Advances in magnetic tweezers for single molecule and cell biophysics
Devrim
Kilinc
and
Gil U.
Lee
*
UCD Nanomedicine Centre, School of Chemistry and Chemical Biology, University College Dublin, Belfield, Dublin 4, Ireland. E-mail: gil.lee@ucd.ie; Fax: +353-1-716-1178; Tel: +353-1-716-2399
First published on 14th November 2013
Abstract
Magnetic tweezers (MTW) enable highly accurate forces to be transduced to molecules to study mechanotransduction at the molecular or cellular level. We review recent MTW studies in single molecule and cell biophysics that demonstrate the flexibility of this technique. We also discuss technical advances in the method on several fronts, i.e., from novel approaches for the measurement of torque to multiplexed biophysical assays. Finally, we describe multi-component nanorods with enhanced optical and magnetic properties and discuss their potential as future MTW probes.
Insight, innovation, integrationMagnetic tweezers is a powerful single molecule biophysics technique with unique properties that enable the application of force or torque to biological specimen. Single molecule measurements provide insight on the mechanics of biological entities towards discovering their structure and function. Recent advances in the method in multiple fronts resulted in sophisticated instruments that can twist, stretch, or bend biological specimen from single molecules to individual cells to organisms. In parallel, new classes of magnetic particles with enhanced magnetic and optical properties are emerging. The integration of these novel magnetic particles into existing measurement modalities will potentially enhance the method towards higher resolution, higher throughput systems that are suitable for diagnostic applications. |
Mechanical manipulation of biological material, from proteins to tissue, is receiving growing interest in biomedical research since it provides quantitative information on how mechanical stimuli regulate a multitude of biological processes from enzyme activity to organ development.1 Among the techniques developed to apply force to biological entities, magnetic tweezers (MTW) has claimed particular attention as it is capable of applying torque, it can be fully integrated with a range of powerful light microscopy imaging modes, and it offers high force, spatial, and temporal sensitivity.2 In this review, we present a brief summary of recent measurements made with MTW and highlight several technical advances that are taking place in MTW instrumentation.
The MTW technique is based on the non-invasive manipulation of magnetic particles, and thereby the biological entity they are attached to, via an externally-imposed magnetic field and/or field gradient. A typical MTW setup uses a light microscope to track the position of a magnetic particle in an external magnetic field. The majority of magnetic particles used in MTW studies are superparamagnetic (SPM) or weakly ferromagnetic i.e., they are composites of 20–90% by weight Fe3O4 or Fe2O3 nanoparticles embedded in a polymeric matrix. The superparamagnetic or ferromagnetic properties of the particles are defined by the size of the nanoparticles they contain.3 When placed in an external magnetic field H, a particle with a magnetic moment m results in magnetic induction B = μ0(H + m/V), where μ0 is the vacuum permeability and V is its volume. The particle is subjected to a torque τ = m × B that tends to align the particle's magnetic moment with the field. If the field has a gradient, ∇H, the particle is subjected to a force F = (m·∇)B directed towards regions with higher field density (illustrated for a SPM particle in Fig. 1A). The force can be expressed as F = Vχ∇(B2/2μ0) where χ is the effective susceptibility of the particle, indicating that the magnetic force scales with the volume and the magnetic susceptibility of the particle. Ferromagnetic nanoparticles are preferred in applications where the external magnetic field is weak and particle size is limited, due to their high saturation magnetization. Spherical SPM particles with 0.1–100 μm diameter and a large selection of chemically modified surfaces are commercially available. We have recently demonstrated the emulsion-templated synthesis that results in SPM beads with six times higher magnetization compared to commercial beads.3,4
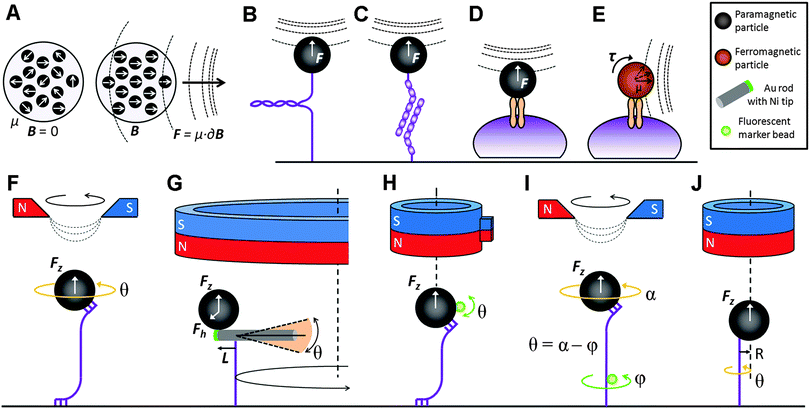 | ||
| Fig. 1 Superparamagnetic beads contain magnetic nanoparticles whose magnetic moments are randomly oriented. In a magnetic field, these domains align with the field gradient and a pulling force is generated (A). Examples of MTW stretch experiments include single molecules (B); inter-molecular bonds (C); and transmembrane proteins (D). Ferromagnetic particles can apply torque on a transmembrane protein when exposed to a magnetic field parallel to the cell surface (E). Torque can be applied to a constrained single molecule by rotating the magnetic field (F). A bead-rod hybrid probe (ref. 34) can apply torque by circling the probe around the magnet centerline (G). A soft torsional trap can also be generated via a side magnet attached to a cylindrical magnet (ref. 36) (H). The twist in the molecule can be measured by the relative rotation of a fluorescent rotor bead attached to it (ref. 25) (I), or by observing the free rotation of the bead about the axis of the molecule due to the point contact between the two (ref. 37) (J). | ||
