Molecular analysis of interactions between dendrimers and asymmetric membranes at different transport stages
XiaoCong
He
ab,
ZhiGuo
Qu
*ab,
Feng
Xu
bc,
Min
Lin
bc,
JiuLing
Wang
d,
XingHua
Shi
d and
TianJian
Lu
b
aKey Laboratory of Thermo-Fluid Science and Engineering of Ministry of Education, School of Energy and Power Engineering, Xi'an Jiaotong University, Xi'an 710049, P.R. China. E-mail: zgqu@mail.xjtu.edu.cn
bBioinspired Engineering and Biomechanics Center, Xi'an Jiaotong University, Xi'an 710049, P.R. China
cThe Key Laboratory of Biomedical Information Engineering of Ministry of Education, School of Life Science and Technology, Xi'an Jiaotong University, Xi'an 710049, P.R. China
dThe State Key Laboratory of Nonlinear Mechanics, Institute of Mechanics, Chinese Academy of Sciences, Beijing 100190, P.R. China
First published on 24th October 2013
Abstract
Studying dendrimer–biomembrane interactions is important for understanding drug and gene delivery. In this study, coarse-grained molecular dynamics simulations were performed to investigate the behaviors of polyamidoamine (PAMAM) dendrimers (G4 and G5) as they interacted with asymmetric membranes from different sides of the bilayer, thus mimicking different dendrimer transport stages. The G4 dendrimer could insert into the membrane during an equilibrated state, and the G5 dendrimer could induce pore formation in the membrane when the dendrimers interacted with the outer side (outer interactions) of an asymmetric membrane [with 10% dipalmitoyl phosphatidylserine (DPPS) in the inner leaflet of the membrane]. During the interaction with the inner side of the asymmetric membrane (inner interactions), the G4 and G5 dendrimers only adsorbed onto the membrane. As the membrane asymmetry increased (e.g., increased DPPS percentage in the inner leaflet of the membrane), the G4 and G5 dendrimers penetrated deeper into the membrane during the outer interactions and the G4 and G5 dendrimers were adsorbed more tightly onto the membrane for the inner interactions. When the DPPS content reached 50%, the G4 dendrimer could completely penetrate through the membrane from the outer side to the inner side. Our study provides molecular understanding and reference information about different dendrimer transport stages during drug and gene delivery.
1. Introduction
With the rapid development of nanotechnology, nanoparticles (NPs) have been widely applied in everyday life and biomedicine in applications such as drug delivery, cancer diagnosis, and therapy.1–4 Of the various NPs, polyamidoamine (PAMAM) dendrimers have attracted attention for use in DNA carrier and drug delivery system applications because of their nanoscale size, convenient preparation and functionalization, and superior drug delivery capability.5 For these applications, PAMAM dendrimers must pass through a biomembrane barrier during the transport process to reach cellular compartments.6 Although passive and endocytosis pathways have been proposed, the exact mechanism of the transport process is only partially understood.7 In cells, dendrimers can interact with biomembranes, such as the membrane of an endosome (membrane-bounded compartment inside cells)8 and a cell membrane.9,10 Therefore, interactions between PAMAM dendrimers and biomembranes at different transport stages should be understood.Biomembranes, one of the most important parts of a biological system, have been considered symmetric in several studies (e.g., in simulations). However, this result may not represent the native biomembrane features. The compositions of the outer leaflet and inner leaflet of the membrane bilayer are different in many natural membranes. A membrane with a different composition in terms of the types and percentage of lipids in the outer and inner leaflets is defined as an “asymmetric membrane” in our study. For example, the human erythrocyte (red blood cell) membrane is a typical asymmetric lipid bilayer,11 which is composed of phosphatidylcholine, sphingomyelin, and glycolipids in the outer leaflet and phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol in the inner leaflet. Furthermore, the dipalmitoyl phosphatidylserine (DPPS) percentage in the inner leaflet of the membrane varies in different membranes,12 which means different levels of asymmetry in our study. This asymmetry may have important functions in biological behaviors. For instance, phosphatidylserine in the inner leaflet of the membrane can transfer from the inner leaflet to the outer leaflet, thereby inducing recognition by macrophages (a cell with defense function) when the cells undergo apoptosis (programmed cell death).13 Nevertheless, the underlying biological functions of asymmetric membranes remain poorly understood. Therefore, the influence of the asymmetry of a lipid bilayer on NP–biomembrane interactions should be considered.
Several simulation studies have been performed to investigate NP–asymmetric membrane interactions. A study regarding the transfer of benzocaine (a small molecule used in pain relievers) from an aqueous solution to the asymmetric lipid bilayer demonstrated that the presence of DPPS can change the preferred position of benzocaine in the lipid bilayer.12 Another study revealed that the potential of the mean force (PMF) curve is asymmetric during peptide–asymmetric membrane interactions.14 There was a local minimal energy position in the outer leaflet of the membrane, and the overall lowest energy position was in the inner membrane leaflet due to the electrostatic attraction between the positively charged peptide and the negatively charged inner leaflet of the membrane. Unfortunately, most existing studies have only focused on small molecules, such as benzocaine and peptides; these small molecules may differ from dendrimers, which are larger and have higher charge density and branched structures. Only a few studies partially focused on dendrimer–asymmetric membrane interactions.8 Tian et al.8 investigated the mechanism by which the G4 dendrimer escapes from endosomes. Their study simulated the interactions between the G4 dendrimer and the tensed membrane under low or neutral pH conditions.
