAs) after 9 hours.33,37 Here, a ratio of siRNAs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CPLAs (1
CPLAs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128) was used throughout the experiments. After forming the nanoplexes with IL-8 siRNAs (CLPA–IL-8), the dynamic light scattering (DLS) data in Fig. 3 shows that the nanoplexes had an average hydrodynamic size of 60–80 nm. Because CPLAs were modified with tertiary amine-based cationic groups, this rendered CLPA–IL-8 positively charged with a zeta potential of +13.1 mV.
128) was used throughout the experiments. After forming the nanoplexes with IL-8 siRNAs (CLPA–IL-8), the dynamic light scattering (DLS) data in Fig. 3 shows that the nanoplexes had an average hydrodynamic size of 60–80 nm. Because CPLAs were modified with tertiary amine-based cationic groups, this rendered CLPA–IL-8 positively charged with a zeta potential of +13.1 mV.
Fig. 4 shows the fluorescent images of MiaPaCa-2 cells treated with different formulations for 4 hours, where the siRNAs were labelled with fluorescent FAM for visualization. The signals from the FAM channel in Fig. 4D demonstrate that the siRNAs were successfully delivered into the cells by conjugating with CPLAs. Fig. 4E shows results from a commercial transfection reagent Oligofectamine conjugated siRNAFAM (Oligo–siRNAFAM) serving as a positive control. In comparison, no FAM fluorescent signal was detected from the cells treated with free siRNAFAM (Fig. 4C), which was most likely due to the fast degradation of the unprotected siRNAs and the fact that the uptake of negatively charged siRNA was impeded by the cell membrane. These results indicate that the use of CPLAs as nanocarriers can effectively protect siRNAs from fast degradation and successfully transport them into the cells across the cell membrane. Parallel experiments were carried out on Panc-1 cells and similar results were observed (data not shown).
The transfection efficiency of siRNAs by CPLA was further quantified by flow cytometry analysis. Fig. 5 shows the representative plots of the fluorescence intensity in MiaPaCa-2 cells treated with different formulations for 4 hours, where Oligo–siRNA was introduced as a positive control. The results were consistent with the fluorescent imaging analysis that almost no fluorescent signal from FAM was detected in cells treated with free siRNAFAM (3.55 ± 1.20%) or CPLAs only (2.45 ± 0.84%). In contrast, cells treated with CPLA–siRNAFAM and Oligo–siRNAFAM showed strong fluorescent signals, which indicated the abundant accumulation of siRNAs. The fraction of cells with strong FAM fluorescent signals in the group treated with CPLA–siRNAFAM was counted to be over 90% (94.72 ± 1.25%) in our experiments, which was significantly higher than those in the blank, CPLAs and siRNAFAM groups (P < 0.001), and comparable with the group treated with Oligo–siRNAFAM (93.94 ± 1.38%). This portion represented in percentage was directly proportional to the delivery efficiency of siRNAs by CPLAs, and thus strongly demonstrates that the CPLAs can be used as efficient transfection reagents for siRNAs.
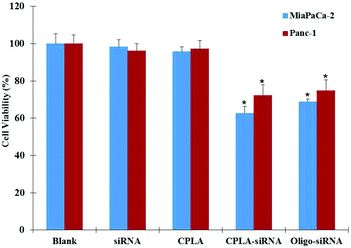
To evaluate the silencing efficiency of the CPLA–siRNA nanoplex, a specific siRNA sequence targeting IL-8 gene was conjugated with CPLAs and delivered to the Panc-1 and MiaPaCa-2 cells. The IL-8 mRNAs expressed by both cells were measured by RT-PCR as shown in Fig. 6A. It shows that the cells treated with CPLAs or free siRNAs exhibited minimal suppression when compared with non-treated negative control. As a comparison, CPLA–siRNA transfected cells have shown a remarkable decrease in the gene expression of IL-8, with knockdown efficiencies of 50.81% and 53.27% for Panc-1 and MiaPaCa-2 cells, respectively (Fig. 6B). Also, cells treated with the CPLA–siRNA nanoplex have shown an obvious decrease in the protein release of IL-8 (Fig. 6C). These results indicated that the CPLAs conjugated IL-8 siRNAs can be successfully released and bind to the targeting mRNA for silencing. As a positive control, the Oligo–siRNA formulation resulted in comparable knockdown efficiencies of 51.19% and 48.82% for Panc-1 and MiaPaCa-2 cells, respectively. In addition to the gene knockdown efficiency, to assess the therapeutic effects of the IL-8 targeted gene therapy, the viability of the cells after treatment were examined. Both Panc-1 and MiaPaCa-2 cells were treated with different formulations and the cell viabilities were evaluated by MTT assays 72 hours post-treatment. As shown in Fig. 7, no significant difference was observed in the cell proliferation ability between the control, CPLAs and free siRNA groups. In contrast, an evident decrease in the cell viability of the CPLA–siRNA treated group was observed, indicating that targeting IL-8 by employing the CPLAs conjugated IL-8 siRNAs is a promising strategy to suppress the proliferation of both the Panc-1 and MiaPaCa-2 pancreatic cancer cells. Results have also shown no difference between the CPLA–siRNA and the positive control Oligo–siRNA groups, which is consistent with the gene knockdown efficiency results.