With the exception of PicoTwist (http://www.picotwist.com/), MTW systems are not commercially available; therefore, researchers typically build their own magnets around a particular microscope. The large variation in the MTW setups found in the literature demonstrates the flexibility of the method, where the configuration is typically built to answer the biophysical question at hand. Permanent magnet assemblies, electromagnets, or combinations thereof are used as field sources. Rare earth permanent magnets have been widely used to build MTW systems as they have saturation magnetization of 1.3 T and are available in a range of shapes. Electromagnets, where the field is generated by passing electrical current through a coil, usually contain para- or ferro-magnetic yokes with a sharpened tip to increase the local field gradient. Electromagnets are capable of generating 0.5 T fields and allow time varying forces to be applied to a sample. Depending on the magnet configuration, the applied force can be perpendicular to the biological sample providing a stretch (Fig. 1B–D),4 or be in parallel with the biological sample providing a twist (Fig. 1E).5 A force calibration step is required for each type of magnetic particle to be used as a MTW probe. The calibration is typically conducted by estimating the drag force acting on a particle from its ultimate velocity in a viscous fluid4 or by measuring the thermal fluctuation of the particle position along the force direction if the particle is tethered to a surface. The latter method is non-trivial and requires a high sampling rate for an accurate force measurement. The reader is referred to a recent protocol describing the construction and calibration of a MTW system.6
Magnetic tweezers in biophysics
MTW has been used to generate force and torque to measure the mechanical properties of biological samples, from individual molecules7 to inter-molecular bonds8 to whole cells.9 Cell stiffness, for example, may indicate the metastatic potential of cancer cells.25 Apart from being a mechanical characterization tool, MTW can also be used to manipulate a biomolecule to discover its function. The function of a biomolecule may be altered with externally applied force or torque through changing its conformation or folding state7 and/or exposing its cryptic sites that are hidden in the native state. The proteolysis rate of the trimeric collagen by matrix metalloproteinase-1 (MMP-1), for example, increases exponentially when the molecule is stretched, suggesting a buried site in the relaxed state that is preferentially cleaved by MMP-1.20 Detailed analysis of protein folding states in response to constant stretching force provides additional insight on the mechanosensitive domains of the cytoskeletal crosslinker protein filamin A and their different conformational states that emerge when unfolded.16A major advantage of MTW over other biophysical methods is that its interference with the specimen is minimal. This feature becomes particularly important in cellular mechanotransduction studies. Integrins, a large family of transmembrane proteins that mechanically link the cytoskeleton with the extracellular matrix, have been the most popular targets for delivering mechanical stimuli to cells. Depending on the rate (or frequency) and magnitude of the applied force, pulling on integrins elicit a wide range of biological responses in different cell types: minute forces (3 pN; cyclic at 1 Hz) are sufficient to hyperpolarize stem cells through stretch-activated ion channels.17 Higher forces (1 nN; cyclic at 1 Hz) are required to identify the roles of talin and α-actinin in focal adhesion initiation and maturation, respectively.19 A rapid stretch (4 nN at 40 nN s−1), however, causes traumatic axonal injury in primary neurons.13 Based on integrin-mediated pulling, these studies demonstrate the flexibility of MTW. It is also worth mentioning recent pulling experiments conducted in developing Xenopus14 and Drosophila22 embryos, to highlight the benign nature of MTW. A more comprehensive list of recent applications of MTW in protein and cell biophysics is given in Table 1. For applications at the nucleic acid level, the reader is referred to a recent review by Bryant and colleagues.27
| Acellular | Transmembrane | Intracellular | |
|---|---|---|---|
| a Molecular handle (measurement conducted or observation made). | |||
| Force | DNA (enzyme activity)7 | TREK-1 K+ channel (differentiation)10 | Unknown (intracellular transport)11 |
| DNA origami (bending rigidity)12 | Integrin (traumatic axonal injury)13 | Unknown (gene expression)14 | |
| Talin,15 filamin A16 (conformational change) | Integrin (ion channel activation)17 | ||
| Microtubule network (stiffness and creep)18 | Integrin (focal adhesion formation)19 | ||
| Collagen trimer (proteolysis rate)20 | PECAM-1 (cell signalling)21 | ||
| apCAM (bond strength)4 | Cadherin (collective cell migration)22 | ||
| SNARE complex (rupture force)8 | Death receptor 4 (receptor clustering)23 | ||
| Torque | DNA (torque spectroscopy)24 | Integrin (metastatic potential)25 | |
| DNA origami (torsional rigidity)12 | Integrin (cell stiffness)9 | — | |
| F1-ATPase rotary motor (function)26 | Cadherin (cell stiffness)5 | ||
Recent technical advances
Higher spatial, temporal, and force resolution