The mechanism of dendrimer–biomembrane interactions in different cellular transport stages is not fully understood. Fig. 1 shows a schematic diagram of the cellular transport process of dendrimers. The processes of cellular uptake and elimination were considered in this study. During the cellular uptake process, dendrimers interact with the outer side of an asymmetric membrane (outer interactions), for which direct penetration and endocytosis pathways have been proposed. Within cells, dendrimers might still interact with biomembranes, such as the membrane of the Golgi, mitochondria, cytoskeletons, nucleus and cell membrane. However, for the endocytosis pathway, the dendrimers are trapped in endosomes at first, and a few dendrimers could escape from the endosomes to interact with other biomembranes. In particular, interactions between the dendrimers and cell membranes during the elimination process involve the inner side of the asymmetric membrane (inner interactions). Another possible elimination process is exocytosis, in which the dendrimer is inside of an endosome and is expelled from cells through fusion of the endosome with the cell membrane. In this case, the dendrimer only interacts with the outer leaflet of the membrane. The present study does not focus on this process.
As reviewed above, the interactions between a charged dendrimer and different sides of the asymmetric membrane during different transport stages have not yet been investigated. The current study used PAMAM dendrimers and an asymmetric lipid bilayer as a model system to perform coarse-grained molecular dynamics (CGMD) simulations. Although the asymmetric membrane of the erythrocyte membrane model was applied in the work of Tian et al.,8 the focus of each study is different. The work of Tian et al.8 focused on the mechanism of endosomal escape; in contrast, the process of cellular uptake and elimination with membranes with different asymmetry levels (in terms of different percentages of DPPS) were considered in the present study. The present investigation could provide further information about the dendrimer transport process as well as about the application of dendrimers in drug and gene delivery.
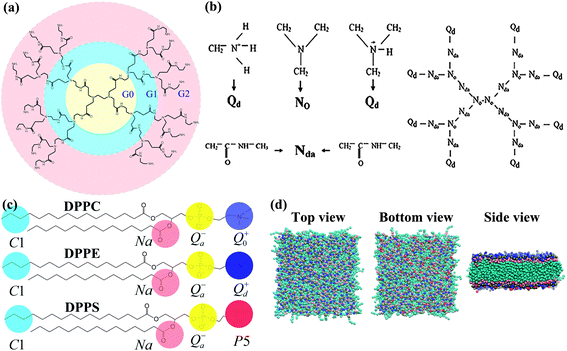
2. Model and methods
The MARTINI coarse-grained force field15 was used during the simulations. This model maps four atoms into one coarse-grained (CG) particle with significantly improved computational efficiency. Four main types of particles are used in this model: polar (P), apolar (C), nonpolar (N), and charged (Q). Four subtypes (d-donor, a-acceptor, da-both, and o-none) are used to distinguish different hydrogen-bonding capabilities, and five levels (1 to 5, polarity from low to high) are used to denote the polarity degree. This CG model is typically used to simulate the lipid membrane and proteins. Because the effective time is approximately 4 times greater than that of atomistic simulations in this CG model, we used effective times in this paper.16Three different types of lipid models were used in our asymmetric membrane simulations: dipalmitoyl phosphatidylcholine (DPPC), dipalmitoyl phosphatidylethanolamine (DPPE), and dipalmitoyl phosphatidylserine (DPPS). The DPPC lipid bilayer15 is a typical lipid model in the MARTINI CG force field, and we substituted DPPC lipids with DPPE and charged DPPS molecules to simulate the asymmetric membrane. The ratios of the different lipids was 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (DPPC
1 (DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE) in the outer leaflet of the membrane and 3
DPPE) in the outer leaflet of the membrane and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (DPPC
2 (DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE
DPPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPS) in the inner leaflet of the membrane. These particular ratios were chosen to mimic the human erythrocyte membrane.8 In the same manner, we assembled two other asymmetric membranes by changing the percentage of DPPS in the inner leaflet to 3
DPPS) in the inner leaflet of the membrane. These particular ratios were chosen to mimic the human erythrocyte membrane.8 In the same manner, we assembled two other asymmetric membranes by changing the percentage of DPPS in the inner leaflet to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (DPPC
5 (DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE
DPPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPS) and 100% DPPS (Table 1), with the percentages calculated from the membrane charge density.17 Each asymmetric membrane contained 480 CG lipids and was equilibrated in a water and ion system for 200 ns in the NPT ensemble.
