Nanoparticles and their applications in cell and molecular biology
Edina C.
Wang
and
Andrew Z.
Wang
*
Department of Radiation Oncology, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA. E-mail: zawang@med.unc.edu
First published on 19th September 2013
Abstract
Nanoparticles can be engineered with distinctive composition, size, shape, and surface chemistry to enable novel techniques in a wide range of biological applications. The unique properties of nanoparticles and their behavior in biological milieu also enable exciting and integrative approaches to studying fundamental biological questions. This review will provide an overview of various types of nanoparticles and concepts of targeting nanoparticles. We will also discuss the advantages and recent applications of using nanoparticles as tools for drug delivery, imaging, sensing, and for the understanding of basic biological processes.
Insight, innovation, integrationNanoparticles are promising tools for applications in biology. Many innovative techniques using nanoparticles are being developed for the detection of low levels of analytes, pathogens, and cells. Nanoparticles are also being explored as delivery vehicles, imaging agents, and to better understand biology at the molecular, cellular, and tissue level. The integration of nanotechnology with biology offers many advantages that can help stimulate further biological and biomedical studies. |
1. Introduction
Nanotechnology is a relatively new branch of science that has found a wide range of applications that range from energy production to industrial production processes to biomedical applications.1 One of the key applications is in biology and biomedical research. Nanoparticles (NPs) can be engineered to possess unique compositions and functionalities to enable novel tools and techniques that have not existed previously in biomedical research. For example, NPs can be used to image biological processes at the cellular level. They can also be utilized to detect analytes at concentrations in the attomolar range. In this review, we aim to discuss the types of NPs and their potential application in biology and biomedical research.2. Types of nanoparticles
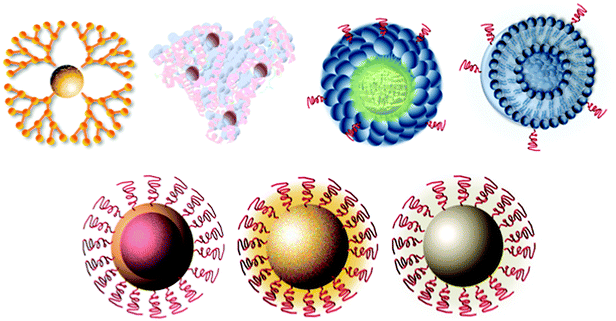
There are many types of NP platforms with differing size, shape, composition, and functionality (Fig. 1). Furthermore, each type of NP can potentially be fabricated using different techniques, such as both nanoprecipitation and lithography for polymeric NPs. While it is not within this manuscript's scope to discuss the differences in NP platforms and their fabrication in detail, we will discuss the major characteristics and functionalities of each NP that are relevant for biological research.2.1 Liposomes
Liposomes were one of the first NP platforms. Liposomes were first described in 1965 as a model of cellular membranes.2 Since then, liposomes have moved from a model in biophysical research to one of the first NP platforms to be applied for gene and drug delivery. Liposomes are spherical vesicles that contain a single or multiple bilayered structure of lipids that self-assemble in aqueous systems.3 Unique advantages imparted by liposomes are their diverse range of compositions, ability to carry and protect many types of biomolecules, as well as their biocompatibility and biodegradability.3,4 These advantages have led to the well-characterized and wide use of liposomes as transfection agents of genetic material into cells (lipofection) in biology research.5 Lipofection generally uses a cationic lipid to form an aggregate with the anionic genetic material. Another major application of liposomes is their use as therapeutic carriers since their design can allow for entrapment of hydrophilic compounds within the core and hydrophobic drugs in the lipid bilayer itself.6 To enhance their circulation half-life and stability in vivo, liposomes have been conjugated with biocompatible polymers such as polyethylene glycol (PEG).3 Liposomes can also be functionalized with targeting ligands to increase the accumulation of diagnostic and therapeutic agents within desired cells. Today, there are twelve clinically approved liposome-based therapeutic drugs.2.2 Albumin-bound NPs
Albumin-bound NPs (nab) use the endogenous albumin pathways to carry hydrophobic molecules in the bloodstream.7 Albumin naturally binds to the hydrophobic molecules with non-covalent reversible binding, avoiding solvent-based toxicity of therapeutics.8 As a result, this platform has been successfully adapted as a drug delivery vehicle. Abraxane, a 130 nm nab paclitaxel, was approved by the FDA in 2005 for the treatment of metastatic breast cancer.9 Abraxane concentrates in cells through albumin receptor (gp60)-mediated transport in endothelial cells. It may also target the albumin-binding protein SPARC (secreted protein acidic and rich in cysteine), which is overexpressed in certain tumors.10 Further understanding of the mechanism of action may lead to better targeting and development of novel therapeutics using the nab platform.2.3 Polymeric NPs
Polymeric NPs formed from biocompatible and biodegradable polymers have been extensively investigated as therapeutic carriers.11 Polymeric NPs are formulated through block-copolymers of different hydrophobicity.12 These copolymers spontaneously assemble into a core–shell micelle formation in an aqueous environment.13 Polymeric NPs have been formulated to encapsulate hydrophilic and/or hydrophobic small molecule drugs, as well as proteins and nucleic acid macromolecules.14 The NP design can allow for slow and controlled release of drug at target sites. Polymeric NPs are usually able to improve the safety and efficacy of the drugs they carry. Functionalizing polymeric NPs with targeting ligands for improved drug delivery has been an important area of investigation since polymeric NPs are unique in their ability to be tailored prior to particle assembly. The incorporation of targeting ligands on the NPs can lead to their increased uptake along with their cargo, leading to enhanced therapeutic outcomes.Another type of polymeric NP is dendrimers. Dendrimers are regularly branched macromolecules made from synthetic or natural elements including amino acids, sugars, and nucleotides.15 They have a central core, interior layers of branches, and an exterior surface.16 The varied combination of these components can yield dendrimers of well-defined size, shape, and branching length/density.17 As a result of their unique design, dendrimers can be developed as sensors as well as drug and gene delivery carriers. Dendrimers can be loaded with small molecules in the cavities of the cores through chemical linkage, hydrogen bond, and/or hydrophobic interaction.18 The exterior surface can also be readily modified to produce chemical functional groups for the attachment of molecular targeting groups, detection and imaging agents, and therapeutic agents.19
2.4 Iron oxide
