Dirty hands: photodynamic killing of human pathogens like EHEC, MRSA and Candida within seconds†
Anja
Eichner
a,
Fernanda Pereira
Gonzales
a,
Ariane
Felgenträger
a,
Johannes
Regensburger
a,
Thomas
Holzmann
b,
Wulf
Schneider-Brachert
b,
Wolfgang
Bäumler
a and
Tim
Maisch
*a
aDepartment of Dermatology, Regensburg University Hospital, 93053 Regensburg, Germany. E-mail: tim.maisch@klinik.uni-regensburg.de
bInstitute for Clinical Microbiology and Hygiene, University of Regensburg, 93053 Regensburg, Germany
First published on 2nd August 2012
Abstract
Hand hygiene is one of the most important interventions for reducing transmission of nosocomial life-threatening microorganisms, like methicillin resistant Staphylococcus aureus (MRSA), enterohemorrhagic Escherichia coli (EHEC) or Candida albicans. All three pathogens have become a leading cause of infections in hospitals. Especially EHEC is causing severe diarrhoea and, in a small percentage of cases, haemolytic-uremic syndrome (HUS) as reported for E. coli 104:H4 in Germany 2011. We revealed the possibility to inactivate very fast and efficiently MRSA, EHEC and C. albicans using the photodynamic approach. MRSA, EHEC and C. albicans were incubated in vitro with different concentrations of TMPyP for 10 s and illuminated with visible light (50 mW cm−2) for 10 and 60 s. 1 μmol l−1 of TMPyP and an applied radiant exposure of 0.5 J cm−2 achieved a photodynamic killing of ≥99.9% of MRSA and EHEC. Incubation with higher concentrations (up to 100 μmol l−1) of TMPyP caused bacteria killing of >5 log10 (≥99.999%) after illumination. Efficient Candida killing (≥99.999%) was achieved first at a higher light dose of 12 J cm−2. Different rise and decay times of singlet oxygen luminescence signals could be detected in Candida cell suspensions for the first time, indicating different oxygen concentrations in the surrounding for the photosensitizer and singlet oxygen, respectively. This confirms that TMPyP is not only found in the water-dominated cell surrounding, but also within the C. albicans cells. Applying a water–ethanol solution of TMPyP on ex vivo porcine skin, fluorescence microscopy of histology showed that the photosensitizer was exclusively localized in the stratum corneum regardless of the incubation time. TMPyP exhibited a fast and very effective killing rate of life-threatening pathogens within a couple of seconds that encourages further testing in an in vivo setting. Being fast and effective, antimicrobial photodynamic applications might become acceptable as a tool for hand hygiene procedures and also in other skin areas.
Introduction
In recent decades, the incidence of infections caused by multidrug-resistant Gram-positive and Gram-negative bacteria as well as Candida albicans has considerably increased not just in terms of morbidity and mortality, but also in terms of health care budgets and prolonging treatment times. At this point, methicillin-resistant Staphylococcus aureus (MRSA) is the most prevalent pathogen in dermatology causing a broad range of pyogenic, community-acquired (CA) and health care-associated (HA), acute and chronic, superficial and deep skin infections which can progress to life-threatening systemic infections. The natural habitat of S. aureus in humans is the nasal cavity.1 Mostly via the hands MRSA colonises or contaminates wider ranges of skin and mucous membranes. In total 20% of the German population are permanently colonised with MRSA and 30–60% intermittently.2 Gram-negative enterohemorrhagic Escherichia coli (EHEC) is an important zoonotic pathogen of humans, causing severe diarrhoea (hemorrhagic colitis) and, in a small percentage of cases, haemolytic-uremic syndrome (HUS). A novel strain of E. coli 104:H4 caused a serious outbreak of food borne illness focused in northern Germany in May through June 2011, but also affected people in many other European countries. The German Robert Koch Institute (RKI) stated that 855 HUS diseases and 2987 EHEC gastroenteritis diseases were diagnosed yielding a total of 3842 cases. Antibiotic therapy has been discouraged after earlier experience indicating a danger of aggravating the disease due to induced or enhanced release of Shiga-toxin and Verotoxin which are critical for the disease and its complications.Within the genera Candida, C. albicans is the most virulent fungal specie, colonizing and infecting predominantly oral mucosal layers.3 Therapeutic options for Candida infections are limited, especially in immunocompromised hosts. Fluconazole, introduced in 1988, has been widely used for treating fungal infections; however, the recently emerged resistance to this drug may limit its use in the future. The use of fluconazol in HIV patients increased azole resistance, since this drug has a fungistatic activity.4
Therefore successful inactivation, decolonisation or sterilization of pathogenic microorganisms is one important goal in a world of increasing multiresistant pathogens in fields like the food industry and medicine.5–8 The use of UV is dangerous and critical because it can damage the DNA structure and induce mutagenesis.9 Chemical agents such as ethylene oxide or hydrogen peroxide for sterilization are limited because of their toxicity to eukaryotic cells.10,11 In clinical applications, a successful decolonisation of MRSA strongly depends on the compliance of the patients and the treatment protocol used. Such a protocol usually consists of a triple therapy for five days of daily nasal mupirocin ointment, chlorhexidine mouth rinse, and full-body wash with chlorhexidine soap.12 Furthermore it has been suggested that hygiene and hand-washing promotion may be one of the most cost-effective interventions for preventing infectious diseases. Recently Judah et al. could demonstrate that 28% of the general adult population have bacteria of faecal origin on their hands in the UK.13 This indicates the importance of hands as routes of transmission of bacteria not only of potential faecal origin but also of multi-resistant pathogens in the broad population and emphasises the development of new techniques for efficient inactivation of such pathogens.
Recently photodynamic treatment of microorganisms has been shown to be very effective in vitro as well as in vivo.14–19 Photodynamic inactivation of microorganisms is based on the concept of positively charged photosensitizers that can attach and/or accumulate in or at the pathogen to induce irreversible damage upon light activation of the photosensitizer.20,21 The absorption of light by a photosensitizer leads to the generation of reactive oxygen species such as singlet oxygen which induces irreversible oxidative damage of the pathogens during illumination.22 The presence of multiple positive charges enables the photosensitizer agent to interact with the negatively charged outer cell wall areas of bacteria, in particular with the negatively charged lipopolysaccarides of Gram-negative bacteria.23 Tetra-cationic porphyrins are very effective against bacteria, like TMPyP (5,10,15,20-tetrakis(1-methylpyridinium-4-yl)-porphyrin tetra p-toluenesulfonate).24 Many light sources have been applied and approved for topical photodynamic tumor treatment in dermatology, such as lasers and incoherent light sources.25 Non-coherent light sources have the advantage of a broad emission spectrum (400 to 1000 nm) in the visible wavelength range, which covers the absorption spectra of many photosensitizers.
As a first step on the way to clinical application, we investigated the inactivation of the pathogens in vitro. MRSA, wildtype E. coli, EHEC and C. albicans were the microbial cells used in this study. By using a non-coherent light source and short incubation times of a few seconds, we aimed to achieve a fast and effective photodynamic inactivation of both bacteria and fungi yielding more than a 3 log10 (≥99.9%) reduction.