DPPS) and 100% DPPS (Table 1), with the percentages calculated from the membrane charge density.17 Each asymmetric membrane contained 480 CG lipids and was equilibrated in a water and ion system for 200 ns in the NPT ensemble.
| System | Membrane | Membrane charge | PAMAM | CG beads | PAMAM charge | Ions |
|---|---|---|---|---|---|---|
| 1 | (outer leaflet) DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE = 9 DPPE = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (inner leaflet) DPPC 1 (inner leaflet) DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE DPPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPS = 3 DPPS = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (DPPS percent = 10%) 2 (DPPS percent = 10%) |
48− | G4 | 250 | 64+ | 16Cl− |
| 2 | (outer leaflet) DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE = 9 DPPE = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (inner leaflet) DPPC 1 (inner leaflet) DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE DPPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPS = 3 DPPS = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (DPPS percent = 10%) 2 (DPPS percent = 10%) |
48− | G5 | 506 | 128+ | 80Cl− |
| 3 | (outer leaflet) DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE = 9 DPPE = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (inner leaflet) DPPC 1 (inner leaflet) DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE DPPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPS = 3 DPPS = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (DPPS percent = 25%) 5 (DPPS percent = 25%) |
120− | G4 | 250 | 64+ | 56Na+ |
| 4 | (outer leaflet) DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE = 9 DPPE = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (inner leaflet) DPPC 1 (inner leaflet) DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE DPPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPS = 3 DPPS = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (DPPS percent = 25%) 5 (DPPS percent = 25%) |
120− | G5 | 506 | 128+ | 8Cl− |
| 5 | (outer leaflet) DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE = 9 DPPE = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (inner leaflet) DPPS only (DPPS percent = 50%) 1 (inner leaflet) DPPS only (DPPS percent = 50%) |
240− | G4 | 250 | 64+ | 176Na+ |
| 6 | (outer leaflet) DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPE = 9 DPPE = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (inner leaflet) DPPS only (DPPS percent = 50%) 1 (inner leaflet) DPPS only (DPPS percent = 50%) |
240− | G5 | 506 | 128+ | 112Na+ |
There are primary amine groups at the branching end of PAMAM dendrimers and tertiary amine groups at the branching point. We mapped the atomic structures of the G4 and G5 PAMAM dendrimers generated using the Material Studio package to the CG structures18 (Fig. 2). The bonded potentials are  and
and  . The equilibrium bond distance Rbond is = 0.5 nm, and the equilibrium angle is θ = 120° or 180°. The force constant of the bonding potential Kbond is = 1250 kJ mol−1 nm−2, and the force constant of the angle potential is Kangle = 150 kJ mol−1 rad−2.16 The CG structures of the G4 and G5 PAMAM dendrimers contain 250 and 506 CG particles, respectively, and 64 and 128 positive charges, respectively. To equilibrate the CG dendrimer model, a long molecular simulation (1 μs) of the CG dendrimers in the CG solvents was performed.8 The radii of gyration of the G4 and G5 dendrimers were 2.01 and 2.56 in CG solvents (Table 2).
. The equilibrium bond distance Rbond is = 0.5 nm, and the equilibrium angle is θ = 120° or 180°. The force constant of the bonding potential Kbond is = 1250 kJ mol−1 nm−2, and the force constant of the angle potential is Kangle = 150 kJ mol−1 rad−2.16 The CG structures of the G4 and G5 PAMAM dendrimers contain 250 and 506 CG particles, respectively, and 64 and 128 positive charges, respectively. To equilibrate the CG dendrimer model, a long molecular simulation (1 μs) of the CG dendrimers in the CG solvents was performed.8 The radii of gyration of the G4 and G5 dendrimers were 2.01 and 2.56 in CG solvents (Table 2).
For the initial positions, the center of mass separation distance in the Z direction between the PAMAM dendrimer and the membrane (Z-distance) was 4 nm for G4 and 5 nm for G5. The dendrimer was placed above and below the bilayer for the dendrimer–outer side interaction and inner side interaction, respectively. The PAMAM dendrimer–lipid membrane system, water and ions were added into a simulation box. The xy size of the simulation box was dependent on the size of the membrane without tension (12.44 nm × 12.72 nm), and the simulation box was 25 nm in the Z direction. The size of the simulation box in the x and y directions was similar to that of the box in the study by Lee et al.,18 in which G3 and G5-membrane interactions with a box (size 12 × 13 × 9 nm3) were simulated. Those researchers compared two methods in their simulations: simulations with an electrostatic cutoff and with particle mesh Ewald summation (PME). The results indicated that the un-acetylated dendrimer did not insert into the membrane using the electrostatic cutoff method. In contrast, with the PME method, dendrimers inserted into the membrane, and pore formation was observed; these results were similar to experimental studies. Therefore, the use of long-range electrostatic interactions with the PME method in our simulation systems is reasonable. After energy minimization, we constrained the lipid bilayer and PAMAM dendrimer with a force constant of 1000 kJ mol−1 nm−2 to perform the constrained molecular dynamics simulation to equilibrate the water and ions for 200 ns. Subsequently, the lipid bilayer and PAMAM dendrimer were released and allowed to interact freely.