Iron oxide NPs are widely studied as passive and active targeting imaging agents since they are mainly superparamagnetic. The superparamagnetic iron oxide NPs (SPIONs) generally have an iron oxide core with a hydrophilic coat of dextran or other biocompatible compound to increase their stability.20,21 The most widely used SPIONs consist of a magnetite (Fe3O4) and/or maghemite (γFe2O3) core. These NPs exhibit size-dependent superparamagnetism, which allows them to become magnetized with the application of an external magnetic field and exhibit zero net magnetization upon removal of the magnetic field. SPIONs have been successfully used as T2-weighted magnetic resonance (MR) contrast agents to track and monitor cells.22 SPIONs have several advantages over conventional gadolinium-chelate contrast agents including decreased toxicity and increased imaging sensitivity and specificity.23 SPIONs can also be degraded to iron and iron oxide molecules that are metabolized, stored in cells as ferritin, and incorporated into hemoglobin.23 Currently, two SPIO agents, ferumoxides (120–180 nm) and ferucarbotran (60 nm), are clinically approved for MRI. SPIONs have also been used in molecular imaging applications such as the detection of apoptosis24 and gene expression.25 SPIONs can be functionalized with optical and radionuclide targeting ligands for multimodal imaging. They can also potentially be used as non-invasive diagnostic tools and as drug delivery vehicles.262.5 Quantum dot
First discovered in 1980, quantum dots (QDs) are semiconductor particles that are less than 10 nm in diameter. QDs display unique size-dependent electronic and optical properties.27 Most QDs studied consist of a cadmium selenide (CdSe) core and a zinc selenide (ZnS) cap.28 The absorption spectra of these particles are very broad and emission is confined to a narrow band. QDs can also emit bright colors, have long lifetimes and high efficiencies and are stable against photobleaching. They can be generated to have various biochemical specificities and can be simultaneously excited and detected. As a result, QDs have several significant advantages over many organic fluorophore dyes for optical applications. They are widely used in biological research as fluorescence imaging tools for applications such as cell labeling and biomolecule tracking.28–30 The small size of quantum dots also enables them to be suitable for biomedical applications such as medical imaging and diagnostics.312.6 Gold
Gold NPs offer many size-and-shape dependent optical and chemical properties, biocompatibility, and facile surface modification.21 Gold NPs can strongly enhance optical processes such as light absorption, scattering, fluorescence, and surface-enhanced Raman scattering (SERS) due to the unique interaction of the free electrons in the NP with light.32 These properties have enabled the realization of gold NPs in many applications such as biochemical sensing and detection, biological imaging, diagnostics, and therapeutic applications. Sensing techniques include the use of gold NPs in colorimetric arrays and the use of gold NPs as substrates in SERS to significantly enhance Raman scattering, allowing for spectroscopic detection and identification of proteins and single molecules at the NP surface.33 Gold NP probes have also been used to detect heart disease and cancer biomarkers.34,35 They can also transform absorbed light into heat and therefore have high potential for infrared phototherapy.363. Targeted nanoparticles
The concept of targeting has become a significant focus in biological research in recent years. In proteomics, targeted proteomics has gained traction as an approach to address specific biological questions by focusing on a subset of proteins of interest.37 In genetics, gene targeting in mice has become a gold standard for determining gene function in mammals.38 In the areas of drug discovery and molecular therapeutics, the development of small molecules and monoclonal antibodies such as imatinib,39 trastuzumab,40 bevacizumab,41 and rituximab42 has considerably changed the treatment of cancer.NPs show much promise in biological applications. In particular, NPs offer many unique properties that enhance or confer advantages over current techniques in biological and biomedical research. As a result, there has been significant interest in applying targeting concepts to NP design. NPs can be targeted by active targeting and passive targeting under in vivo conditions.
3.1 Passive targeting
In the case of passive targeting, NP systems have been successfully developed for cancer therapy by taking advantage of tumor tissue biology. Normal tissue vascular biology has an organized structure while tumor vasculature is irregularly branched and disorganized.43 Tumors also have high vascular density, increased vascular permeability, and impaired lymphatic drainage, an attribute of solid tumors and inflamed tissue.43,44 Together, these features are known as the enhanced permeability and retention (EPR) effect, which allows NPs to accumulate preferentially in tumor tissue.44 NPs have extended retention times in tumor tissue, which results in higher concentrations than in other tissues. Properties that mediate this passive targeting process include particle composition, size, shape, and surface characteristics.45 Thus, NPs can be engineered to better target a particular tissue or cell type by optimizing their physicochemical characteristics.3.2 Active targeting
Active targeting involves the use of targeting ligands for enhanced delivery of NP systems to a specific site. Typical targeting ligands include small molecules, peptides, antibodies and their fragments, and nucleic acids such as aptamers.14 These ligands have all been conjugated to NPs.14Conjugating targeting ligands to NP surfaces can be performed via covalent and non-covalent methods. With covalent conjugation, the same chemical methods for functionalization can be applied to various types of NPs since conjugation of functional groups to the NP surface is dependent on the functional groups on the NP surface and the functional groups of the ligand being conjugated. Certain conjugation techniques are also suitable for specific targeting ligand classes. The maleimide–thiol coupling is commonly used for conjugation of peptides, antibodies and their fragments to NPs. In this reaction, maleimides (maleic acid imides) spontaneously react with sulfhydryl groups at pH 6.5 to 7.5. Thus, NP surfaces incorporated with maleimide modified polymers or lipids can be readily conjugated with targeting ligands engineered with thiol groups or cysteine, which has a thiol side chain. Another commonly used conjugation method is the carbodiimide-mediated amide coupling between carboxyls and amines to form amide bonds. In this reaction, carboxylate groups on a molecule can be converted to active esters by using a variety of carbodiimides such as the water-soluble 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) and N-hydroxy-succinimide (NHS) or sulfo-NHS. The activated ester intermediate can react with a primary amine to form an amide bond. This method is generally preferred for aptamers, which can be modified with amines, and small molecules, which can contain carboxylates or primary amines. Click chemistry reactions are another method used to conjugate functional groups with high yield and selectivity under moderate reaction conditions. One of the most popular reactions with this class is the [3+2] cycloaddition between alkynes and azides.46
Conjugating targeting ligands to the NP surface can facilitate active targeting of NPs to receptors that are present on target cells, leading to enhanced cell internalization and/or specific uptake through receptor-mediated endocytosis.47,48 For NP delivery of therapeutics, these advantages result in higher drug concentrations and reduced systemic toxicity compared to non-targeted NPs and their small molecule counterparts. Thus, there has been intense interest in identifying new disease biomarkers and their ligands for use in targeted drug delivery. Targeted NPs can also identify molecular targets with good affinity and selectivity. These NPs can bind to analytes, pathogens, and biomarkers, amplifying their signal for detection and molecular imaging.