In applications where sub-pN forces are desired, such as measuring the mechanical properties of DNA, MTW experiments are generally conducted in a quasi-static mode, since the accurate analysis of the particle position requires considerable time. By controlling the force through the position of a pair of permanent magnets, and by precisely measuring the height of the magnetic bead, real-time force spectroscopy can be conducted at nm spatial and sub-pN force resolution to reveal the folding of DNA chromatin.28 However, for short tethers, accurate height measurement is only possible with small beads, limiting the applied force. Considerably high, yet accurate forces may be required for a detailed description of protein folding. By quantifying the thermal fluctuations of the particle position along a direction orthogonal to the force direction, the force can be directly determined with high accuracy, overcoming this limitation.29 Increased spatial resolution becomes critically important in high-force applications where the Brownian motion is minimal. In an attempt to increase the spatial resolution, reflection interference contrast microscopy (RICM) has been implemented. RICM relies on the interference between the reflections of the incident light from the bead surface and from the transparent substratum. By coating magnetic beads and glass surfaces with 50 nm TiO2 and Au layers, respectively, the instrumental resolution has been reduced to <0.2 nm.30 The spatial resolution in a MTW experiment can also be reduced by employing sophisticated bead tracking algorithms. The vertical distance of the magnetic bead from a reference surface has been traditionally measured by analyzing the diffraction pattern of the transmitted light. A novel tracking algorithm is able to reduce the lateral resolution to less than 1 nm at 30× magnification (to <0.3 nm at 100×);31 an achievement that is particularly important for tracking multiple beads simultaneously. The temporal resolution of MTW has also recently been improved. By combining a superluminescent diode illuminator with a high speed CMOS camera, a recent MTW instrument is capable of measuring the stochastic folding/unfolding events of a DNA hairpin. Operating at 702 Hz, it can resolve a 11 ms residence of the hairpin in the folded state; which had been previously unresolved due to lack of sufficient temporal resolution.32 These improvements now define the current boundaries of the MTW method for single molecule applications as well as for multiplexed measurements.Measuring torque independent of force
Traditional MTW can be used to apply or measure torque on a molecule that is rotationally constrained, through multiple contacts at the magnetic bead and substrate ends of the molecule, by rotating the magnetic field and hence the probe (Fig. 1F).33 However, this method has several important limitations: (i) torque in the range of 10 pN nm, typically exerted by a single double-stranded (ds) DNA molecule, cannot be measured; (ii) twisting torque cannot be decoupled from stretching force; and (iii) torque exerted by a molecule cannot be measured without applying an external torque to that molecule. Recent years have witnessed a range of MTW designs that aimed to overcome these limitations. A hybrid structure, composed of a magnetic bead attached to the tiny Ni segment of a NiPt nanorod, increased the torque resolution considerably (Fig. 1G).34 By circling the probe around the magnet center, a 10 kb dsDNA molecule has been successfully overwound and unwound at ∼1 pN stretching force. A soft torsional trap is generated due to the moment of the horizontal force (Fh × L) and the torque is calculated by multiplying the trap stiffness kBT/〈δθ2〉 with θ.Several alternative approaches have been considered in order to decouple torque from the stretching force. “Soft MTW” is based on the principle that in a fast rotating field, the bead rotation cannot follow the field rotation due to the viscous drag force. However, this instrument is limited to very low stretching forces.35 “Magnetic torque tweezers” (MTT) utilizes a cylindrical permanent magnet to apply a vertical magnetic field to a vertically-aligned DNA tether and a small marker bead attached to the magnetic bead to measure its angular orientation (Fig. 1H). A side magnet attached to the cylindrical magnet distorts the magnetic field such that a soft torsional trap is created.36 In “rotor bead tracking”, the twist in the dsDNA molecule is measured by tracing a fluorescent marker bead that is attached to the molecule (Fig. 1I).24 The twist (θ) is determined by subtracting the rotation of the rotor bead (φ) from the rotation of the magnetic bead (α). “Freely-orbiting MTW” repeats the cylindrical magnet configuration of MTT and demonstrates that the bead rotates freely about the axis of the molecule (at radius R), provided that the tether has a single contact point with the bead (Fig. 1J).37 This design was further improved to include Helmholtz coils that induce an additional magnetic field in the horizontal plane. The horizontal component results in a torsional trap whose stiffness can be controlled by changing the Helmholtz field amplitude. The so-called “electromagnetic torque tweezers” instrument is able to exert stretching forces from 10 fN to 10 pN and its torsional stiffness ranges from zero to 1000 pN nm rad−1.38 In summary, the rapid developments in the field of torque tweezers resulted in a series of fine instruments with broad ranges of force and torque.