We performed the MD simulation for 800 ns for each case under periodic boundary conditions to ensure system equilibration. The Berendsen thermostat was used to control the group temperature at 323 K.19 In the NPT ensemble, Berendsen pressure coupling was used to control the group pressure at 1 bar. The cutoff was 1.2 nm for van der Waals (vdW) interactions, and to reduce the cut-off noise, the L–J potential was smoothly shifted to zero between 0.9 and 1.2 nm. We used the particle mesh Ewald summation (PME) method20 for the electrostatic interactions. The umbrella sampling method21 and the weighted histogram analysis method22 were used to calculate the PMF, and the force constant of the harmonic potential was 1000 kJ mol−1 nm−2. The initialized conformations with a 0.1 nm step size were produced by acceleration using an external force. For each window, we performed long runs of 400 ns. All the simulations used the GROMACS 4.5.4 package.23 All the simulation results were represented using Visual Molecular Dynamics (VMD) software, version 1.9.24
3. Results and discussion
3.1. Interactions between the G4 dendrimer and asymmetric membranes
To understand the mechanism of the interaction between the G4 dendrimer and the asymmetric membranes, interactions of the G4 dendrimer with the outer and inner sides of an asymmetric membrane were simulated with DPPS percentages ranging from 10% to 50%. We observed significant differences between the interaction of G4 with the outer side of the membrane (outer interaction) and with the inner side of the membrane (inner interaction) for different asymmetry levels (Fig. 3). In the case of the outer interaction with 10% DPPS, the G4 dendrimer could insert into the membrane when an equilibrated state was achieved (Fig. 3a). In contrast, with the symmetric membrane, the charged G4 dendrimer will just stay on the outer leaflet of the symmetric membrane because of a high energy barrier.25 Such a difference between the symmetric and asymmetric membrane is attributed to the electrostatic attraction between the inner leaflet of the membrane and the charged dendrimer. For example, Ma et al.25 showed that electrostatic interactions play a major role in the G4 dendrimer–membrane interactions because the electrostatic potential energy is 50 times lower than that of van der Waals (vdW) energy during equilibrium. Therefore, the negatively charged DPPS in the inner leaflet of the asymmetric membrane attracted the positively charged dendrimer for insertion into the asymmetric membrane, as in the case of current simulation. Experimental research26 also indicated the importance of electrostatic attraction between a cationic dendrimer–gene complex and a positively charged cell membrane. The results showed that the gene transfection efficiency was higher when the dendrimer–gene complex surface was cationic. In addition, the DPPS lipid molecules could move to the upper leaflet of the membrane though the inserted dendrimer (Fig. 3a side view), a result that was also observed in another recent study.8 This process could reduce the asymmetry of the membrane.As for the inner interaction, the G4 dendrimer could only be adsorbed on the inner leaflet of the membrane (Fig. 3b) because of the electrostatic attraction between the positively charged dendrimer and the negatively charged inner leaflet of the membrane. As shown in Fig. 3b, the dendrimer, which adsorbed onto the inner leaflet of the membrane, was in a flat conformation. This result was consistent with the simulation results in the study by Kelly et al.27 The study indicated that dendrimers retained a spherical shape when interacting with a gel-phase lipid bilayer and changed to a flat shape when interacting with a fluid-phase membrane. In addition, the DPPS lipids were distributed around the dendrimer due to the electrostatic attraction.
To investigate the effects of the asymmetry levels of the membrane on the interaction between the G4 dendrimer and the membrane, we assembled two other asymmetric membranes (Table 1) with 25% and 50% DPPS. From the snapshots of the G4–asymmetric membrane interactions in the equilibrium state, we observed that the equilibrium states in the case with 25% DPPS were similar to the case with 10% DPPS (Fig. 3c and d). However, for the interactions with 50% DPPS, the dendrimer could completely penetrate through the membrane in the outer interaction (Fig. 3e) while the dendrimer was still adsorbed on the inner leaflet of the membrane in the inner interaction (Fig. 3f).
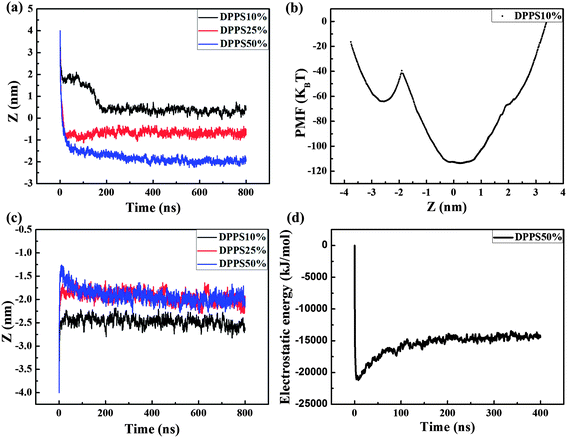
To further assess the interaction, we quantified both the center of mass separation distance in the Z direction between the PAMAM dendrimer and the membrane (Z-distance) and the PMF of these interactions, as shown in Fig. 4. We observed that with increasing membrane asymmetry levels, the Z-distance became lower in the membrane as the DPPS percent was increased from 10% to 50%, as shown in Fig. 4a. The increased charge density in the inner leaflet of the membrane may account for the change in the Z-distance. The enhanced electrostatic attraction leads to the improved penetration of the dendrimer from the outer side of the membrane. We also observed that the Z-distance (DPPS percent: 10%) decreased rapidly to 1.83 nm at an early stage, followed by a further gradual decrease to the equilibrated location at approximately 0.36 nm (Fig. 4a, black line). These results demonstrated that a metastable state was observed at a Z-distance of 1.83 nm, indicating the rapid binding of the dendrimer with the head groups of the membrane. Because of the electrostatic attraction between the dendrimer and the inner leaflet of the membrane, the dendrimer began to insert into the membrane and eventually reached equilibrium around the membrane center. From the variation in PMF of this interaction, we observed an asymmetry in the PMF curve (Fig. 4b). The potential energy trap around the membrane center was much lower than the energy trap at a Z-distance of −2.49 nm. The Z-distance (0.36 nm) with the lowest energy was the preferred position for the G4 dendrimer. To penetrate through the inner leaflet of the membrane, the G4 dendrimer must overcome a high energy barrier of 74.21 kBT.