The biocompatibility of the nanosized gene carriers is of great concern for biomedical applications. In order to evaluate the toxicity of CPLA, cell viability studies were performed on Panc-1 and MiaPaCa-2 cells. Fig. 8 shows that both cell lines maintained over 80% viability across a wide range of dosages up to 160 μg mL−1, 24 or 48 hours after exposure. These results indicate that the CPLA formulation has a high biocompatibility and we propose that the nanoformulation may be safely used for in vivo studies.
The aggressiveness of pancreatic cancer has made it one of the most deadly cancers around the world.38 RNAi based gene therapy has brought great promises for patients fighting against it. Lots of efforts have been made in identifying effective gene targets for pancreatic cancer and several have been found, such as K-Ras, P53, LSM1 and IL-8. However, an ideal risk-free transfection reagent with high gene silencing efficiency is still in urgent need. Our results here have provided a reasonable alternative. Although the complete suppression of the cancer cell proliferation is still challenging, the development of this new biodegradable CPLA formulation has shed a light on the key factor for gene transfection. With these promising results, we believe that further development in therapeutic strategies incorporating RNAi may achieve better results and help us move forward in the battle.
4. Conclusion
In conclusion, we have proposed a biodegradable nanoformulation for RNAi based pancreatic cancer gene therapy. The tumor overexpressed chemokine IL-8 was chosen as the gene target. The nanoformulation was based on CPLA with ternary amine-based cationic groups. Specific siRNAs targeting the IL-8 gene were conjugated with the CPLA and delivered into two pancreatic cancer cell lines with high transfection efficiency over 90%. The IL-8 gene expression was successfully suppressed and more importantly, the proliferation of both the cell lines was inhibited after 72 hours of transfection. MTS assays have shown that CPLA is highly biocompatible. Our results show that CPLA can serve as a great gene delivery platform for cancer treatment incorporating new therapeutic strategies.Acknowledgements
The study was supported by the start-up grant (M4080141.040) from Nanyang Technological University, Tier 1 Academic Research Funds (M4010360.040 RG29/10) from Singapore Ministry of Education and partially from the Singapore Ministry of Education under Tier 2 Research Grant MOE2010-T2-2-010 (4020020.040 ARC2/11), and the grants from National Natural Science Foundation of China (NSFC) (no. 81301318) and U.S. National Science Foundation (CBET-1133737).References
- A. Jemal, R. Siegel, J. Xu and E. Ward, Ca-Cancer J. Clin., 2010, 60, 277–300 CrossRef PubMed.
- A. Jemal, T. Murray, A. Samuels, A. Ghafoor, E. Ward and M. J. Thun, Ca-Cancer J. Clin., 2003, 53, 5–26 CrossRef.
- J. Ferlay, H. R. Shin, F. Bray, D. Forman, C. Mathers and D. M. Parkin, Int. J. Cancer, 2010, 127, 2893–2917 CrossRef CAS PubMed.
- D. M. Parkin, F. Bray, J. Ferlay and P. Pisani, Ca-Cancer J. Clin., 2005, 55, 74–108 CrossRef.
- G. Antoniou, P. Kountourakis, K. Papadimitriou, V. Vassiliou and D. Papamichael, Cancer Treat. Rev., 2014, 40, 78–85 CrossRef CAS PubMed.
- A. S. Paulson, C. H. Tran, M. A. Tempero and A. M. Lowy, Gastroenterology, 2013, 144, 1316–1326 CrossRef CAS PubMed.
- S. Torgerson and L. A. Wiebe, Oncology, 2013, 27, 183–190 Search PubMed.
- H. Chang, Cancer Gene Ther., 2007, 14, 677–685 CrossRef CAS PubMed.
- W. Lv, C. Zhang and J. Hao, World J. Gastroenterol., 2006, 12, 4636–4639 CAS.
- A. Strillacci, C. Griffoni, M. C. Valerii, G. Lazzarini, V. Tomasi and E. Spisni, J. Biomed. Biotechnol., 2010, 2010, 828045 Search PubMed.