Magnetic tweezers as a complementary biophysical technique
MTW method is inherently based on microscopy. In contrast to traditional MTW systems that align the molecule vertically, transverse designs enable the application of forces in the focal plane such that inter-molecular processes, such as, protein exchange on a DNA tether, could be directly visualized via fluorescence microscopy.39 Evanescent field illumination has recently been integrated to MTW systems. Frustrated total internal reflectance (f-TIR) can be used to detect the presence of magnetic nanoparticles on a sensor surface;40 whereas total internal reflection fluorescence (TIRF) can be used to determine the vertical position of a fluorescent magnetic probe at nm resolution.41,42 Single molecule Förster resonance energy transfer (FRET) is an indicator of the distance between acceptor and donor molecules, and therefore can be used to measure molecular folding events. The folding of a guanine/cytosine repeat has been characterized within the context of B-DNA to Z-DNA transition under torsional stress, by flanking it between the acceptor and donor of a FRET pair.43 Similarly, G-quadruplex (GQ) folding and unfolding in telomere DNA has been studied by flanking GQ with two dsDNA molecules tagged with FRET fluorophores.44Due to its flexible design, MTW can be combined with other force probing techniques. In a delicate experimental setting, the interaction between two DNA molecules, one horizontally stretched between two optical tweezers (OT) probes and the other vertically held between the substrate and a magnetic bead.45 The horizontally-aligned molecule can be pushed towards the vertically-aligned molecule, which, in turn, can be stretched and twisted to reveal the nature of the interaction. Another study combining MTW with OT used the F1-ATPase, a rotary motor protein which is an essential part of the ATP synthesis machinery, as a reel to wind individual dsDNA molecules.46 One end of the dsDNA molecule was tethered to the γ-subunit of F1-ATPase, which was rotated via a MTW probe, and the other end was tethered to an OT probe to measure the stretching force. The bending stiffness of the DNA molecule could then be estimated from the measured bending diameter and the stretching force.
At the cellular level, MTW method has been used in combination with traction force microscopy, a technique where cells are cultured on an array of elastic micropillars and the force applied on each pillar is measured by its deformation.47 Smooth muscle cells exhibited contractile force reinforcement when stimulated with magnetic nanorods that were either internalized or attached to their membranes. Interestingly, the response of the cells depended on the actuation frequency, but not on the magnitude of the force or torque. This observation could only be made by using MTW, since it controls frequency and magnitude independently. Cumulatively, these studies display the flexibility of the MTW method in the sense that it can be combined with other biophysical techniques.
Multiplexing magnetic tweezers
MTW force and torque application can be multiplexed as long as highly uniform magnetic fields are imposed on magnetic particles with uniform size and magnetism. Such uniform fields are usually delivered by a pair of permanent magnets at the expense of the field gradient, resulting in low forces. Parallel vertical pulling has been utilized to study the cleavage of DNA by type III restriction enzymes,48 the proteolysis of collagen by MMP-1,20 and the unbinding of cell adhesion molecules4 under external force. Similarly, with the help of microcontact printing, dense arrays of DNA-bead tethers can be formed in order to apply force and torque to hundreds of molecules simultaneously.49 As an alternative, the so-called “imaginary” MTW has been proposed which relies on the highly-uniform magnetic repulsion forces (10−4–1 pN range) that are applied to polymer particles embedded in ferrofluid.50 Multiplexed MTW is most suited for parallel assays involving proteins and cells towards biosensor applications. For a more comprehensive review of high-throughput MTW studies, the reader is referred to a recent review by De Vlaminck and Dekker.51Novel particles for enhanced magnetic tweezers
Despite these recent advances in the MTW method, the vast majority of experiments are based on commercial magnetic beads. Novel magnetic particles exhibiting control of both shape and element composition have recently emerged as alternative tools for enhanced MTW.52,53 Cylindrical magnetic particles of metallic47,54 and polymeric origin55 have recently been employed in the MTW context. These so-called nanowires or nanorods are of particular interest as they can combine multiple metallic elements54 or alloys53 into a single structure, potentially creating isolated molecular binding sites for targeting applications. Compared to a commercial 1 μm diameter spherical bead (Dynabeads MyOne), a 2 μm long, 100 nm diameter pure iron rod provides 1.3× higher stretching force; yet, its rotational drag coefficient (fr) is 54× smaller. This is particularly important in torque measurements where one needs to use small beads to achieve high torque sensitivity (fr ∼ r3) at the expense of the stretching force (also ∼r3).Apart from their enhanced magnetic properties, multi-component nanorods can also be decorated with gold segments that have strong plasmonic properties. We have recently characterized the optical signature of FeAu nanorods using transmission polarization microscopy.54 60 nm diameter rods with a 191 nm Au segment were visualized despite their sub-wavelength dimensions (Fig. 2A). Larger rods (295 nm in diameter a with 1056 nm Au segment) were visualized in both bright field (Fig. 2B) and dark field modes, where the latter was achieved by using an orthogonally-aligned polarizer–analyzer pair. Sequential images in Fig. 2 show how the scattered light intensity depends on the orientation of the rod where θ is the angle between the polarization axis and the rod's longitudinal axis. The gold segment exhibits an orientation-dependent intensity that peaks at 45° and 135° angles (Fig. 2C). Combined with the strong magnetization provided by the Fe segment (1600 emu cm−3 saturation magnetization),54 these rods are excellent candidates for intracellular MTW experiments.