4. Nanoparticle applications in biology
4.1 Nanoparticles as sensors
NPs have been employed in sensors for a variety of applications including detecting analytes at very low concentrations, detecting and separating pathogens, detecting and capturing cells, and detecting molecular and cellular functions.NP platforms such as gold NPs have been extensively used as sensors due to their surface chemistry. One method of using the unique surface chemistry of gold NPs for signal transduction amplification is the bio-barcode method developed by the Mirkin group. This method has been applied for protein50 and DNA51 target detection. For target DNA detection, PCR-like sensitivity can be achieved. This technique has been reported for the detection of cytokines at a concentration of 30 aM (attomolar)52 and for the detection of a soluble pathogenic biomarker for Alzheimer's disease at a concentration around 100 aM.53 More recently, the group used this method to detect prostate-specific antigen (PSA), a commonly investigated cancer biomarker that can indicate the presence of prostate cancer when expressed at elevated levels.54 In the bio-barcode assay approach, DNA-functionalized gold NPs (30 nm) are conjugated to PSA-specific antibodies to generate PSA gold NP probes (Fig. 2).55,56 The DNA strands are the bio barcodes. A magnetic microparticle (MMP) functionalized with monoclonal antibodies to PSA is mixed with the PSA target protein. After a washing step, the PSA gold NP probes are added to the MMP-bound PSA. After further separation by magnetic field application and wash steps, the PSA-specific DNA barcodes are released into solution and analyzed using the scanometric assay, which takes advantage of gold NP catalyzed silver enhancement. This assay achieved a sensitivity of 330 fg mL−1 of PSA.
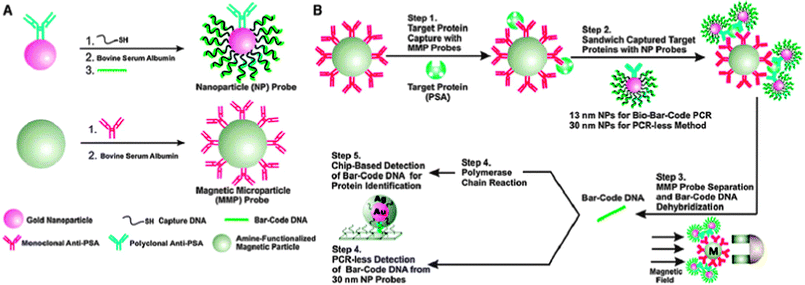 | ||
| Fig. 2 The bio-barcode method. (A) Development of the NP probe. (B) Method of detection using prostate specific antigen-conjugated gold NP probes. Reproduced from ref. 56. | ||
The DNA bio-barcode method combines the ability to conjugate a large number of DNA strands onto gold NP surfaces and the detection of these DNA strands using techniques such as PCR amplification or scanometric assays. Another approach for sensing that uses optical properties of the NP surface has been developed by the Rotello group.57 They found that anionic fluorescent polymers may reversibly bind and dissociate from functionalized cationic gold NPs through non-covalent chemical interactions. When the fluorescent polymer is bound to the NP, its fluorescence is quenched. When it is dissociated from the NP, the fluorescence is restored. This phenomenon is the basis of a “chemical nose” system.58 In this system, the target protein is detected according to its different fluorescence responses upon binding to different types of surface-modified gold NPs. A fluorescence response pattern is generated and then analyzed through statistical methods. This process can detect, quantify, and distinguish different molecular targets from each other. This process was initially developed for the detection and sensing of proteins,58 but has also been adapted for the detection and sensing of bacteria,59 for the detection of physicochemical differences between healthy, cancerous and metastatic human breast cells, and for the differentiation of isogenic healthy and transformed cells (Fig. 3).60,61 “Chemical nose” sensing also provides the advantage of not using antibodies for detection since the identity of the target analyte does not need to be known in order to detect the analyte.