- Z. Binkhathlan and A. Alshamsan, Ther. Delivery, 2012, 3, 1117–1130 CrossRef CAS.
- F. L. Tan and J. Q. Yin, Front. Biosci., 2005, 10, 1946–1960 CrossRef CAS PubMed.
- W. E. Holmes, J. Lee, W. J. Kuang, G. C. Rice and W. I. Wood, Science, 1991, 253, 1278–1280 CAS.
- C. Murdoch, P. N. Monk and A. Finn, Cytokine, 1999, 11, 704–712 CrossRef CAS PubMed.
- L. Xu and I. J. Fidler, Oncol. Res., 2000, 12, 97–106 CAS.
- D. J. Waugh and C. Wilson, Clin. Cancer Res., 2008, 14, 6735–6741 CrossRef CAS PubMed.
- A. Browne, R. Sriraksa, T. Guney, N. Rama, S. Van Noorden, E. Curry, H. Gabra, E. Stronach and M. El-Bahrawy, Cytokine, 2013, 64, 413–421 CrossRef CAS PubMed.
- S. Desai, S. Laskar and B. N. Pandey, Cell. Signalling, 2013, 25, 1780–1791 CrossRef CAS PubMed.
- A. Yuan, J. J. Chen, P. L. Yao and P. C. Yang, Front. Biosci., 2005, 10, 853–865 CrossRef CAS PubMed.
- Z. Shi, W. M. Yang, L. P. Chen, D. H. Yang, Q. Zhou, J. Zhu, J. J. Chen, R. C. Huang, Z. S. Chen and R. P. Huang, Breast Cancer Res. Treat., 2012, 135, 737–747 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS PubMed.
- L. B. Couto and K. A. High, Curr. Opin. Pharmacol., 2010, 10, 534–542 CrossRef CAS PubMed.
- R. G. Vile, A. Tuszynski and S. Castleden, Mol. Biotechnol., 1996, 5, 139–158 CrossRef CAS.
- S. Lehrman, Nature, 1999, 401, 517–518 CrossRef CAS PubMed.
- Q. Liu and D. A. Muruve, Gene Ther., 2003, 10, 935–940 CrossRef CAS PubMed.
- J. Y. Sun, V. Anand-Jawa, S. Chatterjee and K. K. Wong, Gene Ther., 2003, 10, 964–976 CrossRef CAS PubMed.
- A. Pathak, S. Patnaik and K. C. Gupta, Biotechnol. J., 2009, 4, 1559–1572 CrossRef CAS PubMed.
- A. Wongrakpanich, V. B. Joshi and A. K. Salem, Pharm. Dev. Technol., 2013, 18, 1255–1258 CrossRef CAS PubMed.
- Y. Chen and L. Huang, Expert Opin. Drug Delivery, 2008, 5, 1301–1311 CrossRef CAS PubMed.
- Y. Hattori, Yakugaku Zasshi, 2010, 130, 917–923 CrossRef CAS.
- S. P. Wong, O. Argyros and R. P. Harbottle, Methods Mol. Biol., 2012, 891, 133–167 CAS.
- C. Fu, X. Sun, D. Liu, Z. Chen, Z. Lu and N. Zhang, Int. J. Mol. Sci., 2011, 12, 1371–1388 CrossRef CAS PubMed.
- G. Lin, R. Hu, W. C. Law, C. K. Chen, Y. Wang, C. H. Li, Q. T. Nguyen, C. K. Lai, H. S. Yoon, X. Wang, G. Xu, L. Ye, C. Cheng and K. T. Yong, Small, 2013, 9, 2757–2763 CrossRef CAS PubMed.
- S. Akhtar and I. Benter, Adv. Drug Delivery Rev., 2007, 59, 164–182 CrossRef CAS PubMed.
- I. R. Gilmore, S. P. Fox, A. J. Hollins and S. Akhtar, Curr. Drug Delivery, 2006, 3, 147 CrossRef CAS.
- Y. Omidi, A. J. Hollins, M. Benboubetra, R. Drayton, I. F. Benter and S. Akhtar, J. Drug Targeting, 2003, 11, 311–323 CrossRef CAS PubMed.
- C. H. Jones, C. K. Chen, M. Jiang, L. Fang, C. Cheng and B. A. Pfeifer, Mol. Pharmaceutics, 2013, 10, 1138–1145 CrossRef CAS PubMed.
- C. Bosetti, P. Bertuccio, E. Negri, C. La Vecchia, M. P. Zeegers and P. Boffetta, Mol. Carcinog., 2012, 51, 3–13 CrossRef CAS PubMed.
Footnotes |
| † These authors contributed equally to this work. |
| ‡ Designed the biomaterial. |
| This journal is © The Royal Society of Chemistry 2014 |