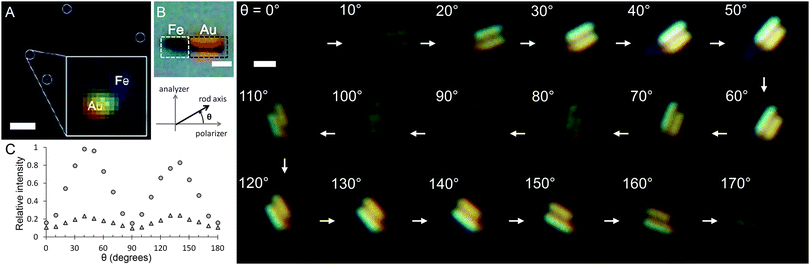 | ||
| Fig. 2 Optical signature of iron–gold composite nanorods (ref. 54). Iron and gold segments of 60 nm diameter nanorods (white circles) can be identified with transmission polarization microscopy (A; scale bar = 10 μm; marked area is 40× magnified). Bright-field image of a 295 nm diameter nanorod (B; scale bars = 500 nm). A series of dark-field images of the same rod (right panel) obtained by orthogonally aligning the polarizer and the analyzer. Scattered light intensity from small (triangles) and large (circles) rods changes as a function of the angle between the polarizer and the rod axis (C). | ||
Magnetic nanoparticles developed for drug delivery or theranostic applications, such as FePt capsules56 and Fe2O3 tubes,57 may also be optimized for MTW use. Other potential candidates are the soft composite materials with programmable shape changes: stripes that twist or bend,52 and tubes that collapse upon exposure to weak magnetic fields58 can be further exploited for their use as MTW probes. Regardless of the application mode, novel magnetic particles with enhanced magnetic and other properties provide an unexplored territory for the future of MTW method.
Conclusions
Magnetic tweezers has proved to be a powerful biophysical tool with unique abilities to apply torque independent from force or controlling hundreds of probes simultaneously. Recent efforts to advance the method focused on improving its resolution limits, combining it with other biophysical methods, and multiplexing. MTW is able measure the viscoelastic properties of biological samples from proteins to cells. Measuring cell stiffness is a promising new tool in medical diagnostics.9,25 Highly-sensitive, cell-based MTW assays may be developed in the near future by integrating recent technical advances into existing biophysical measurements. In parallel, the particle synthesis field now offers new classes of magnetic particles with enhanced magnetic properties, a variety of geometries, and asymmetric features leading to multi-functionality. We expect these novel particles to replace the traditional MTW probes, e.g., spherical SPM beads, in the near future, expanding the potential uses of the MTW method and pushing its current boundaries.Acknowledgements
This work was supported by Science Foundation Ireland (08/RP1/B1376 and 08/IN1/B2072), by the Programme for Research in Third-Level Institutions funded by the Irish Higher Education Agency and co-funded by the European Regional Development Fund, and by a Marie Curie Intra-European Fellowship (FP7-People-2010-IEF-273420).Notes and references
- B. D. Hoffman, C. Grashoff and M. A. Schwartz, Dynamic molecular processes mediate cellular mechanotransduction, Nature, 2011, 475, 316–323 CrossRef CAS PubMed.
- L. B. Oddershede, Force probing of individual molecules inside the living cell is now a reality, Nat. Chem. Biol., 2012, 8, 879–886 CrossRef CAS PubMed.
- J. J. O'Mahony, M. Platt, D. Kilinc and G. Lee, Synthesis of superparamagnetic particles with tunable morphologies: the role of nanoparticle-nanoparticle interactions, Langmuir, 2013, 29, 2546–2553 CrossRef CAS PubMed.