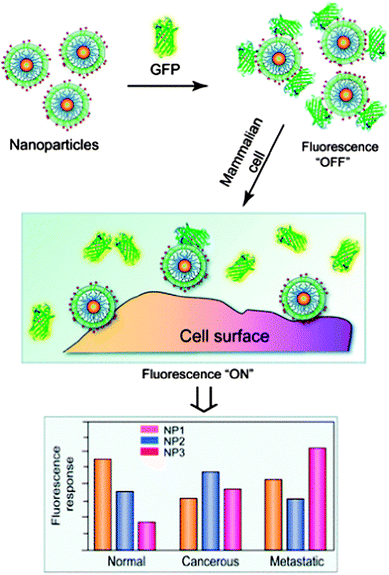 | ||
| Fig. 3 Interactions between NP–GFP complexes and cell surfaces generate different fluorescence patterns. Reproduced from ref. 61. | ||
Gold NPs can also enhance surface-enhanced Raman scattering (SERS) for detection and identification of biomolecules at the NP surface. The SERS technique has been frequently used to detect specific analytes through their unique vibrational spectra. The narrow width of SERS spectra allows for multiple analyte detection within complex mixtures, including detection down to the single-molecule level.33 Thus, SERS techniques have been used for ultrasensitive detection of biomolecules such as glucose,62 hemoglobin,63 bacteria,64 and viruses.65 Using this approach, a group demonstrated that gold NPs encoded with Raman reporters and conjugated with single-chain variant fragment (ScFv) antibodies can target cancer biomarkers such as epidermal growth factor receptors (EGFR) in vitro and in vivo (Fig. 4).66,67 Recently, investigators also demonstrated a SERS system based on a thin-film NP array self-assembled at the liquid–liquid interphase for multi-phase trace analyte detection.68 The group was able to detect single phase analytes as well as multiple analytes dissolved in water and organic phases.
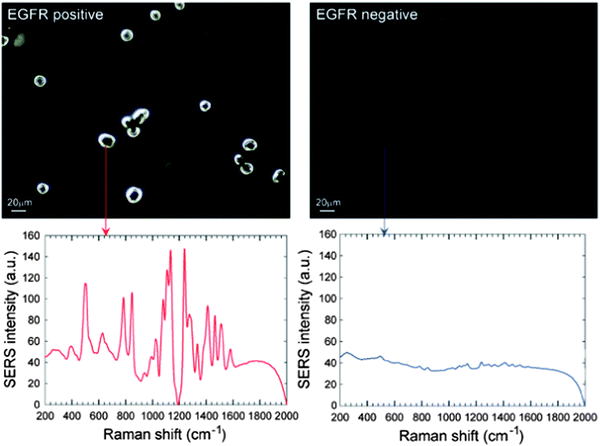 | ||
| Fig. 4 Surface-enhanced Raman scattering (SERS) spectra and the correlated surface plasmon imaging of single cancer cells tagged with ScFv-conjugated gold NPs. Reproduced from ref. 67. | ||
Nanosensors have also been developed that exploit the magnetic properties of magnetic NPs such as SPIONs. There is considerable interest in magnetic NPs since they have strong magnetic properties and most biological samples exhibit negligible magnetic susceptibility.69 SPIONs have been used as magnetic tags for many different types of sensors including giant magnetoresistive (GMR) biosensors, which are based on the binding of magnetic particles to a sensor surface.70 The magnetic fields of the particles can alter the magnetic fields of the sensor, resulting in changes in the electrical resistance of the sensor. Recently, Wang and colleagues at Stanford University have demonstrated the use of this technique for a protein detection assay in which an array of GMR sensors is used to detect binding events of proteins to arrays of surface-bound antibodies with the use of SPIONs as magnetic tags.71 In this assay, the target antigen is between two antibodies, one bound to the sensor and the other tagged with a SPION. The presence or absence of the magnetized NP is detected by the underlying sensor. The group demonstrated the assay to be matrix insensitive to various biological fluids, but still capable of detecting proteins down to attomolar concentrations and over an extensive range of concentrations. Using a similar strategy, they have also demonstrated the detection of cancer-associated proteins in 50% serum at sub-picomolar concentrations using MACS particles (commercialized SPIONs for cell separation).72 Recently, they have used streptavidin functionalized antiferromagnetic NPs to detect DNA with high sensitivity (10 pM).73
The superparamagnetic property of IONPs is also the basis of magnetic relaxation switching. The Weissleder group has studied magnetic relaxation switches consisting of 3–5 nm SPIONs coated with 10 nm thick dextran and stabilized by crosslinking.74 Targeted SPIONs are developed by functionalizing them with amino groups for the attachment of a range of sulfhydryl-bearing molecules.75 The nanoswitches can undergo reversible assembly (clustering and declustering) in the presence of a molecule that is recognized by the ligands immobilized on the NP, resulting in a change in the transverse magnetic relativity (1/T2) of surrounding water protons. The changes in relaxation rates as a result of NP assembly can be used to detect a variety of biological targets including DNA and proteins at the low femtomole level (0.5–10 fmol).76 Recently, the Weissleder group developed a portable chip-based diagnostic magnetic resonance (DMR) system for rapid and quantitative detection of biological targets.77 The DMR also uses IONPs as sensors to amplify molecular interactions caused by assemblies of SPIONs. They demonstrated proof of concept by detecting with high sensitivity the presence of proteins in parallel and by detecting bacteria. Compared to the benchtop NMR relaxometer, the DMR system had a mass detection limit improved by two orders of magnitude (detection limit of 15 fmol).
Magnetic NP relaxation sensors have also been used in assays in which the basis of the assay is the relaxation of the magnetic moments within magnetic NPs.78 Brownian relaxation is the dominant mechanism for these NP biosensors. In this method, NPs form aggregates upon recognition of target analytes, leading to a larger hydrodynamic size and thus slower Brownian relaxation responses than individual NPs. Using this principle, a group recently detected a bacterial antibody at the sensitivity of 0.3 nM with an AC susceptometer.79
The Weissleder group has also used antibody-coated NPs in a microfluidic device to detect bacteria.82 They also reported that core–shell NPs with Fe metal cores have enhanced sensitivity compared to IONPs for the detection of bacterial cells. Recently, the group also developed an assay based on a magnetic barcoding strategy that did not require antibodies and could detect single-gene mutations.83 In this approach, PCR-amplified mycobacterial genes are sequence-specifically captured on polymeric beads that are modified with complementary DNA, labeled by SPIONs, and detected by NMR. The platform could detect M. tuberculosis and identify drug-resistance strains from sputum samples within 2.5 h. The investigators also developed a similar system that uses rRNA as a target marker for NP labeling.84 The approach used a universal and specific nucleic acid probe that detects 16S rRNA, which is abundant in and common to many bacterial species. The device was sensitive enough to detect as few as 1–2 E. coli bacteria in 10 mL of blood and accurately estimate bacterial load.