Material and methods
Microorganisms
Biochemical analysis and resistance testing of each bacterial strain were done with a VITEK 2 system (bioMérieux, Nuertingen, Germany), according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS, Wayne, PA). A clinical strain of enterohemorrhagic E. coli (EHEC) O104:H4 isolated during the 2011 outbreak in Germany was used as well as wildtype E. coli (ATCC 25922) as the respective control strain. The EHEC strain HUSECO41 (O104:H4) is characterized as follows: shiga toxine stxt1: neg; shiga toxine sxt2: pos; intimin eae: neg; enterohaemolysine hly: neg; heavy metal resistance terD: pos; extended-spectrum β-lactamase (ESBL) producer: pos. The bacterial strains methicillin-sensitive S. aureus (MSSA; ATCC 25923), methicillin-resistant S. aureus (MRSA: ATCC BAA-44 and ATCC 43300) and E. coli (ATCC 25922) were purchased from LGC Standards GmbH (Wesel, Germany). All bacteria were grown aerobically at 37 °C in Mueller-Hinton broth (Gibco Life Technologies GmbH, Eggenstein, Germany).The bacteria of an overnight bacteria culture (5 ml) were harvested by centrifugation (200 g, 15 min), washed with phosphate-buffered saline (PBS; Biochrom, Berlin, Germany) at pH 7.4 containing 0.027 mol l−1 KCl and 0.14 mol l−1 NaCl, and suspended in PBS at an optical density of 0.6 at 600 nm, which corresponded to ∼108 bacteria ml−1, for use in the phototoxicity experiments.
The fungus C. albicans (ATCC-MYA-273, LGC Standards GmbH, Germany) was cultured overnight at 37 °C on a shaker platform (200 rpm) in Sabouraud dextrose broth (SDB) (Sigma Chemical Co., St. Louis, MO). When the cultures reached the stationary phase of growth, the cells were harvested by centrifugation (200 g, 5 min) and washed once with Dulbecco's phosphate-buffered saline (PBS without Ca2+ and Mg2+, PAA Laboratories GmbH, Austria). Then cells were resuspended in PBS, corresponding to approximately 5 × 107 cells ml−1.
Photosensitizer TMPyP
TMPyP (5,10,15,20-tetrakis(1-methylpyridinium-4-yl)-porphyrin tetra p-toluenesulfonate) was purchased from Sigma Aldrich (Taufkirchen, Germany), purity 97%. TMPyP was dissolved in distilled water at a stock concentration of 1 mmol l−1, passed through a 0.22 μm pore-size filter and stored at 4 °C until use. Dilutions were done in PBS-buffer (PAA Laboratories GmbH, Pasching, Austria).Light source and irradiation parameters
A non-coherent light source (Omnicure Series 2000, igb-tech, Friedelsheim, Germany) was used in this study and had the following nominative excitation filter set of 400–500 nm (Fig. 1). The effective radiant exposure of the light source was calculated as follows (see eqn (a)): | (a) |
 | ||
| Fig. 1 Spectral emission of the light source and absorption spectra of TMPyP. The absorption spectrum of 10 μmol l−1 TMPyP (blue) is shown in the range between 350 and 650 nm (Soret band and Q-bands I–IV) and the emission spectra of the used light source in the same range between 300–700 nm (red). The emission wavelength data of the light source were normalized to its maxima at 435.7 nm. OD: optical density of TMPyP. | ||
The tip of the lamp was fixed on a tripod at a constant distance (9 cm) from the samples. For measuring the spectra, the emitted spectrum was recorded with a spectrometer (270M, Jobin Yvon, Longjumeau, France) with 300 grid-lines mm−1 and a spectral resolution of approximately 0.4 nm. The detection range was 300 to 1000 nm. The recorded spectral data were corrected regarding the spectral sensitivity of the spectrometer. The absorption spectrum of 10 μmol l−1 TMPyP (Sigma-Aldrich, Taufkirchen, Germany) was measured in aqua dest.
Detection of singlet oxygen generated by TMPyP
The ability of TMPyP to generate singlet oxygen was qualitatively evaluated using a frequency-doubled Nd:YAG laser (PhotonEnergy, Ottensoos, Germany) as previously described at a wavelength of 532 nm.26,27 Singlet oxygen luminescence generated by 50 μmol l−1 of TMPyP in H2O was detected with an IR-sensitive photomultiplier (R5509-42, Hamamatsu Photonics Deutschland GmbH, Herrsching, Germany) at different wavelengths from 1200 to 1400 nm using a monochromator (Horiba, Yobin Yvon Inc. USA) in front of the multiplier.Singlet oxygen luminescence in C. albicans incubated with TMPyP
The C. albicans cell solution used for the singlet oxygen luminescence detection diluted in PBS had a concentration of 5 × 106 ml−1. C. albicans was incubated with TMPyP gaining a final concentration of 1 μmol l−1, 10 μmol l−1, and 100 μmol l−1 in the solution with a volume of 2 ml of each sample. A PBS concentration of 50% was used in each sample. Immediately after mixing cells with the photosensitizer, the samples were illuminated in acrylic cuvettes (Sarstedt, Nümbrecht, Germany) during magnetic stirring. The laser beam at λ = 420 nm excited the porphyrin TMPyP in its Soret band with a frequency of f = 1 kHz and a laser power of P = 360 mW. The singlet oxygen luminescence at 1270 nm (FWHM = 30 nm, LOT Oriel GmbH, Darmstadt, Germany) was detected in the near-backward direction with respect to the excitation beam using an infrared sensitive photomultiplier. The oxygen concentration of the solution surrounding the cells during the measurement was regulated with a gas-flow into the solution which was measured with a needle sensor (MICROX TX, PreSens GmbH, Regensburg, Germany). In order to determine the rise and decay times of the luminescence signals, the Levenberg–Marquardt-algorithm of Mathematica (Wolfram Research, Champaign, USA) was used.Phototoxicity assay of bacteria
A total of ∼108 bacterial cells per ml were placed in a 96-well microtiter plate (100 μl per well) and incubated with different concentrations of TMPyP (0 μmol l−1, 1 μmol l−1, 10 μmol l−1 and 100 μmol l−1) for 10 s in the dark. Immediately at the end of the incubation period, the bacteria were illuminated with 50 mW cm−2 for 10 or 60 s corresponding to a radiant exposure of 0.5 J cm−2 or 3 J cm−2, respectively. Controls were neither sensitized with TMPyP nor exposed to the light source or were incubated with a photosensitizer only or illuminated only. After illumination the survival of the bacteria was determined by counting the numbers of CFU per ml using the Miles, Misra and Irwin technique.28 Serially diluted aliquots of treated and untreated (no photosensitizer, no light) cells were plated on Mueller-Hinton agar, and the number of CFU per ml was counted after 24 h of incubation at 37 °C.Phototoxicity assay of C. albicans
Candida cell suspensions of 5 × 107 cells ml−1 were incubated in the dark with different concentrations of the photosensitizer (0 to 5 μmol l−1) for 1 min under continuous agitation. A total of 200 μl of solution was transferred to a sterile flat-bottomed 96-well polystyrene plate and illuminated for 4 min (50 mW cm−2, 12.0 J cm−2). Illumination was done from the bottom side of the 96-well plate to avoid refraction of the light in the cell culture media. Control wells were neither sensitized with a photosensitizer nor exposed to the light source or were incubated with a photosensitizer only. Immediately after light exposure, ten-fold serial dilutions in SBD broth were carried out and aliquots of 0.1 ml were seeded onto Sabouraud dextrose agar (SDA) for 48 h at 37 °C for determining the number of colony forming units per ml (CFU ml−1).Normal human epidermal keratinocytes (NHEK) cell culture
NHEK cells were purchased from Gibco Life Technologies (Eggenstein, Germany) and seeded into a T75 cell culture flask with 10 ml of EpiLife medium (Gibco Life Technologies) with supplement HKGS (human keratinocyte growth supplement, Gibco Life Technologies). Cells were incubated at 37 °C in a humidified atmosphere with 5% CO2 (v/v). The medium was replaced every 2 days. Cells were used for experiments after reaching 80% confluence up to the 6th passage. Cells were washed once with 10 ml PBS (Biochrom, Berlin, Germany) and removed from the cell culture flask bottom with 2 ml 0.1% trypsin–EDTA solution (Gibco Life Technologies, Eggenstein, Germany). Cells were seeded into 96 well plates (∼10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) and incubated at 37 °C and 5% CO2 overnight. On the next day, cells were incubated with 0.5, 1, 10 or 100 μmol l−1 TMPyP, respectively, and illuminated for 10 or 60 s with 50 mW cm−2. Afterwards TMPyP solution was removed from each well and cells were incubated with 100 μl of fresh medium overnight at 37 °C and 5% CO2. Aliquots of treated and untreated (no photosensitizer with light, photosensitizer without light, and no photosensitizer/no light) cells were used as control values.