In the inner interactions, the Z-distance was −2.49 nm for the case with 10% DPPS in the membrane and −1.97 nm for the cases with 25% and 50% DPPS (Fig. 4c). The result indicated that the dendrimer was adsorbed on the membrane more tightly due to the stronger electrostatic attraction when the DPPS percentage increased from 10% to 25%. As the DPPS percentage was further increased to 50%, the Z-distance was the same as for the case with 25% DPPS. This result was observed because the stronger electrostatic attraction was not able to make the dendrimer overcome the high energy barrier in the −1.97 nm of Z-distance. The Z-distance peaked at approximately −1.28 nm during the first dozens of ns and then fell to −1.97 nm in the equilibrium state (Fig. 4c, blue line). This behavior occurred because the electrostatic attraction between the positively charged dendrimer and the negatively charged lipids played a major role in the interactions at first,28 as confirmed by the rapid decrease in the electrostatic energy in the first several ns (Fig. 4d). The electrostatic attraction induced the movement of more charged lipids to interact with the charged dendrimer branches; thus, the membrane bent. At a Z-distance of −1.97 nm, the dendrimer eventually reached an equilibrium of all the forces, including the bonded interactions, electrostatic interactions, and vdW interactions.
To analyze the effect of dendrimers in the dendrimer–biomembrane systems on the membrane structures, the radial distribution functions (RDFs) of the lipid head groups, tail groups, and ions at the dendrimer surface and the order parameters of lipid tails were quantified. Fig. 5a shows the RDFs during the outer interactions of the G4–membrane (10% DPPS) system. There was a large phosphate peak (Fig. 5a, red line) at 0.5 nm, which indicated that the positively charged G4 dendrimer terminals strongly interacted with the negatively charged phosphate groups of the lipid head groups. Similar results have also been observed experimentally;29 a strong interaction between the G4 dendrimer and the polar head groups of the lipids was found through Raman spectroscopy. The highest peak at 0.5 nm was from the chlorine ion and was also caused by the electrostatic attraction. The RDFs for the inner interactions of the G4–membrane (10% DPPS) system are shown in Fig. 5b. Similar large peaks were observed for the phosphate groups and ions at 0.5 nm. However, comparing the RDFs for the outer interaction with those of the inner interaction indicated that the peaks of the glycerol, choline and phosphate groups at 0.5 nm were much higher for the outer interaction than for the inner interaction. A possible reason is that the dendrimer inserted into the membrane during the outer interaction; thus, the dendrimer terminals sufficiently interacted with the lipid head groups in both the outer and inner leaflet of the membrane. In contrast, the dendrimer was just adsorbed on the inner leaflet of the membrane during the inner interactions.
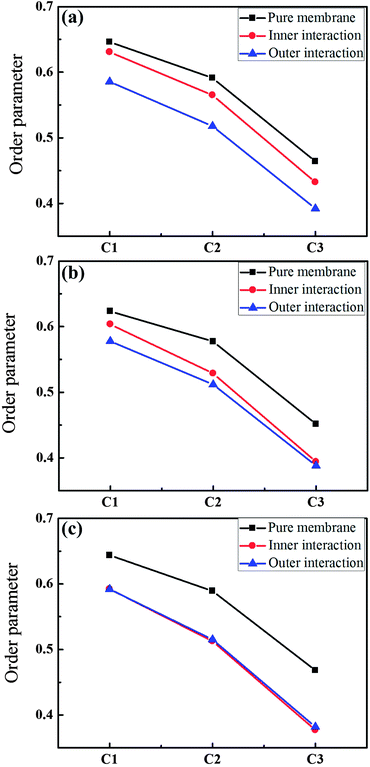
The order parameters of the phospholipid tail beads (C1, C2, C3) were obtained in the pure membrane and dendrimer–membrane systems. The expression for calculating the order parameter is  , where θz is the angle between the molecular vector and the Z axis. The molecular vector is from Cn+1 to Cn−1. The order parameter was calculated by averaging over last 200 ns of the simulations for the phospholipid tails. Fig. 6a indicates that the introduction of the G4 dendrimer during the outer and inner interactions decreased the lipid order relative to that for the pure membrane lipids. This result may explain the experimental observation30,31 that some concentrations of dendrimers (G2, G3, G4) could lead to a change in the shape of red blood cells that was accompanied by hemolysis. The shape change and hemolysis were attributed to the perturbation to the membrane caused by the dendrimer. Furthermore, the lipid disorder during the outer interactions of the inserting mechanism was more significant than that during the inner interaction of the adsorption mechanism. Comparing Fig. 6a–c revealed that the lipids became more disordered with increased membrane asymmetry levels during the inner interactions. The stronger electrostatic attraction in the membrane with higher asymmetry levels (25% or 50% DPPS) induced stronger adsorption of the dendrimer to the inner leaflet, resulting in more perturbations of the lipids. The overlap of red and blue lines in Fig. 6c also confirmed that the conformation of the adsorbed dendrimer on the inner leaflet after completely penetrating through the membrane during the outer interaction was similar to that for the inner interactions in the G4–membrane (50% DPPS) system.