- D. Kilinc, A. Blasiak, J. J. O'Mahony, D. M. Suter and G. U. Lee, Magnetic tweezers-based force clamp reveals mechanically distinct apCAM domain interactions, Biophys. J., 2012, 103, 1120–1129 CrossRef CAS PubMed.
- H. Tabdili, M. Langer, Q. Shi, Y. C. Poh, N. Wang and D. Leckband, Cadherin-dependent mechanotransduction depends on ligand identity but not affinity, J. Cell Sci., 2012, 125, 4362–4371 CrossRef CAS PubMed.
- T. Lionnet, J. F. Allemand, A. Revyakin, T. R. Strick, O. A. Saleh, D. Bensimon and V. Croquette, Single-molecule studies using magnetic traps, Cold Spring Harb. Protoc., 2012, 2012, 34–49 Search PubMed.
- M. Manosas, S. K. Perumal, V. Croquette and S. J. Benkovic, Direct observation of stalled fork restart via fork regression in the T4 replication system, Science, 2012, 338, 1217–1220 CrossRef CAS PubMed.
- D. Min, K. Kim, C. Hyeon, Y. Hoon Cho, Y. K. Shin and T. Y. Yoon, Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism, Nat. Commun., 2013, 4, 1705 CrossRef PubMed.
- R. J. Saphirstein, Y. Z. Gao, M. H. Jensen, C. M. Gallant, S. Vetterkind, J. R. Moore and K. G. Morgan, The focal adhesion: a regulated component of aortic stiffness, PLoS One, 2013, 8, e62461 CAS.
- J. M. Kanczler, H. S. Sura, J. Magnay, R. O. Oreffo, D. Green, J. P. Dobson and A. J. El Haj, Controlled differentiation of human bone marrow stromal cells using magnetic nanoparticle technology, Tissue Eng., Part A, 2010, 16, 3241–3250 CrossRef CAS PubMed.
- J. Mahowald, D. Arcizet and D. Heinrich, Impact of external stimuli and cell micro-architecture on intracellular transport states, ChemPhysChem, 2009, 10, 1559–1566 CrossRef CAS PubMed.
- D. J. Kauert, T. Kurth, T. Liedl and R. Seidel, Direct mechanical measurements reveal the material properties of three-dimensional DNA origami, Nano Lett., 2011, 11, 5558–5563 CrossRef CAS PubMed.
- M. A. Hemphill, B. E. Dabiri, S. Gabriele, L. Kerscher, C. Franck, J. A. Goss, P. W. Alford and K. K. Parker, A possible role for integrin signaling in diffuse axonal injury, PLoS One, 2011, 6, e22899 CAS.
- A. Kumar and G. V. Shivashankar, Mechanical force alters morphogenetic movements and segmental gene expression patterns during drosophila embryogenesis, PLoS One, 2012, 7, e33089 CAS.
- A. del Rio, R. Perez-Jimenez, R. Liu, P. Roca-Cusachs, J. M. Fernandez and M. P. Sheetz, Stretching single talin rod molecules activates vinculin binding, Science, 2009, 323, 638–641 CrossRef CAS PubMed.
- H. Chen, S. Chandrasekar, M. P. Sheetz, T. P. Stossel, F. Nakamura and J. Yan, Mechanical perturbation of filamin A immunoglobulin repeats 20-21 reveals potential non-equilibrium mechanochemical partner binding function, Sci. Rep., 2013, 3, 1642 CAS.
- G. R. Kirkham, K. J. Elliot, A. Keramane, D. M. Salter, J. P. Dobson, A. J. El Haj and S. H. Cartmell, Hyperpolarization of human mesenchymal stem cells in response to magnetic force, IEEE Trans. Nanobiosci., 2010, 9, 71–74 CrossRef PubMed.
- Y. Yang, M. Bai, W. S. Klug, A. J. Levine and M. T. Valentine, Microrheology of highly crosslinked microtubule networks is dominated by force-induced crosslinker unbinding, Soft Matter, 2013, 9, 383–393 RSC.
- P. Roca-Cusachs, A. del Rio, E. Puklin-Faucher, N. C. Gauthier, N. Biais and M. P. Sheetz, Integrin-dependent force transmission to the extracellular matrix by alpha-actinin triggers adhesion maturation, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E1361–E1370 CrossRef CAS PubMed.
- A. S. Adhikari, J. Chai and A. R. Dunn, Mechanical load induces a 100-fold increase in the rate of collagen proteolysis by MMP-1, J. Am. Chem. Soc., 2011, 133, 1686–1689 CrossRef CAS PubMed.
- C. Collins, C. Guilluy, C. Welch, E. T. O'Brien, K. Hahn, R. Superfine, K. Burridge and E. Tzima, Localized tensional forces on PECAM-1 elicit a global mechanotransduction response via the integrin-RhoA pathway, Curr. Biol., 2012, 22, 2087–2094 CrossRef CAS PubMed.