Many groups have also utilized small molecule functionalized NPs to label bacteria. One group developed a magnetic glyco-NP based system that could detect E. coli strains in 5 minutes and enable up to 88% removal from the sample by exploiting the bacterial interaction with carbohydrates on mammalian cell surfaces (Fig. 5).85,86 Another group reported the use of vancomycin-modified SPIONs in a magnetic capture assay for various Gram-positive and Gram-negative bacteria.87 They also demonstrated that as size and ligand coverage on the NP increase, the time required for efficient labeling to the bacteria with the NPs decreases.
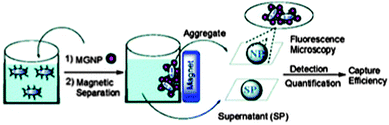 | ||
| Fig. 5 Depiction of bacteria detection by magnetic glyco-NPs (MGNPs). Reproduced from ref. 86. | ||
Optical biosensing of bacteria has also been employed using both metallic NPs and QDs. The bio-barcode assay approach50 provides amplification and the possibility of simultaneously detecting many different targets in one sample. Using this method, Bacillus subtilis double-stranded genomic DNA was detected at a 2.5 fM concentration.88 Similarly, Salmonella enteritidis was detected at 0.2 fM.89 QDs have also been used as pathogen sensors. Edgar et al. reported a technique that combined in vivo biotinylation of engineered host-specific bacteriophage and attachment of the phage to streptavidin-coated QDs.90 The method provides specific detection of E. coli among several different bacterial strains and can detect as few as 10 bacteria per mL of the experimental samples.
NP immunomagnetic techniques are some of the more commonly used methods for the identification and capture of CTCs. These techniques involve the use of magnetic NPs to target and isolate CTCs using a ligand–receptor based mechanism. Currently, the CellSearch device, which uses an immunomagnetic technique, is the only FDA-approved test for CTC assessment.92,93 CellSearch is based on a positive epithelial cell adhesion molecule (EpCAM) selection of the CTCs and utilizes iron NPs coated with a polymer layer carrying biotin analogues and conjugated with anti-EpCAM, for the capture of CTCs in vitro. This system can also be used for the labeling and identification of leukocytes using an anti-CD45-APC antibody as the NP targeting ligand. Recently, a group also demonstrated that IONPs functionalized with anti-HER2 or anti-HER2/neu could be used to separate 73.6% of HER2/neu over-expressing cancer cells that were spiked in 1 mL of blood.94 The receptor–ligand interactions resulted in the preferential capture of the cancer cells. In another preclinical study, in vivo CTC detection approach was achieved using PEGylated magnetic NPs.95 Investigators dually targeted CTCs in vivo with magnetic NPs conjugated with plasminogen activator (uPA) and folate-targeted nanotubes for subsequent detection using photoacoustic flow cytometry.
The incorporation of polymers, which can enable targeted detection and surface capturing, in other organic and inorganic NP platforms can potentially lead to novel NP sensors for CTC detection.96 Recently, a group used targeted polymer coated gold NPs and the SERS technique to directly measure CTCs in the presence of white blood cells (Fig. 6).97,98 EGF peptides were conjugated to the polymer coated gold NPs that were encoded with QSY reporters. The NPs successfully identified CTCs in the peripheral blood of 19 patients with squamous cell carcinoma of the head and neck, with a sensitivity range of 1 to 720 CTCs per mL of whole blood.
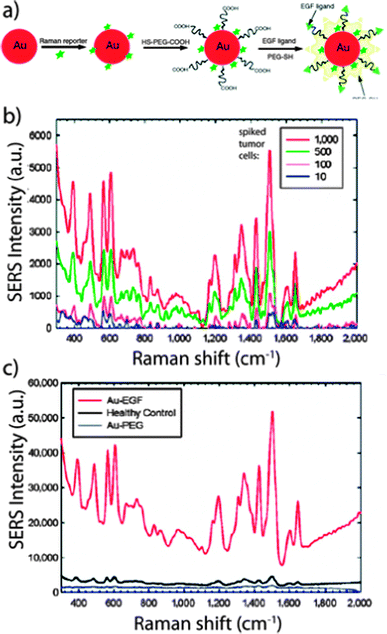 | ||
| Fig. 6 Surface-enhanced Raman scattering (SERS) detection of circulating tumor cells. (A) Schematic of EGF peptide-conjugated NPs. (B) SERS spectra of different numbers of cancer cells spiked into the white blood cell sample. (C) SERS spectra of a blood sample incubated with targeted and non-targeted NPs. Reproduced from ref. 98. | ||
Other techniques for identification and separation of cells include the use of biomimetic nanotechnology, which takes advantage of naturally occurring processes. The Hong group has investigated biomimetic approaches to develop microfluidic devices for the identification and separation of cells. These devices exploit the natural process of cell rolling, which results from adhesive interactions between selectin molecules expressed on the endothelial venules and glycoprotein receptors on the cancer cells.99 Recently, the cell rolling process using E-selectin was applied to CTC detection for enhanced surface sensitivity and specificity.100 Seventh-generation (G7) poly(amidoamine) (PAMAM) dendrimers were also used to engineer cell capture surfaces for simultaneous binding of multiple ligands to multiple receptors (multivalent binding). The biomimetic combinations of dendrimer-mediated multivalent effect and cell rolling significantly enhanced the surface capture of CTCs.