000 cells per well) and incubated at 37 °C and 5% CO2 overnight. On the next day, cells were incubated with 0.5, 1, 10 or 100 μmol l−1 TMPyP, respectively, and illuminated for 10 or 60 s with 50 mW cm−2. Afterwards TMPyP solution was removed from each well and cells were incubated with 100 μl of fresh medium overnight at 37 °C and 5% CO2. Aliquots of treated and untreated (no photosensitizer with light, photosensitizer without light, and no photosensitizer/no light) cells were used as control values.
Cell viability assay
To evaluate the effects of illumination with different concentrations of TMPyP on NHEK cells, cell viability was estimated directly by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) test as described by Mosmann.29 Briefly, 10 μl MTT solution (stock solution of 5 mg ml−1 in PBS, stored at −20 °C) was added to each well containing the treated/untreated keratinocytes. For blank values 10 μl MTT was added to 100 μl medium. After incubation for 4 h at 37 °C and 5% CO2, 100 μl SDS solution (20%) was added to each well and plates were again incubated at 37 °C and 5% CO2 overnight. The absorbance of the produced formazan was recorded using an EMax endpoint ELISA microplate reader (Molecular Devices, USA) at 540 nm. Results were presented as percent viability to the control values.Preparation of porcine skin and tissue penetration and localisation of TMPyP
Fresh skin from six month old female pigs was obtained from a local slaughter house. The excised skin was washed with distilled water, epilated with a dry razor and the adipose tissue beneath the dermis was removed with a scalpel. Then the skin was cut under sterile conditions into 4 × 4 cm2 pieces, placed in a Petri dish and embedded with Hepes-agar (145 mM NaCl, 5 mM KCl, 1 mM MgSO4, 10 mM Hepes, 10 mM glucose, 5% agarose, pH 7.5). A 100 μmol l−1 TMPyP water–ethanol (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) formulation was applied on ex vivo porcine skin for 5, 15, 60 min, and 4 h under occlusion. Thereafter, samples were frozen in liquid nitrogen and kept at −80 °C; one tissue sample was frozen without incubation of TMPyP and served as a control. Serial cryostat sections (5 μm) were cut upwards from dermis to epidermis to avoid smearing and contamination. At different time points the distribution of TMPyP was examined by fluorescence microscopy (Zeiss Axiotech, Goettingen, Germany) using a dual band filter set (Omega Optical, Brattleboro, VT, USA) for excitation (405 ± 40 nm) and emission (600 nm long pass). Fluorescence images were superimposed onto the corresponding light microscopy image with an image processing program (Image-Pro Plus 5.0; Media Cybernetics, Silver Spring, MD, USA).
1 v/v) formulation was applied on ex vivo porcine skin for 5, 15, 60 min, and 4 h under occlusion. Thereafter, samples were frozen in liquid nitrogen and kept at −80 °C; one tissue sample was frozen without incubation of TMPyP and served as a control. Serial cryostat sections (5 μm) were cut upwards from dermis to epidermis to avoid smearing and contamination. At different time points the distribution of TMPyP was examined by fluorescence microscopy (Zeiss Axiotech, Goettingen, Germany) using a dual band filter set (Omega Optical, Brattleboro, VT, USA) for excitation (405 ± 40 nm) and emission (600 nm long pass). Fluorescence images were superimposed onto the corresponding light microscopy image with an image processing program (Image-Pro Plus 5.0; Media Cybernetics, Silver Spring, MD, USA).