- G. F. Weber, M. A. Bjerke and D. W. DeSimone, A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration, Dev. Cell, 2012, 22, 104–115 CrossRef CAS PubMed.
- M. H. Cho, E. J. Lee, M. Son, J. H. Lee, D. Yoo, J. W. Kim, S. W. Park, J. S. Shin and J. Cheon, A magnetic switch for the control of cell death signalling in in vitro and in vivo systems, Nat. Mater., 2012, 11, 1038–1043 CrossRef CAS PubMed.
- F. C. Oberstrass, L. E. Fernandes and Z. Bryant, Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 6106–6111 CrossRef CAS PubMed.
- M. F. Coughlin, D. R. Bielenberg, G. Lenormand, M. Marinkovic, C. G. Waghorne, B. R. Zetter and J. J. Fredberg, Cytoskeletal stiffness, friction, and fluidity of cancer cell lines with different metastatic potential, Clin. Exp. Metastasis, 2013, 30, 237–250 CrossRef CAS PubMed.
- R. Watanabe, R. Iino and H. Noji, Phosphate release in F1-ATPase catalytic cycle follows ADP release, Nat. Chem. Biol., 2010, 6, 814–820 CrossRef CAS PubMed.
- Z. Bryant, F. C. Oberstrass and A. Basu, Recent developments in single-molecule DNA mechanics, Curr. Opin. Struct. Biol., 2012, 22, 304–312 CrossRef CAS PubMed.
- M. Kruithof, F. T. Chien, A. Routh, C. Logie, D. Rhodes and J. van Noort, Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30 nm chromatin fiber, Nat. Struct. Mol. Biol., 2009, 16, 534–540 CAS.
- H. Chen, H. Fu, X. Zhu, P. Cong, F. Nakamura and J. Yan, Improved high-force magnetic tweezers for stretching and refolding of proteins and short DNA, Biophys. J., 2011, 100, 517–523 CrossRef CAS PubMed.
- K. Kim and O. A. Saleh, A high-resolution magnetic tweezer for single-molecule measurements, Nucleic Acids Res., 2009, 37, e136 CrossRef PubMed.
- M. T. van Loenhout, J. W. Kerssemakers, I. De Vlaminck and C. Dekker, Non-bias-limited tracking of spherical particles, enabling nanometer resolution at low magnification, Biophys. J., 2012, 102, 2362–2371 CrossRef CAS PubMed.
- B. M. Lansdorp, S. J. Tabrizi, A. Dittmore and O. A. Saleh, A high-speed magnetic tweezer beyond, 10,000, frames per second, Rev. Sci. Instrum., 2013, 84, 044301 CrossRef PubMed.
- C. Breme and F. Heslot, Mapping of single-base differences between two DNA strands in a single molecule using holliday junction nanomechanics, PLoS One, 2013, 8, e55154 CAS.
- A. Celedon, I. M. Nodelman, B. Wildt, R. Dewan, P. Searson, D. Wirtz, G. D. Bowman and S. X. Sun, Magnetic tweezers measurement of single molecule torque, Nano Lett., 2009, 9, 1720–1725 CrossRef CAS PubMed.
- F. Mosconi, J. F. Allemand and V. Croquette, Soft magnetic tweezers: a proof of principle, Rev. Sci. Instrum., 2011, 82, 034302 CrossRef PubMed.
- J. Lipfert, J. W. Kerssemakers, T. Jager and N. H. Dekker, Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments, Nat. Methods, 2010, 7, 977–980 CrossRef CAS PubMed.
- J. Lipfert, M. Wiggin, J. W. Kerssemakers, F. Pedaci and N. H. Dekker, Freely orbiting magnetic tweezers to directly monitor changes in the twist of nucleic acids, Nat. Commun., 2011, 2, 439 CrossRef PubMed.
- X. J. Janssen, J. Lipfert, T. Jager, R. Daudey, J. Beekman and N. H. Dekker, Electromagnetic torque tweezers: a versatile approach for measurement of single-molecule twist and torque, Nano Lett., 2012, 12, 3634–3639 CrossRef CAS PubMed.
- J. S. Graham, R. C. Johnson and J. F. Marko, Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA, Nucleic Acids Res., 2011, 39, 2249–2259 CrossRef CAS PubMed.
- D. M. Bruls, T. H. Evers, J. A. Kahlman, P. J. van Lankvelt, M. Ovsyanko, E. G. Pelssers, J. J. Schleipen, F. K. de Theije, C. A. Verschuren, T. van der Wijk, J. B. van Zon, W. U. Dittmer, A. H. Immink, J. H. Nieuwenhuis and M. W. Prins, Rapid integrated biosensor for multiplexed immunoassays based on actuated magnetic nanoparticles, Lab Chip, 2009, 9, 3504–3510 RSC.