4.2 Nanoparticles as imaging agents
NPs have been explored as novel labels and contrast agents in molecular imaging. The unique properties of NPs can enable sensitive and specific monitoring of molecular targets as well as of cell responses associated with diseases such as cancer and cardiovascular diseases.101Inorganic NPs such as QDs are among the most promising fluorescent labels for cellular imaging. QDs can emit light of specific wavelengths and also be tuned to emit in the NIR region of the spectrum, in which tissue autofluorescence is reduced and excitation light penetration increased.110 Monofunctionalized QDs have been used to track individual proteins in cells111 and receptors involved in cell movement during development and metastasis.112 QDs have been used for imaging of specific tumor biomarkers, such as the use of arginine–glycine–aspartic acid (RGD) peptide-conjugated NIR QDs for the targeting of integrin αvβ3.113 Targeted QDs have also been studied for their capability for multiplex imaging, which involves simultaneous imaging of many molecular targets using different QDs that use different emission wavelengths. Recently, QDs have been applied for multiplex molecular imaging of lymph nodes,114 embryonic stem cells,115 as well as tumor cells and vasculature.116,117
Gold NPs have been studied as a non-invasive modality for in vivo cancer imaging using small organic molecules as near-infrared (NIR) SERS reporters. Recently, antibody conjugated gold NPs were used in conjunction with a sensitive and stable cyanine reporter that was developed and screened from a combinatorial library of SERS reporters for the detection of HER2-positive tumors in xenograft models.118 Other recent in vivo imaging efforts have been directed towards the use of several targeted SERS gold NPs for multiplexed imaging in mouse models.119,120
Magnetic NPs are one of the more well studied NP platforms for targeted molecular imaging. Magnetic NP imaging systems have shown potential for real-time visualization of biological events, such as cell migration/trafficking,121–123 enzyme activities,124 and other biological interactions at the molecular and cellular level.25 Magnetic NPs have also shown promising use as contrast agents in magnetic resonance imaging (MRI), a biomedical technique based on nuclear magnetic resonance of various interacting nuclei. SPIONs, formed from iron oxide crystals coated with dextran or carboxydextran, are widely used MRI contrast agents for cancer imaging.20 When injected into patients, SPIONs have been shown to remain in the tumors 24 hours after injection compared with 1 hour for gadolinium-based MR agents.125 This difference is due to easier uptake of the NP by the tumor and lower diffusivity of the NP out of the tumor.
Many studies have investigated the use of SPIONs for the targeted detection of tumors and their metastases.126–129 SPIONs have also been targeted to cancer cells without using targeting ligands. In one study, it was demonstrated that a recombinant human heavy-chain ferritin protein shell containing IONPs targeted tumor cells overexpressing transferrin receptor 1.130 The iron oxide core also catalyzed the oxidation of peroxidase substrates in the presence of hydrogen peroxide to produce a color reaction that is used to visualize tumor tissues. In another study, investigators assembled SPIONs and targeting peptides on a modified viral scaffold to increase the number of SPIONs reaching tumor cells.129 Glutamic acid residues were introduced into the protein coat of M13, a virus that infects bacteria. The negatively charged residues promoted the electrostatic assembly of NPs along the M13 coat's filamentous structure. The viral coat was also rendered to display a peptide that targets SPARC glycoprotein, which is overexpressed in various cancers. Compared to traditional approaches where NPs are directly functionalized with targeting ligands, this approach may amplify the contrast when performing MR imaging.
Novel NPs with advanced magnetic properties have also been pursued for visualizing biological events. One such class is metal-doped ferrite NPs with a composition of MFe2O4, where M is +2 cation of Mn, Fe, Co or Ni, to tune specific magnetic properties.127 MnFe2O4 NPs were found to be non-toxic in vitro and possess the highest magnetic susceptibility, suggesting that they may make better MRI probes. When these NPs were conjugated with antibodies, they showed enhanced MRI sensitivity for the detection of cancer markers. Other NP platforms have been combined with SPIONs including dendrimers (magnetodendrimers)123 and liposomes (magnetoliposomes).131 These SPIONs have been used for applications such as monitoring the migration of cells or visualizing bone marrow in vivo.132
4.3 Nanoparticles as delivery vehicles
NPs offer a potential solution to the challenges in siRNA delivery. Cationic lipid or polymer NPs have been used to transport anionic nucleic acids into cells due to their ability to form a condensed complex with nucleic acids.136 This stabilizes and protects them from enzymatic degradation. Cationic materials can also help NPs escape sequestration in endosomes/lysosomes. Groups such as the nitrogens in the cationic polymer polyethyleneimine (PEI), which become protonated in the pH environment of the endosomes/lysosomes, can facilitate endosomal escape by increasing Cl− influx in response to protonation at acidic pH. The result is an increase in osmotic pressure and swelling, which leads to organelle burst and delivery of the siRNA NPs. This phenomenon is referred to as the “proton sponge effect”. However, a recent study showed that siRNA delivery in a cationic lipid NP system was substantially reduced as approximately 70% of the internalized siRNA was recycled to the extracellular media due to the exit of the lipid NPs from late endosomes/lysosomes.137 Thus, siRNA delivery might be improved by designing novel NP vehicles that can escape the recycling pathways. Active targeting methods of non-endocytic uptake of NP delivery of nucleic acids have also been explored using fusogenic peptides and cell penetrating peptides.138 To better understand siRNA NP interactions with biological systems, siRNA probes have been used to study intracellular trafficking139 as well as assembly and disassembly of siRNA NPs.140
Recently, NPs have been used to deliver siRNA to silence genes in immune cells since these cells can have pivotal roles in homeostasis and disease.141 In one study, NPs were used to selectively silence Cyclin D1 (CyD1), a cell cycle-regulatory molecule, in leukocytes in vivo to determine the exact role of the molecule in gut inflammation.141 NPs were loaded with CyD1 siRNA and functionalized with antibodies to β7 integrin. The study revealed that these targeted NPs silenced CyD1 in leukocytes and reversed experimentally induced colitis in mice by suppressing leukocyte proliferation and T helper cell 1 cytokine expression. Another recent report also described the use of lipid NPs for the in vivo delivery of siRNA to silence disease genes in immune cells.142 The study demonstrated siRNA-mediated silencing in myeloid cell types of nonhuman primates and established the feasibility of targeting multiple gene targets in rodent myeloid cells. The therapeutic potential was validated using siRNA targeting tumor necrosis factor-α (TNFα). Another study used NPs to explore the immunological mechanisms triggering nonalcoholic steatohepatitis (NASH).142 The investigators found that TNFα produced by Kupffer cells can trigger the development of NASH through the enhanced production of chemokines IP-10 and MCP-1. Moreover, TNFα silencing in myeloid cells reduced the chemokine production and prevented the development of NASH, suggesting potential of TNFα as a novel therapeutic target in NASH.