Statistical methods
All results are shown as medians, including the 25% and 75% quartiles, which were calculated from the values of at least 3 independent experiments, each experiment was conducted in triplicate, with Prism 4 for Windows (GraphPad Software Inc., San Diego, CA, USA). The calculation was referred to untreated controls, which were neither incubated with TMPyP nor illuminated, so-called dark control (black dotted horizontal line). In Fig. 3–5, medians on or below the dotted horizontal line in red or green represent ≥99.9% efficacy or ≥99.999% of bacteria killing corresponding to at least more than three magnitudes or five magnitudes of log10 reduction compared to matching untreated controls (bacteria, but no light and no photosensitizer). A reduction of at least three magnitudes of log10 of viable median numbers of bacteria was stated as biologically relevant with regard to the guidelines of hand hygiene.30 Percentage of phototoxicity was calculated as follows (eqn (b)):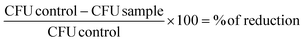 | (b) |
Results
Overlap of the TMPyP absorption spectrum with the emission spectra of the non-coherent light source
Generally, light absorption by TMPyP for wavelengths above 700 nm is very low and the emission wavelengths of the light source were detected between 400–500 nm. Therefore, the spectral overlap of TMPyP absorption and emission of the light source was considered in the range of 400–500 nm, which is shown in Fig. 1. The emission spectrum of the light source closely matches the Soret absorption peak of TMPyP (Fig. 1).Singlet oxygen generation and detection
The singlet oxygen generation by TMPyP upon excitation with visible light is shown in Fig. 2A. The combination of the spectral and time-resolved distribution of the singlet-oxygen luminescence signals detected by the laser/photomultiplier-system confirms the generation of singlet oxygen by TMPyP resulting in a characteristic fingerprint of singlet oxygen, which is in good agreement with already published data for singlet oxygen in aqueous solutions.27 | ||
| Fig. 2 Singlet oxygen fingerprint and singlet oxygen signals of TMPyP in C. albicans. (a) Combination of time resolved measurement of singlet oxygen and wavelength scan by summing up the luminescence signals of singlet oxygen at different wavelengths from 1200 nm to 1400 nm generated by 50 μmol l−1 TMPyP in H2O. (b) Detection of singlet oxygen luminescence in C. albicans cells. A 2 ml suspension of 5 × 106 cells per ml with 10 μmol l−1 TMPyP in 50% PBS was irradiated with laser light at 420 nm with 10k laser pulses. The signal reveals a mono-exponential rise and a bi-exponential decay component. | ||
Singlet oxygen detection in suspension
Detection of singlet oxygen luminescence and its corresponding rise and decay times in a suspension containing the investigated pathogens is a crucial factor to achieve an efficient inactivation of microorganisms. Due to the high cell concentrations the planktonic solutions were flooded with 0.1 l min−1 O2 and 0.4 l min−1 N2 in order to keep a constant amount of oxygen in the solution comparable to air-saturated water, in order to compensate the oxygen consumption of the microorganisms at this high cell concentration.Pure TMPyP with a concentration of 10 μmol l−1 shows a rise and decay time of tR = (2.0 ± 0.2) μs and tD = (3.5 ± 0.4) μs in H2O, which is in accordance with already published data.27 Here tD is the decay of singlet oxygen in H2O, tR is the decay of the triplet-T1-state of TMPyP at an oxygen concentration of [O2] = 2.7 × 10−4 M. In C. albicans planktonic solution only a decay time was detected after the incubation of 1 μmol l−1 TMPyP (tD = (9.2 ± 0.9) μs), whereas there was a rise and decay part for the luminescence signal with 10 μmol l−1 (tR = (2.0 ± 0.2) μs, tD = (4.8 ± 0.5) μs) and 100 μmol l−1 (tR = (2.1 ± 0.2) μs, tD = (4.2 ± 0.4) μs) TMPyP (Fig. 2B). For a 10 μmol l−1 concentration of TMPyP the signal shows decay in a multi-exponential manner and was fitted as a bi-exponential fit in the first part (count 5–700), and a mono-exponential fit in the second part (count 800–1500) (Fig. 2B) with a tD2 = (22 ± 2) μs. The different rise and decay parts indicate different oxygen concentrations in the surrounding for the photosensitizer and singlet oxygen, respectively. This confirms that TMPyP is not only found in the water-dominated cell surrounding, but also within the C. albicans cells. In accordance with already published data we could detect as well luminescence signals of singlet oxygen in bacteria suspensions containing S. aureus or E. coli (data not shown).22
Phototoxicity of TMPyP-sensitized MRSA
Two MRSA strains (BAA-44 and 43 300, respectively) were illuminated with different TMPyP concentrations for 10 or 60 s in vitro. After 10 s of incubation, the cells were exposed to a light dose of 50 mW cm−2 for 10 s (0.5 J cm−1). Hence, both MRSA strains exhibit a ≥3 log10 (≥99.9%) killing rate when TMPyP of 0.5 μmol l−1 was used. Incubation with higher TMPyP concentrations (1, 10 or 100 μmol l−1) revealed a high inactivation efficacy of ≥5 log steps (≥99.999%) upon light activation with 0.5 J cm−1 (Fig. 3A and C). | ||
| Fig. 3 Photosensitized inactivation of MRSA. Survival of MRSA strains BAA-44 (A and B) or 43300 (C and D) incubated with different TMPyP concentrations for 10 s in the dark and followed by illumination with 50 mW cm−2 for (A and C) 10 s (0.5 J cm−2) and (B and D) 60 s (3 J cm−2). Control: bacteria alone or incubated with TMPyP only, but not irradiated. Bars represent the median, including the 25% and 75% quartiles, of three independent experiments. Values on or below the dotted horizontal line represent ≥99.9% (red) or ≥99.999% (green) efficacy of bacteria killing, which was referred to the untreated controls (bacteria alone). | ||
In addition, illumination for 60 s with 50 mW cm−1 (3 J cm−1) and different TMPyP concentrations showed similar results as mentioned above. After incubation of 10 s with TMPyP, both BAA44 and 43 300 strains were reduced by ≥3 log steps (≥99.9% killing rate) when incubated with 0.5 μmol l−1 TMPyP and illuminated for 60 s. The efficient reduction of viable cells was higher when both MRSA strains were exposed to 1, 10 or 100 μmol l−1 TMPyP and a light dose of 3 J cm −2. In this case the killing rate was ≥5 log steps, which means that 99.999% of viable cells were inactivated (Fig. 3B and D).
Incubation of all tested strains without a photosensitizer and with or without light revealed no killing effect even when cells were exposed to a maximal radiant exposure of 3 J cm−2 (Fig. 3A–D).
Phototoxicity of TMPyP-sensitized wildtype and enterohaemorrhagic E. coli
The EHEC strain O104:H4 and wildtype E. coli were incubated with different concentrations of TMPyP for only 10 s. Immediately after incubation, the bacteria were illuminated with 50 mW cm−2 for 10 s (0.5 J cm−2) or 60 s (3 J cm−2). Incubation of wildtype E. coli with TMPyP caused a biologically relevant decrease in CFU ml−1 upon illumination (Fig. 4). A TMPyP concentration of 0.5 μmol l−1 already exhibited a killing efficacy of ∼2 magnitudes of log10 reduction at a radiant exposure of 0.5 J cm−2 (Fig. 4A). Incubation with higher concentrations of TMPyP (10 μmol l−1 or 100 μmol l−1) showed a further decrease in bacterial survival of ≥5 log10 orders (killing efficacy ≥99.999%) upon light activation with 0.5 J cm−2. In addition, E. coli was already killed to more than 99.999% at 1 μmol l−1 of TMPyP when the radiant exposure was increased to 3 J cm−2 (Fig. 4B). | ||
| Fig. 4 Photosensitized inactivation of E. coli. Survival of E. coli incubated with different TMPyP concentrations for 10 s in the dark and followed by illumination with 50 mW cm−2 for (A) 10 s (0.5 J cm−2) and (B) 60 s (3 J cm−2). Control: bacteria alone or incubated with TMPyP only, but not irradiated. Bars represent the median, including the 25% and 75% quartiles, of three independent experiments. Values on or below the dotted horizontal line represent ≥99.9% (red) or ≥99.999% (green) efficacy of bacteria killing, which was referred to the untreated controls (bacteria alone). | ||
In order to investigate whether the observed growth reduction of E. coli was independent of the antibiotic resistance pattern, an EHEC strain was photosensitized with TMPyP for 10 s and illuminated under conditions identical to those used for wildtype E. coli (Fig. 5). Compared to the un-irradiated control group a biologically relevant decrease of 5 log10 (≥99.999% reduction) in CFU per ml of EHEC was detected after incubation (1 μmol l−1 TMPyP) and irradiation with 0.5 J cm−2 (Fig. 5A).
 | ||
| Fig. 5 Photosensitized inactivation of EHEC. Survival of EHEC incubated with different TMPyP concentrations for 10 s in the dark and followed by illumination with 50 mW cm−2 for (A) 10 s (0.5 J cm−2) and (B) 60 s (3 J cm−2). Control: bacteria alone or incubated with TMPyP only, but not irradiated. Bars represent the median, including the 25% and 75% quartiles, of three independent experiments. Values on or below the dotted horizontal line represent ≥99.9% (red) or ≥99.999% (green) efficacy of bacteria killing, which was referred to the untreated controls (bacteria alone). | ||
Furthermore, illumination with 3 J cm−1 of TMPyP-sensitized EHEC at concentrations identical to those in the experiments mentioned before revealed a greater decrease in CFU per ml already at a TMPyP concentration of 0.5 μmol l−1 compared to an applied radiant exposure of 0.5 J cm−1 (Fig. 5Aversus 5B). Furthermore efflux systems may influence photodynamic efficacy in bacteria. Therefore a fluoroquinolone resistant E. coli strain was photosensitized. Photodynamic killing efficacy was not influenced using such an efflux pump overexpression strain, indicating that such an antibiotic resistant phenotype is a target for the photodynamic process (data not shown).