, where θz is the angle between the molecular vector and the Z axis. The molecular vector is from Cn+1 to Cn−1. The order parameter was calculated by averaging over last 200 ns of the simulations for the phospholipid tails. Fig. 6a indicates that the introduction of the G4 dendrimer during the outer and inner interactions decreased the lipid order relative to that for the pure membrane lipids. This result may explain the experimental observation30,31 that some concentrations of dendrimers (G2, G3, G4) could lead to a change in the shape of red blood cells that was accompanied by hemolysis. The shape change and hemolysis were attributed to the perturbation to the membrane caused by the dendrimer. Furthermore, the lipid disorder during the outer interactions of the inserting mechanism was more significant than that during the inner interaction of the adsorption mechanism. Comparing Fig. 6a–c revealed that the lipids became more disordered with increased membrane asymmetry levels during the inner interactions. The stronger electrostatic attraction in the membrane with higher asymmetry levels (25% or 50% DPPS) induced stronger adsorption of the dendrimer to the inner leaflet, resulting in more perturbations of the lipids. The overlap of red and blue lines in Fig. 6c also confirmed that the conformation of the adsorbed dendrimer on the inner leaflet after completely penetrating through the membrane during the outer interaction was similar to that for the inner interactions in the G4–membrane (50% DPPS) system.
These results could be further confirmed by the snapshots of the interactions between the G4 dendrimer and the asymmetric membrane with 50% DPPS (Fig. 7). For instance, the dendrimer could completely penetrate the membrane from the outer side to the inner side (Fig. 7a). The G4 dendrimer was adsorbed onto the membrane rapidly and began to insert into the membrane at 2 ns; after 150 ns, the dendrimer has completely passed the outer head groups of the membrane. During the inner interaction, the dendrimer induced bending of the membrane at 10 ns and reached equilibrium at 100 ns (Fig. 7b).
3.2. Interactions between the G5 dendrimer and asymmetric membranes
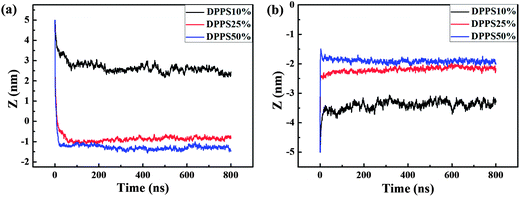
Compared with the G4 dendrimer, the G5 dendrimer has 64 more positively charged beads and a larger size. The G5 dendrimer is widely used in the drug delivery field, particularly for its nanoscale size that is similar to the biological components of transthyretin (a protein that transports thyroxine and retinol).32 To investigate the response of an asymmetric membrane to the G5 dendrimer, we simulated the interaction of the G5 dendrimer with asymmetric membranes from both sides of the membrane. During the outer interaction, the Z-distance in the equilibrated state in the membrane became lower with increasing membrane asymmetry levels. The Z-distance was 2.55 nm in the membrane with 10% DPPS, whereas it was approximately −0.86 nm and −1.32 nm in the membrane with 25% DPPS and 50% DPPS, respectively (Fig. 8a). This was because the stronger electrostatic attraction induced deeper penetration of the dendrimer into the membrane. Pore formation was observed during the outer interaction of the G5–membrane (10% DPPS) system. This observation was similar to the findings reported in previous numerical and experimental studies of the interactions between an un-acetylated G5 dendrimer and a DPPC lipid bilayer,18 a DMPC lipid bilayer33 and cell membranes (KB and Rat2).34 The experimental results indicated that the pore formation during the interactions of the dendrimer–DMPC lipid bilayer was caused by the removal of lipid molecules from the supported substrate by the dendrimers, resulting in a composite of dendrimers and lipids.33During the inner interaction, similar to the G4 dendrimer, the G5 dendrimer was also adsorbed tightly onto the inner leaflet of the membrane. With increasing DPPS percentage in the inner leaflet of the membrane, the center of mass of the dendrimer moved towards the membrane center (Fig. 8b). The corresponding Z-distance was −3.35 nm, −2.14 nm, and −1.97 nm for a DPPS percentage of 10%, 25% and 50%, respectively (Fig. 8b), which was further from the membrane center (0 nm) compared to that for the G4 dendrimer. For instance, the Z-distance was approximately −3.35 nm for G5, whereas it was −2.49 nm for G4 when the DPPS percent was 10%. This result indicated that the adsorption of the G5 dendrimer was not as tight as that of the G4 dendrimer because the G5 dendrimer was larger (256 more CG particles) than the G4 dendrimer. Furthermore, the G5 dendrimer had 128 positively charged particles while negatively charged DPPS in the membrane had only 48. Therefore, electrostatic attraction was not strong enough for tight adsorption of the G5 dendrimer on the membrane.
The increased membrane asymmetry could also create obvious disorder in the lipids in the G5–membrane systems, as shown in Fig. 9. The effects of the G5 dendrimer during the outer and inner interactions were similar to those of the G4 dendrimer, whereas, the order parameters during the outer and inner interactions for the G5 dendrimer were both lower than those for the G4 dendrimer. For instance, the order parameter of C3 in Fig. 9a (blue line) was below 0.3, whereas it was approximately 0.4 in the G4–membrane systems. These results indicated that the G5 dendrimers induced more perturbations to the lipids than did the G4 dendrimers.