NPs have also been used as vehicles to deliver siRNA in plant cells to study cellular pathways at the single cell level.143 One group used amine-conjugated polymeric NPs as vehicles to deliver siRNA targeting specific genes in the cellulose biosynthesis pathway. They found that NtCesA-1, a factor involved in cell wall synthesis in whole plants, also plays an essential role in cell wall regeneration in isolated protoplasts.
The ability to easily modify and functionalize NPs has resulted in recent interest in using these vehicles to deliver agents to subcellular organelles. Targeted NPs can bind to targets localized on the cell surface and enter the cell through endocytosis. However, if the target is located intracellularly, NPs and their cargo may not be able to reach the target of interest due to intracellular sequestration of the NP or due to the lack of subcellular targeting capabilities. In particular, NPs carrying oligonucleotides need to escape the endosome and then be targeted to be effective. Tools for effective subcellular targeting are emerging for targeted delivery to the nucleus,146 cytosol,147 mitochondria,148 endosomes,149 and lysosomes.150 In general, two approaches are being investigated for the design of NPs for subcellular targeting: (1) passive targeting of NPs to a particular organelle by varying NP characteristics such as size, shape, and composition151 and (2) active targeting of NPs to the organelle of interest by functionalizing NP surfaces with targeting ligands directed towards the organelle. These approaches have been applied with varying levels of success.
Hurdles to successful sub-cellular targeting include biological barriers specific to the target organelle. For example, NPs targeted to the nucleus must enter the cell membrane, escape endosomal–lysosomal pathways, possess a way to interact with the nuclear pore complex, and be small enough to cross the nuclear membrane.146 Targeting ligands such as the nuclear localization signal (NLS) have been used to enhance nuclear delivery by the active transport mechanism.152 In one study, the localization of NLS conjugated gold NPs at the nucleus of a cancer cell damaged the DNA.152 In contrast, in another study, NLS conjugated gold NPs were not able to target the nucleus of cells from outside the plasma membrane because they were unable to enter the cells or were trapped in the endosomes. Instead, NPs conjugated with both NLS and receptor-mediated endocytosis (RME) peptides reached the nucleus.153
For NPs targeted to the mitochondria, biological barriers include intracellular transport to the mitochondria and outer and inner mitochondrial membranes and toxicity.154 Thus far, most studies have primarily developed metal oxide or liposomal NPs for delivery to the mitochondria. Delivery to the mitochondria has also been based on electrostatic interactions between the NP and the organelle. The mitochondria have a negative membrane potential compared to other cellular membranes, which can lead to the accumulation of lipophilic cations.155 This concept was utilized in the fabrication of stearyl triphenyl phosphonium (STPP) targeted liposomes with anti-cancer agents ceramide156 and scarleol157 to target the mitochondria. STPP was chosen since it exhibits both cationic and lipophilic properties. Another group used a polymeric NP system to deliver mitochondria-acting therapeutics to their destination.158 The NPs were synthesized with a lipophilic triphenylphosphonium (TPP) cation, which is known to cross into the mitochondrial matrix space. Through in vitro screening of a library of NPs with varying charge and size, the group identified an optimized targeted NP that improved the efficacy and decreased the toxicity for cancer, Alzheimer's disease, and obesity compared with non-targeted NPs or the small molecule therapeutics.158
4.4 Effects of biology on NPs
Understanding NP interactions with biological systems is necessary in developing effective NPs as sensing, imaging and drug delivery agents. Thus, studies have increasingly focused on the biological effects of NP properties. Properties of NPs such as size, shape, functional group, surface charge, and composition are all factors that have been shown to affect NP interactions with biological systems in vitro and in vivo.102 However, less work has been done to study the effects of biology on NPs. It is known that NPs are rapidly cleared by the cells of the reticuloendothelial system (RES)/mononuclear phagocyte system (MPS) in the body.103 To extend the circulation time of NPs in vivo, uncharged hydrophilic polymers such as PEG are often conjugated to the surface of particles for “stealthing”. Yet, antibodies against PEG can develop after the first administration of PEGylated particles. Understanding the biomolecular interactions of nanoparticles and the MPS is critical for the development of alternative methods for extending NP circulation times in vivo.Recently, it has been shown that the uptake of NPs by cells in vitro can be influenced by the cell cycle phase.104 The study found that NPs internalized by cells are not exported from cells, but are split between daughter cells when the cell divides. The results may have implications for clearance or accumulation of NPs in vivo. Another recent study demonstrated that the global immune status, such as the balance of Th1–Th2 cytokines and M1–M2 macrophages, can also affect the NP clearance process.105 The investigators showed that mouse strains that are prone to Th1 immune responses cleared NPs at a slower rate than Th2-prone mice. Macrophages isolated from the Th1 strains also took up fewer particles in vitro than macrophages from Th2 strains. The results were confirmed in human monocyte-derived macrophages, suggesting that global immune regulation may affect NP clearance in humans.