Overall, all bacterial samples that were incubated without a photosensitizer showed no impairment of growth, with or without illumination. Obviously, the maximal radiant exposure applied (3 J cm−2) had no antibacterial effect (Fig. 4 and 5).
Phototoxicity of TMPyP-sensitized Candida
Using the same parameters as used for bacterial cells, the best conditions found to photokill C. albicans cells were when the cells were 10 and 60 s with 10 μmol l−1 of TMPyP and an illumination with 3 J cm−2. Under these conditions, the cells suffered only a 2 log10 reduction in cell survival (data not shown). Increasing the dose light to 12 J cm−2, 1 μmol l−1 of TMPyP caused the same killing effect (≥99.999%) as for the other bacteria tested (Fig. 6). | ||
| Fig. 6 Photosensitized inactivation of C. albicans. Survival of C. albicans incubated with different TMPyP concentrations for 60 s in the dark and followed by illumination with 50 mW cm−2 for 240 s (12 J cm−2). Control: C. albicans alone or incubated with TMPyP only, but not irradiated. | ||
Cell toxicity of TMPyP and light
The phototoxicity of different concentrations of TMPyP against human keratinocytes (NHEKs) was tested after illumination. As shown in Table 1, incubation of NHEKs with TMPyP yielded reduced cell viability upon illumination. However, NHEKs incubated with 10 μM and illuminated with 0.5 J cm−2 or 3 J cm−2 did show a decrease in viability to 77.2% and 38.2% respectively. Importantly the same parameters induced a killing efficacy of 5 log10 steps (>99.999%) (Fig. 3–5).Tissue penetration and localization of TMPyP
In order to find appropriate parameters to inactivate microorganisms without harming the surrounding tissue, the penetration and localisation of TMPyP was investigated using an ex vivo porcine skin model. As shown by fluorescence microscopy of skin slices, the distribution of the used TMPyP water–ethanol formulation was restricted to the stratum corneum without accumulation in deeper parts of the epidermis or dermis after 60 min of the incubation (Fig. 7). Longer incubation (up to 4 h) did not further enhance penetration efficacy of TMPyP as determined by fluorescence microscopy (data not shown). | ||
Fig. 7 Tissue penetration and localisation of TMPyP. 100 μl TMPyP (100 μmol l−1), diluted in a water–ethanol (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) formulation, was incubated for 60 min on the porcine skin under occlusion. Red: fluorescence signals of TMPyP (bar 100 μm). 1 v/v) formulation, was incubated for 60 min on the porcine skin under occlusion. Red: fluorescence signals of TMPyP (bar 100 μm). | ||
Discussion
Since the dramatic worldwide increase of antibiotic resistance both inside and outside of health care settings, new strategies to control infection are of interest. At present, a wide variety of antibiotics, antiseptics and disinfectants are in use where bacteria achieve multiple drug resistance.31–33 Various studies have shown that photodynamic inactivation of bacteria with light in the visible wavelength range and porphyrin-based photosensitizers, like TMPyP, exhibit significant photosensitizing activities against a broad range of pathogens only in the presence of light and oxygen. A disadvantage might be the long time treatments, which includes the incubation and irradiation procedure.34,35 The results of the present study show that the use of a commercially available light source (400–500 nm) in combination with the photosensitizer TMPyP is able to inactivate multiresistant bacteria, like MRSA and EHEC, but also Candida albicans fast and effectively at the same time.Using a low TMPyP concentration of 1 μmol l−1, the short treatment time, which includes 10 s for incubation and 10 s for irradiation, was effective in killing 5 log10 of viable bacteria. Furthermore no thermal damage of bacteria was observed, because irradiation alone did not affect viability as compared to untreated controls (no light and no photosensitizer). Thus, bacteria killing was solely based on photodynamic action. Firstly, it is important that the emission spectrum of the used light source sufficiently matches the absorption spectra of TMPyP. In our study, the Soret band of TMPyP absorbs effectively the emitted light of the non-coherent light source. Therefore the short illumination time of 10 s was sufficient to run the photodynamic process using TMPyP, which generated reactive oxygen species such as singlet oxygen via the type-II mechanism. In accordance with already published data, here in this study a characteristic fingerprint of singlet oxygen was detected upon excitation of TMPyP at 532 nm 27. The quantum yield of singlet oxygen of TMPyP is ∼77%, which is high compared to other known photosensitizers,36–38 like methylene blue 52%39 or photofrin 36%.40 Taking these facts into account, the efficient generation of singlet oxygen by TMPyP and the interaction of singlet oxygen with the energy-producing membrane area systems of bacteria led to a dissipation of the membrane potential and the striking antimicrobial effect.34
Besides antimicrobial PDT, copper ions and silver ions are used as alternative disinfectants where the use of standard disinfectants such as chlorine or ethylene dioxide may result in the formation of toxic by-products or cause corrosion of surfaces. Both microbial tolerance and resistance to metal ions have been reported, because resistance factors for metals tend to be present on the same plasmids as many antibiotic resistance genes.41,42 Active efflux of metal cations is a frequently utilized strategy of bacteria to get resistance by lowering the intracellular concentration to subtoxic concentrations.43 Therefore, occurrence of resistance to antibiotics and metal ions is possible. In addition such multiple drug resistance is spread within the bacterial community and this situation is leading to pathogens potentially resistant to any available antibiotic.44 In general, photodynamic killing efficacy was not influenced using a porphyrin-based photosensitizer against either a fluoroquinolon resistant E. coli strain (efflux pump overexpression) or its wildtype.
So far, no bacteria strain is known which has developed resistance against the photodynamic process. Grinholc et al. could demonstrate that various porphyrin-based photosensitizers tested executed their antibacterial activity with no change in the antibiotic resistance pattern of the studied strains.45 So far, only the bactericidal effect of photodynamic inactivation was strain-dependent, which means that different photodynamic parameters (applied radiant exposure, concentration, and incubation time) are necessary to achieve complete eradication of different methicillin-resistant and methicillin-susceptible S. aureus strains tested.46 Furthermore, the strain dependent differences of bactericidal photodynamic inactivation could not be correlated either to the levels of photosensitizer uptake or the pharmacological inhibition of the efflux pump.47 Therefore other factors might be influencing such observed differences in the photodynamic efficacy such as aggregation of the photosensitizer, concentration of antioxidant enzymes or cellular repair proteins, but all these factors are not responsible to diminish TMPyP induced photosensitization, because up to now there is a lack of selection of photo-resistant bacteria after multiple photodynamic treatments.48 Thus, the advantage of such a very short photodynamic treatment time of seconds is that it might not allow bacteria to react on the photodynamic process to attain resistance.