Furthermore, we observed a transient pore formation during the inner interaction between the G5 dendrimer and the membrane with 50% DPPS. The transient pore formation occurred within approximately 2.5 ns and disappeared after 11 ns (Fig. 10). Subsequently, the DPPS lipids (16 DPPS lipids in the equilibrated state) moved to the outer leaflet, decreasing the asymmetry. With time, the Z-distance returned to 1.97 nm due to the equilibration of the hydrophobic and electrostatic interactions. Metastable pore formation was also observed in the simulation study of Tian et al.,8 in which the critical tension was lowered in the G4 dendrimer-tensed membrane interaction (0.81 nm2 area per lipid). A possible reason for the transient pore formation in our simulation is that the G5 dendrimer has more charged beads than does the G4, leading to significant membrane bending. When the outer leaflet cannot withstand the pressure induced by membrane bending, pores are formed. Furthermore, to investigate the box-size effect on transient pore formation, a system with a larger box size (18 × 18 × 25 nm3) was simulated. The pore also formed in this system with a larger box size (Fig. 11). However, the pore formed within approximately 2.375 ns and disappeared at approximately 3.75 ns. The duration (1.375 ns) in the larger box was shorter than that (8.5 ns) in the small box. The reason was that the dendrimer concentration (ratio of dendrimer and lipid)16 was decreased in the system with the larger box. The low concentration of dendrimer could induce lower membrane bending-induced tension; thus, the pore disappeared rapidly.
 | ||
| Fig. 11 Snapshots of the transient pore formation during the inner interaction of the G5-membrane (50% DPPS) system with the larger simulation box: (a) side view and (b) top view. To investigate the box-size effects on the phenomena of transient pore formation in Fig. 10, a larger simulation system (box size: 18 × 18 × 25 nm3) was utilized. The pore formed at approximately 2.375 ns and disappeared at approximately 3.75 ns. | ||
4. Conclusion
In the present study, coarse-grained molecular dynamics simulations were performed to investigate the interaction of charged PAMAM dendrimers (G4 and G5) with asymmetric membranes from different sides of the bilayer. When the charged dendrimers interacted with the outer side of the asymmetric membrane (with 10% DPPS), the G4 dendrimer could insert into the membrane in the equilibrated state while the G5 dendrimer induced pore formation in the membrane. During the interaction with the inner side of the asymmetric membrane, the G4 and G5 dendrimers only adsorbed onto the membrane. With increasing membrane asymmetry, the G4 and G5 dendrimers penetrated deeper into the membrane during the outer interactions. Furthermore, the G4 dendrimer could completely penetrate from the outer side to the inner side of the membrane with 50% DPPS. As for the inner interactions, the G4 and G5 dendrimers adsorbed more tightly onto the membrane, and the G5 dendrimer induced transient pore formation in the membrane with 50% DPPS. This study shows that an asymmetric membrane can promote dendrimer penetration during the outer interaction and adsorb dendrimers onto the inner leaflet of the membrane, improving the drug delivery efficiency during the inner interaction. These results can provide molecular understanding and suggestions regarding dendrimer-based drug and gene delivery.Acknowledgements
This study was partially supported by the National Natural Science Foundation of China (Grant no. 51322604, 11272327 and 11372243), the National 111 Project of China (Grant no. B06024), the Major International Joint Research Program of China (Grant no. 11120101002), the Key Project of Chinese Ministry of Education (Grant no. 313045), the South Wisdom Valley Innovative Research Team Program, and the International Science & Technology Cooperation Program of China (Grant no. 2013DFG02930). F. X. was also partially supported by the China Young 1000-Talent Program and the Program for New Century Excellent Talents in University (Grant no. NCET-12-0437).References
- D. Choi, J. H. Moon, H. Kim, B. J. Sung, M. W. Kim, G. Y. Tae, S. K. Satija, B. Akgun, C. J. Yu, H. W. Lee, D. R. Lee, J. M. Henderson, J. W. Kwong, K. L. Lam, K. Y. C. Lee and K. Shin, Soft Matter, 2012, 8, 8294–8297 RSC.
- J. Zupanc, D. Drobne, B. Drasler, J. Valant, A. Iglic, V. Kralj-Iglic, D. Makovec, M. Rappolt, B. Sartori and K. Kogej, Carbon, 2012, 50, 1170–1178 CrossRef CAS PubMed.
- D. E. Warschawski, A. A. Arnold, M. Beaugrand, A. Gravel, É. Chartrand and I. Marcotte, Biochimica et Biophysica Acta, 2011, 1808, 1957–1974 CrossRef CAS PubMed.
- Z. G. Qu, X. C. He, M. Lin, B. Y. Sha, X. H. Shi, T. J. Lu and F. Xu, Nanomedicine, 2013, 8, 1743–5889 CrossRef PubMed.
- M. J. Cloninger, Curr. Opin. Chem. Biol., 2002, 6, 742–748 CrossRef CAS.
- L. Monticelli, E. Salonen, P. C. Ke and I. Vattulainen, Soft Matter, 2009, 5, 4433 RSC.
- A. Akesson, C. V. Lundgaard, N. Ehrlich, T. G. Pomorski, D. Stamou and M. Cardenas, Soft Matter, 2012, 8, 8972–8980 RSC.