- R. Liu, S. Garcia-Manyes, A. Sarkar, C. L. Badilla and J. M. Fernandez, Mechanical characterization of protein L in the low-force regime by electromagnetic tweezers/evanescent nanometry, Biophys. J., 2009, 96, 3810–3821 CrossRef CAS PubMed.
- P. M. Oliver, J. S. Park and D. Vezenov, Quantitative high-resolution sensing of DNA hybridization using magnetic tweezers with evanescent illumination, Nanoscale, 2011, 3, 581–591 RSC.
- M. Lee, S. H. Kim and S. C. Hong, Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4985–4990 CrossRef CAS PubMed.
- X. Long, J. W. Parks, C. R. Bagshaw and M. D. Stone, Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy, Nucleic Acids Res., 2012, 41, 2746–2755 CrossRef PubMed.
- I. De Vlaminck, M. T. van Loenhout, L. Zweifel, J. den Blanken, K. Hooning, S. Hage, J. Kerssemakers and C. Dekker, Mechanism of homology recognition in DNA recombination from dual-molecule experiments, Mol. Cell, 2012, 46, 616–624 CrossRef CAS PubMed.
- H. You, R. Iino, R. Watanabe and H. Noji, Winding single-molecule double-stranded DNA on a nanometer-sized reel, Nucleic Acids Res., 2012, 40, e151 CrossRef CAS PubMed.
- Y. C. Lin, C. M. Kramer, C. S. Chen and D. H. Reich, Probing cellular traction forces with magnetic nanowires and microfabricated force sensor arrays, Nanotechnology, 2012, 23, 075101 CrossRef PubMed.
- K. Van Aelst, J. Tóth, S. P. Ramanathan, F. W. Schwarz, R. Seidel and M. D. Szczelkun, Type III restriction enzymes cleave DNA by long-range interaction between sites in both head-to-head and tail-to-tail inverted repeat, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 9123–9128 CrossRef CAS PubMed.
- I. De Vlaminck, T. Henighan, M. T. J. Van Loenhout, I. Pfeiffer, J. Huijts, J. W. J. Kerssemakers, A. J. Katan, A. Van Langen-Suurling, E. Van Der Drift, C. Wyman and C. Dekker, Highly parallel magnetic tweezers by targeted DNA tethering, Nano Lett., 2011, 11, 5489–5493 CrossRef PubMed.
- Y. Yang, R. M. Erb, B. J. Wiley, S. Zauscher and B. B. Yellen, Imaginary magnetic tweezers for massively parallel surface adhesion spectroscopy, Nano Lett., 2011, 11, 1681–1684 CrossRef CAS PubMed.
- I. De Vlaminck and C. Dekker, Recent advances in magnetic tweezers, Annu. Rev. Biophys., 2012, 41, 453–472 CrossRef CAS PubMed.
- J. W. Tavacoli, P. Bauer, M. Fermigier, D. Bartolo, J. Heuvingh and O. du Roure, The fabrication and directed self-assembly of micron-sized superparamagnetic non-spherical particles, Soft Matter, 2013, 9, 9103–9110 RSC.
- Y. Zhang, Q. Wang, B. Ashall, D. Zerulla and G. U. Lee, Magnetic-plasmonic dual modulated FePt-Au ternary heterostructured nanorods as a promising nano-bioprobe, Adv. Mater., 2012, 24, 2485–2490 CrossRef CAS PubMed.
- Y. Zhang, M. DaSilva, B. Ashall, G. Doyle, D. Zerulla, T. D. Sands and G. U. Lee, Magnetic manipulation and optical imaging of an active plasmonic single-particle Fe-Au nanorod, Langmuir, 2011, 27, 15292–15298 CrossRef CAS PubMed.
- P. Schroeder, J. Schotter, A. Shoshi, M. Eggeling, O. Bethge, A. Hutten and H. Bruckl, Artificial cilia of magnetically tagged polymer nanowires for biomimetic mechanosensing, Bioinspiration Biomimetics, 2011, 6, 046007 CrossRef CAS PubMed.
- T. Fuchigami, Y. Kitamoto and Y. Namiki, Size-tunable drug-delivery capsules composed of a magnetic nanoshell, Biomatter, 2012, 2, 313–320 CrossRef PubMed.
- N. Kang, J. H. Park, J. Choi, J. Jin, J. Chun, I. G. Jung, J. Jeong, J. G. Park, S. M. Lee, H. J. Kim and S. U. Son, Nanoparticulate iron oxide tubes from microporous organic nanotubes as stable anode materials for lithium ion batteries, Angew. Chem., Int. Ed., 2012, 51, 6626–6630 CrossRef CAS PubMed.
- R. Fuhrer, C. M. Schumacher, M. Zeltner and W. J. Stark, Soft iron/silicon composite tubes for magnetic peristaltic pumping: frequency-dependent pressure and volume flow, Adv. Funct. Mater., 2013, 23, 3845–3949 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2014 |