Recently the use of bacteriophages has been investigated as a new tool to control pathogenic microorganisms.49–52 A mixture of bacteriophages was used to inactivate EHEC dependent on increasing values of incubation time, temperature, and multiplicity of infection units.49 At temperatures above 12 °C there was more than a 3 log10 CFU reduction within 10 min of applying bacteriophages on EHEC. Lower temperatures lead to a lesser efficacy of inactivation. However other studies demonstrated that bacteriophages were able to inactivate Salmonella enteritidis or Campylobacter jejuni under certain conditions in the absence of or very limited bacterial growth at low temperatures.50,53,54 In the case of photodynamic inactivation, temperature is not a limitation factor to achieve maximal inactivation of up to 5 log10. This could be an advantage for the photodynamic approach to be effective in killing pathogens where low temperatures play a restrictive role regarding food storage to prevent contamination. However outbreaks by EHEC can be sometimes caused by contaminated foods or clinical devices with a low number of bacteria (e.g. <200 CFU g−1).55 Here the levels of E. coli 104:H4 used in our study were 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 1
000 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 higher than those found in naturally contaminated environments. Nevertheless the combination of TMPyP and blue light was successful to inactivate such high numbers of CFU of EHEC. E. coli 104:H4 cells were grown to stationary phase and then treated by the photodynamic process. This was done to simulate a more rigorous condition as found usually in food processing settings in which EHEC could have been in better growing conditions because of access to nutrients.
000 higher than those found in naturally contaminated environments. Nevertheless the combination of TMPyP and blue light was successful to inactivate such high numbers of CFU of EHEC. E. coli 104:H4 cells were grown to stationary phase and then treated by the photodynamic process. This was done to simulate a more rigorous condition as found usually in food processing settings in which EHEC could have been in better growing conditions because of access to nutrients.
It is also known that bacteria are more susceptible to the photodynamic oxidative burst during exponential phase growth.56 Here we demonstrate that the photodynamic process is able to kill EHEC under stationary phase conditions very efficiently. However inactivation of midexpontial phase growing EHEC would be possibly still better under these conditions. It is imperative that further work is conducted to determine whether other pathogenic bacteria, such as Salmonella sp., and Listeria monocyteogenes, which are relevant in contamination of food processing equipment and facilities, can be inactivated by the photodynamic process.
The susceptibility of C. albicans to photodynamic inactivation with TMPyP and blue light was also tested. A higher concentration of TMPyP to kill C. albicans cells when compared to the concentration needed to kill bacterial cells was necessary. The difference in the susceptibility to photodynamic killing between Gram-positive and Gram-negative bacteria and fungi may be due to the presence of a nuclear membrane in fungal cells protecting the nucleus and acting as a barrier either to the penetration of photosensitizers or the reactive oxygen species formed during the treatment. Besides, C. albicans cells are larger than bacterial cells, presenting more targets to be damaged, consequently the amount of reactive oxygen species necessary to kill yeasts is much greater than the amount necessary to kill bacteria.57,58 These results are in agreement with the data report by Demidova et al., which shows that C. albicans was the hardest to inactivate when compared with a Gram-positive and Gram-negative strain.57
Tissue penetration and localization of TMPyP were evaluated using a porcine skin model concerning the safety of a potential topical application of TMPyP in the future. No fluorescence signals of TMPyP were detected inside the epidermis or dermis which implies a “therapeutic window” for an efficient decolonisation of pathogens on the stratum corneum without side effects for the underlying tissue. Such an ex vivo skin model can be used as a good test for human skin based on many similarities in behaviour, renewal, physiological and permeability properties.59,60 Inherent washing and cleaning of skin surfaces of patients and health care workers may be partially responsible for current problems associated with hospital-acquired infections.61
In this study, singlet oxygen was measured directly by its luminescence inside living Candida albicans cells for the first time. Different rise and decay times were measured indicating different oxygen concentrations in the surrounding for the photosensitizer and singlet oxygen, respectively. This confirms that TMPyP is not only found in the water-dominated cell surrounding, but also within the C. albicans cells. This is in accordance with previous data where we could detect as well luminescence signals of singlet oxygen in bacteria suspensions containing S. aureus or E. coli.22 This observation is important to optimise the efficacy of a given photosensitizer for inactivation of multiresistant pathogens.
Besides medical application of photodynamic based antimicrobial technology, this procedure should be also feasible in environmental technology. Industrial disinfection needs processes with short application times along with simplicity, efficacy and low costs. Here we could show a photodynamic killing efficacy of up to 5 log10 steps within seconds. Considering safety, the use of light activating molecules is safe due to the used wavelengths being part of the visible light spectrum and coupled with the fact that photodynamic inactivation occurs only when the light is switched on does not require additional chemical pre-treatments. The photodynamic process for microbial inactivation makes it very attractive for a range of potential decontamination applications, like horizontal surfaces and items, where liquid disinfectants cannot be used, because protection of corrosive material is needed.
Conclusion
Overall, the results of this study clearly demonstrate for the first time that TMPyP in combination with visible light is able to induce a photodynamic process to inactivate multi-resistant bacteria like MRSA and EHEC as well as C. albicans very fast and efficiently. It should be emphasized that TMPyP has no clinical approval to be used as a medical drug. However, these results should encourage the development of similar porphyrin molecules with similar properties that can be approved for use in humans in the near future. The rapid inactivation procedure within seconds suggests that it may be useful as an effective tool for clinical and industrial purposes where savings in time is a critical point to achieve efficient inactivation of microorganisms on applied surfaces.Acknowledgements
The excellent technical assistance of Ewa Kowalewski and Francesco Santarelli is gratefully acknowledged. No conflict of interest is declared.References
- J. Kluytmans, A. van Belkum and H. Verbrugh, Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks, Clin. Microbiol. Rev., 1997, 10, 505–520 CAS.
- R. Kock, A. Mellmann, F. Schaumburg, A. W. Friedrich, F. Kipp and K. Becker, The epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) in Germany, Dtsch. Arztebl. Int., 2011, 108, 761–767 Search PubMed.
- Y. H. Samaranayake, B. P. Cheung, N. Parahitiyawa, C. J. Seneviratne, J. Y. Yau, K. W. Yeung and L. P. Samaranayake, Synergistic activity of lysozyme and antifungal agents against Candida albicans biofilms on denture acrylic surfaces, Arch. Oral Biol., 2009, 54, 115–126 CrossRef CAS.
- D. M. Arana, C. Nombela and J. Pla, Fluconazole at subinhibitory concentrations induces the oxidative- and nitrosative-responsive genes TRR1, GRE2 and YHB1, and enhances the resistance of Candida albicans to phagocytes, J. Antimicrob. Chemother., 2010, 65, 54–62 CrossRef CAS.