- W.-d. Tian and Y.-q. Ma, Soft Matter, 2012, 8, 6378–6384 RSC.
- L. Y. T. Chou, K. Ming and W. C. W. Chan, Chem. Soc. Rev., 2011, 40, 233–245 RSC.
- K. Kitchens and H. Ghandehari, in Nanotechnology in Drug Delivery, ed. M. Villiers, P. Aramwit and G. Kwon, Springer, New York, 2009, vol. X, ch. 14, pp. 423–459 Search PubMed.
- A. J. Verkleij, R. F. A. Zwaal, B. Roelofsen, P. Comfurius, D. Kastelijn and L. L. M. van Deenen, Biochimica et Biophysica Acta, 1973, 323, 178–193 CrossRef CAS.
- R. D. Porasso, W. F. Drew Bennett, S. D. Oliveira-Costa and J. J. López Cascales, J. Phys. Chem. B, 2009, 113, 9988–9994 CrossRef CAS PubMed.
- S. K. Sahu, S. N. Gummadi, N. Manoj and G. K. Aradhyam, Arch. Biochem. Biophys., 2007, 462, 103–114 CrossRef CAS PubMed.
- Z.-l. Li, H.-m. Ding and Y.-q. Ma, Soft Matter, 2013, 9, 1281–1286 RSC.
- S. J. Marrink, A. H. de Vries and A. E. Mark, J. Phys. Chem. B, 2003, 108, 750–760 CrossRef.
- H. Lee and R. G. Larson, J. Phys. Chem. B, 2008, 112, 7778–7784 CrossRef CAS PubMed.
- J. Lin, H. Zhang, Z. Chen and Y. Zheng, ACS Nano, 2010, 4, 5421–5429 CrossRef CAS PubMed.
- H. Lee and R. G. Larson, J. Phys. Chem. B, 2006, 110, 18204–18211 CrossRef CAS PubMed.
- Y. Li, X. Chen and N. Gu, J. Phys. Chem. B, 2008, 112, 16647–16653 CrossRef CAS PubMed.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
- G. M. Torrie and J. P. Valleau, J. Comput. Phys., 1977, 23, 187–199 CrossRef.
- S. Kumar, J. M. Rosenberg, D. Bouzida, R. H. Swendsen and P. A. Kollman, J. Comput. Chem., 1992, 13, 1011–1021 CrossRef CAS.
- D. Van Der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark and H. J. C. Berendsen, J. Comput. Chem., 2005, 26, 1701–1718 CrossRef CAS PubMed.
- A. D. W. Humphrey and K. Schulten, J. Mol. Graphics, 1996, 33 CrossRef.
- W.-d. Tian and Y.-q. Ma, Soft Matter, 2012, 8, 2627–2632 RSC.
- J. D. Eichman, A. U. Bielinska, J. F. Kukowska-Latallo and J. R. Baker Jr, Pharm. Sci. Technol. Today, 2000, 3, 232–245 CrossRef CAS.
- C. V. Kelly, P. R. Leroueil, B. G. Orr, M. M. Banaszak Holl and I. Andricioaei, J. Phys. Chem. B, 2008, 112, 9346–9353 CrossRef CAS PubMed.
- J. M. Nascimento, O. L. Franco, M. D. L. Oliveira and C. A. S. Andrade, Chem. Phys. Lipids, 2012, 165, 537–544 CrossRef CAS PubMed.
- K. Gardikis, S. Hatziantoniou, K. Viras, M. Wagner and C. Demetzos, Int. J. Pharm., 2006, 318, 118–123 CrossRef CAS PubMed.
- D. M. Domański, B. Klajnert and M. Bryszewska, Bioelectrochemistry, 2004, 63, 189–191 CrossRef PubMed.
- N. Malik, R. Wiwattanapatapee, R. Klopsch, K. Lorenz, H. Frey, J. W. Weener, E. W. Meijer, W. Paulus and R. Duncan, J. Controlled Release, 2000, 65, 133–148 CrossRef CAS.
- P. R. Leroueil, S. Hong, A. Mecke, J. R. Baker, B. G. Orr and M. M. Banaszak Holl, Acc. Chem. Res., 2007, 40, 335–342 CrossRef CAS PubMed.
- A. Mecke, S. Uppuluri, T. M. Sassanella, D.-K. Lee, A. Ramamoorthy, J. R. Baker Jr, B. G. Orr and M. M. Banaszak Holl, Chem. Phys. Lipids, 2004, 132, 3–14 CrossRef CAS PubMed.
- S. Hong, A. U. Bielinska, A. Mecke, B. Keszler, J. L. Beals, X. Shi, L. Balogh, B. G. Orr, J. R. Baker and M. M. Banaszak Holl, Bioconjugate Chem., 2004, 15, 774–782 CrossRef CAS PubMed.
- P. K. Maiti, T. Çaǧın, S.-T. Lin and W. A. Goddard, Macromolecules, 2005, 38, 979–991 CrossRef CAS.
- L.-T. Yan and X. Yu, ACS Nano, 2009, 3, 2171–2176 CrossRef CAS PubMed.
- I. Lee, B. D. Athey, A. W. Wetzel, W. Meixner and J. R. Baker, Macromolecules, 2002, 35, 4510–4520 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2014 |