- S. Mathur and R. Singh, Antibiotic resistance in food lactic acid bacteria–a review, Int. J. Food Microbiol., 2005, 105, 281–295 CrossRef CAS.
- J. R. Bower, Foodborne diseases: Shiga toxin producing E. coli (STEC), Pediatr. Infect Dis. J., 1999, 18, 909–910 CrossRef CAS.
- P. C. Appelbaum, The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus, Clin. Microbiol. Infect., 2006, 12(Suppl 1), 16–23 Search PubMed.
- J. Y. Ang, E. Ezike and B. I. Asmar, Antibacterial resistance, Indian J. Pediatr., 2004, 71, 229–239 Search PubMed.
- R. M. Chapple, B. Inglis and P. R. Stewart, Lethal and mutational effects of solar and UV radiation on Staphylococcus aureus, Arch. Microbiol., 1992, 157, 242–248 CrossRef CAS.
- W. M. Snellings, C. S. Weil and R. R. Maronpot, A two-year inhalation study of the carcinogenic potential of ethylene oxide in Fischer 344 rats, Toxicol. Appl. Pharmacol., 1984, 75, 105–117 CrossRef CAS.
- J. A. Imlay, S. M. Chin and S. Linn, Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivoin vitro, Science, 1988, 240, 640–642 CAS.
- B. V. Krishna and A. P. Gibb, Use of octenidine dihydrochloride in methicillin-resistant Staphylococcus aureus decolonisation regimens: a literature review, J. Hosp. Infect., 2010, 74, 199–203 CrossRef CAS.
- G. Judah, P. Donachie, E. Cobb, W. Schmidt, M. Holland and V. Curtis, Dirty hands: bacteria of faecal origin on commuters’ hands, Epidemiol. Infect., 2010, 138, 409–414 Search PubMed.
- F. Gad, T. Zahra, K. P. Francis, T. Hasan and M. R. Hamblin, Targeted photodynamic therapy of established soft-tissue infections in mice, Photochem. Photobiol. Sci., 2004, 3, 451–458 RSC.
- N. Komerik, H. Nakanishi, A. J. MacRobert, B. Henderson, P. Speight and M. Wilson, In vivo killing of Porphyromonas gingivalis by toluidine blue-mediated photosensitization in an animal model, Antimicrob Agents Chemother., 2003, 47, 932–940 CrossRef CAS.
- T. Maisch, J. Wagner, V. Papastamou, H. J. Nerl, K. A. Hiller, R. M. Szeimies and G. Schmalz, Combination of 10% EDTA, Photosan, and a blue light hand-held photopolymerizer to inactivate leading oral bacteria in dentistry in vitro, J. Appl. Microbiol., 2009, 107, 1569–1578 CrossRef CAS.
- A. S. Garcez, S. C. Nunez, M. R. Hamblin and M. S. Ribeiro, Antimicrobial effects of photodynamic therapy on patients with necrotic pulps and periapical lesion, J. Endod., 2008, 34, 138–142 Search PubMed.
- A. S. Garcez, S. C. Nunez, J. L. Lage-Marques, M. R. Hamblin and M. S. Ribeiro, Photonic real-time monitoring of bacterial reduction in root canals by genetically engineered bacteria after chemomechanical endodontic therapy, Braz Dent. J., 2007, 18, 202–207 Search PubMed.
- X. Ragas, T. Dai, G. P. Tegos, M. Agut, S. Nonell and M. R. Hamblin, Photodynamic inactivation of Acinetobacter baumannii using phenothiazinium dyes: in vitroin vivo studies, Lasers Surg. Med., 2011, 42, 384–390.
- Y. Nitzan, R. Dror, H. Ladan, Z. Malik, S. Kimel and V. Gottfried, Structure–activity relationship of porphines for photoinactivation of bacteria, Photochem. Photobiol., 1995, 62, 342–347 CAS.
- E. Alves, L. Costa, C. M. Carvalho, J. P. Tome, M. A. Faustino, M. G. Neves, A. C. Tome, J. A. Cavaleiro, A. Cunha and A. Almeida, Charge effect on the photoinactivation of Gram-negative and Gram-positive bacteria by cationic meso-substituted porphyrins, BMC Microbiol., 2009, 9, 70 CrossRef.
- T. Maisch, J. Baier, B. Franz, M. Maier, M. Landthaler, R. M. Szeimies and W. Baumler, The role of singlet oxygen and oxygen concentration in photodynamic inactivation of bacteria, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 7223–7228 CrossRef CAS.
- M. Merchat, G. Bertolini, P. Giacomini, A. Villanueva and G. Jori, Meso-substituted cationic porphyrins as efficient photosensitizers of gram-positive and gram-negative bacteria, J. Photochem. Photobiol., B., 1996, 32, 153–157 CrossRef CAS.
- M. Salmon-Divon, Y. Nitzan and Z. Malik, Mechanistic aspects of Escherichia coli photodynamic inactivation by cationic tetra-meso(N-methylpyridyl)porphine, Photochem. Photobiol. Sci., 2004, 3, 423–429 RSC.
- L. Brancaleon and H. Moseley, Laser and non-laser light sources for photodynamic therapy, Lasers Med. Sci., 2002, 17, 173–186 CrossRef CAS.
- J. Regensburger, T. Maisch, A. Felgentrager, F. Santarelli and W. Baumler, A helpful technology–the luminescence detection of singlet oxygen to investigate photodynamic inactivation of bacteria (PDIB), J. Biophotonics., 2010, 3, 319–327 CrossRef CAS.
- J. Baier, T. Fuß, C. Pöllmann, C. Wiesmann, K. Pindl, R. Engl, D. Baumer, M. Maier, M. Landthaler and W. Bäumler, Theoretical and experimental analysis of the luminescence signal of singlet oxygen for different photosensitizers, J. Photochem. Photobiol. B: Biol., 2007, 87, 163–173 CrossRef CAS.
- A. A. Miles, S. S. Misra and J. O. Irwin, The estimation of the bactericidal power of the blood, J. Hyg. (Lond)., 1938, 38, 732–749 Search PubMed.
- T. Mosmann, Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays, J. Immunol. Methods., 1983, 65, 55–63 CrossRef CAS.
- J. M. Boyce and D. Pittet, Guideline for Hand Hygiene in Health-Care Settings. Recommendations of the Healthcare Infection Control Practices Advisory Committee and the HIPAC/SHEA/APIC/IDSA Hand Hygiene Task Force, Am. J. Infect. Control., 2002, 30, S1–S46.
- S. B. al-Masaudi, M. J. Day and A. D. Russell, Antimicrobial resistance and gene transfer in Staphylococcus aureus, J. Appl. Bacteriol., 1991, 70, 279–290 CAS.
- F. Baquero, J. F. Barrett, P. Courvalin, I. Morrissey, L. Piddock and W. J. Novick, Epidemiology and mechanisms of resistance among respiratory tract pathogens, Clin. Microbiol. Infect., 1998, 4(Suppl 2), S19–S26 Search PubMed.
- L. Birosova and M. Mikulasova, Development of triclosan and antibiotic resistance in Salmonella enterica serovar Typhimurium, J. Med. Microbiol., 2009, 58, 436–441 CrossRef CAS.
- K. Komagoe, H. Kato, T. Inoue and T. Katsu, Continuous real-time monitoring of cationic porphyrin-induced photodynamic inactivation of bacterial membrane functions using electrochemical sensors, Photochem. Photobiol. Sci., 2011, 10, 1181–1188 RSC.
- T. Maisch, C. Bosl, R. M. Szeimies, N. Lehn and C. Abels, Photodynamic effects of novel XF porphyrin derivatives on prokaryotic and eukaryotic cells, Antimicrob. Agents Chemother., 2005, 49, 1542–1552 CrossRef CAS.
- T. Breitenbach, M. K. Kuimova, P. Gbur, S. Hatz, N. B. Schack, B. W. Pedersen, J. D. Lambert, L. Poulsen and P. R. Ogilby, Photosensitized production of singlet oxygen: spatially-resolved optical studies in single cells, Photochem. Photobiol. Sci., 2009, 8, 442–452 RSC.
- R. F. Pfeltz, V. K. Singh, J. L. Schmidt, M. A. Batten, C. S. Baranyk, M. J. Nadakavukaren, R. K. Jayaswal and B. J. Wilkinson, Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds, Antimicrob. Agents Chemother., 2000, 44, 294–303 CrossRef CAS.
- V. Gottfried, D. Peled, J. W. Winkelman and S. Kimel, Photosensitizers in organized media: singlet oxygen production and spectral properties, Photochem. Photobiol., 1988, 48, 157–163 CrossRef CAS.
- Y. Usui, Determination of quantum yield of Singlet Oxygen Formation by Photosensitization, Chem. Lett., 1973, 7, 743–744 CrossRef.
- R. Pottier, A. Lachaine, M. Pierre and J. C. Kennedy, A new electronic absorbance band in concentrated aqueous solutions of hematoporphyrin IX detected by photoacoustic spectroscopy, Photochem. Photobiol., 1988, 47, 669–674 CrossRef CAS.
- I. J. Davis, H. Richards and P. Mullany, Isolation of silver- and antibiotic-resistant Enterobacter cloacae from teeth, Oral Microbiol. Immunol., 2005, 20, 191–194 CrossRef CAS.
- J. T. Trevors, Copper resistance in bacteria, Microbiol. Sci., 1987, 4, 29–31 Search PubMed.
- S. Silver, Bacterial resistances to toxic metal ions–a review, Gene, 1996, 179, 9–19 CrossRef CAS.
- M. M. Huycke, D. F. Sahm and M. S. Gilmore, Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future, Emerg. Infect. Dis., 1998, 4, 239–249 Search PubMed.
- M. Grinholc, B. Szramka, K. Olender and A. Graczyk, Bactericidal effect of photodynamic therapy against methicillin-resistant Staphylococcus aureus strain with the use of various porphyrin photosensitizers, Acta Biochim. Pol., 2007, 54, 665–670 CAS.
- M. Grinholc, B. Szramka, J. Kurlenda, A. Graczyk and K. P. Bielawski, Bactericidal effect of photodynamic inactivation against methicillin-resistant and methicillin-susceptible Staphylococcus aureus is strain-dependent, J. Photochem. Photobiol., B, 2008, 90, 57–63.
- M. Grinholc, M. Richter, J. Nakonieczna, G. Fila and K. P. Bielawski, The connection between agr and SCCmec elements of Staphylococcus aureus strains and their response to photodynamic inactivation, Photomed. Laser Surg., 2011, 29, 413–419 CrossRef CAS.
- G. Jori, C. Fabris, M. Soncin, S. Ferro, O. Coppellotti, D. Dei, L. Fantetti, G. Chiti and G. Roncucci, Photodynamic therapy in the treatment of microbial infections: basic principles and perspective applications, Lasers Surg. Med., 2006, 38, 468–481 CrossRef.
- S. Viazis, M. Akhtar, J. Feirtag and F. Diez-Gonzalez, Reduction of Escherichia coli O157:H7 viability on hard surfaces by treatment with a bacteriophage mixture, Int. J. Food Microbiol., 2011, 145, 37–42 CrossRef.
- D. Goode, V. M. Allen and P. A. Barrow, Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages, Appl. Environ. Microbiol., 2003, 69, 5032–5036 CrossRef CAS.
- M. Cislo, M. Dabrowski, B. Weber-Dabrowska and A. Woyton, Bacteriophage treatment of suppurative skin infections, Arch. Immunol. Ther. Exp. (Warsz), 1987, 35, 175–183 Search PubMed.
- S. Guenther, D. Huwyler, S. Richard and M. J. Loessner, Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods, Appl. Environ. Microbiol., 2009, 75, 93–100 CrossRef CAS.
- R. Modi, Y. Hirvi, A. Hill and M. W. Griffiths, Effect of phage on survival of Salmonella enteritidis during manufacture and storage of cheddar cheese made from raw and pasteurized milk, J. Food Prot., 2001, 64, 927–933 CAS.
- B. Leverentz, W. S. Conway, Z. Alavidze, W. J. Janisiewicz, Y. Fuchs, M. J. Camp, E. Chighladze and A. Sulakvelidze, Examination of bacteriophage as a biocontrol method for salmonella on fresh-cut fruit: a model study, J. Food Prot., 2001, 64, 1116–1121 CAS.
- J. P. Nataro, C. A. Bopp, P. I. Fields, J. B. Kaper and N. A. Strockbine, Escherichia, Shigella, and Salmonella, ASM Press, Washington, DC, USA, 9th edn., 2007 Search PubMed.
- F. Gad, T. Zahra, T. Hasan and M. R. Hamblin, Effects of growth phase and extracellular slime on photodynamic inactivation of gram-positive pathogenic bacteria, Antimicrob. Agents Chemother., 2004, 48, 2173–2178 CrossRef CAS.
- T. N. Demidova and M. R. Hamblin, Effect of cell-photosensitizer binding and cell density on microbial photoinactivation, Antimicrob. Agents Chemother., 2005, 49, 2329–2335 CrossRef CAS.
- B. Zeina, J. Greenman, W. M. Purcell and B. Das, Killing of cutaneous microbial species by photodynamic therapy, Br. J. Dermatol., 2001, 144, 274–278 CrossRef CAS.
- W. Meyer, R. Schwarz and K. Neurand, The skin of domestic mammals as a model for the human skin, with special reference to the domestic pig, Curr. Probl. Dermatol., 1978, 7, 39–52 Search PubMed.
- G. A. Simon and H. I. Maibach, The pig as an experimental animal model of percutaneous permeation in man: qualitative and quantitative observations–an overview, Skin Pharmacol. Appl. Skin Physiol., 2000, 13, 229–234 CrossRef CAS.
- M. Whitby, M. L. McLaws and M. W. Ross, Why healthcare workers don't wash their hands: a behavioral explanation, Infect Control Hosp. Epidemiol., 2006, 27, 484–492 Search PubMed.
Footnote |
| † This article is published as part of a themed issue on current topics in photodermatology. |
| This journal is © The Royal Society of Chemistry and Owner Societies 2013 |