Due to their small sizes and the EPR effect, NPs are often passively or actively targeted to cancer tumors.106 Yet the clearance and distribution of NPs can also greatly depend on the tumor vasculature. Jain and colleagues have suggested that leaky and poorly organized blood vessels of cancerous tumors can result in an increase in the interstitial fluid pressure inside tumors, reducing blood supply to them and thus impairing delivery of agents to the tumors.107 They showed that repairing the abnormal vessels in mammary tumors, by blocking vascular endothelial growth factor receptor-2, improves the delivery of smaller NPs (12 nm), while hindering the delivery of larger NPs (125 nm). The role of other proteins such as TGF-β in normalizing tumor vasculature for NP delivery in tumors has also been explored.108 TGF-β blockade was found to significantly decrease tumor growth and metastasis in mice. It also increased the recruitment and incorporation of perivascular cells into tumor blood vessels, increased the fraction of perfused vessels, and decreased the collagen I content of the tumor interstitial matrix. The investigators found that as a result of the vessel and interstitial matrix normalization, TGF-β blockade improved the intratumoral uptake of NP therapeutics, leading to better control of tumor growth.
4.5 Nanoparticles to study biological processes
The unique properties of NPs have enabled their use as promising tools to study biological processes. Many innovative techniques using NPs are being developed to activate cell signaling pathways, to induce protein production, and to improve upon current techniques used in molecular and cellular biology research. NPs such as QDs have been extensively studied for many biological applications that use fluorescence. Some of their uses include immunostaining of fixed cells and tissues, membrane proteins and cytoskeleton filaments.31,159 Recently, QDs have also been used to visualize the molecular dynamics of individual molecules in live cells. One group visualized EGF-bound receptor movements by tagging a small fraction of individual EGFR molecules with a conjugate of one CdSe QD linked to one anti-EGFR antibody Fab fragment (antiEGFR-Fab).160 The individual antiEGFR-Fab-QD-bound EGFRs (EGFR-Fab-QD) were then visualized by total internal reflection fluorescence microscopy.NP platforms can also enable the local perturbation of protein activities in cells at a subcellular scale. In particular, magnetic NPs can be coated with a biocompatible surface layer that can be functionalized with ligands that target specific cell-surface receptors, which then can be activated remotely by applied magnetic fields. In a recent study, investigators used this approach to study how NP mediated activation of specific signaling pathways can lead to changes in cellular responses.161 The magnetic NPs are attached with active signaling proteins, and can be displaced by magnetic forces into different locations of the cell. Once these protein-conjugated NPs are inserted into the cells, they bind partner proteins at their surfaces and locally stimulate signal transduction pathways. This strategy was applied to members of the Rho-GTPases, a set of molecular switches known to regulate cell morphology. NP mediated Rac1 signal was found to induce actin polymerization in protrusive areas of cells while no NP induced actin polymerization was observed in the other areas of the cell, suggesting that Rac1 associates with another partner to polymerize actin in the regions of high membrane activity. The investigators demonstrated that the NP-mediated activation of signaling pathways could also lead to a local alteration of cellular morphology and remodeling of the actin cytoskeleton. Thus, the strategies used in this study could be used to enhance understanding of how other biomolecules are spatially modulated and integrated at the cellular level.
The use of magnetic NPs for controlling cell signaling pathways was demonstrated in another study by Cheon and colleagues.162 The group showed that functionalized magnetic NPs could turn on apoptosis cell signaling when a magnetic field is applied. The magnetic switch developed consisted of zinc-doped IONPs conjugated with a targeting antibody for death receptor 4 (DR4) of DLD-1 colon cancer cells. When a magnetic field was applied to aggregate DR4-conjugated IONPs bound to cell surface receptors, an apoptosis signaling pathway was promoted. This magnetic control of apoptosis was demonstrated in vivo using zebrafish as a model organism. IONPs can also be used to activate cells and remotely regulate protein production.163,164 Stanley and colleagues used anti-His conjugated IONPs to bind to a modified epitope tagged TRPV1 channel. The temperature sensitive TRPV1 channel was activated with local heating of bound anti-His IONPs, resulting in activation of a Ca2+-sensitive promoter that stimulated the synthesis and release of bioengineered insulin. This result along with lowered blood glucose was confirmed in vivo with mice expressing the bioengineered insulin gene. The investigators further showed that cells can be engineered to synthesize genetically encoded ferritin NPs and inducibly release insulin. The use of IONPs in noninvasive techniques for cell manipulation potentially provides a useful tool for basic biology research.
Other inorganic NP platforms have also shown potential as tools to regulate cellular activities with spatial and temporal control. Recently, infrared-absorbing gold NPs were used as optical switches of gene interference and were remotely controlled using light.165 The technique involved functionalizing the NPs with double-stranded oligonucleotides and at specific times and intracellular locations, NIR illumination was used to photothermally heat the gold NPs, causing the double-stranded oligonucleotides to denature at their melting temperature and the antisense oligonucleotides to be released from the carriers.
On a macroscopic scale, the spatial organization of cells in cell culture can be important in basic biology research. Since cellular activities in conventional 2D cell culture differ from those found in vivo, many efforts have focused on developing 3D cell culture for a more physiologically relevant microenvironment. Recently, Pasqualini and colleagues reported an innovative approach to 3D tissue culture based on the magnetic levitation of cells with a hydrogel consisting of gold NPs, IONPs, and M13-derived phage particles that display integrin-targeting ligands.163 By spatially controlling the magnetic field, the authors were able to control the geometry of the cell mass and produce concentrated clustering of different cell types in co-culture. They also found that magnetically levitated human glioblastoma cells showed similar protein expression profiles to those observed in human tumor xenografts. The results indicate that magnetic levitation of cells may be a useful method for recapitulating in vivo protein expression without the use of a specific medium, engineered scaffolds or matrices.
5. Conclusions and future directions
Nanotechnology and NPs have a wide range of applications in biology. As mentioned above, nanotechnology enables the creation of devices on the same scale as individual cells and biomolecules, creating a unique approach to imaging, sensing, drug delivery and characterizing basic biological processes. Since the interest in biological application of NPs is very recent, there is reason for high optimism for more innovative and exciting applications of NPs in biology.While a significant number of genes and proteins integral to biological processes have been identified, the manner in which many of these biological units and processes assemble and integrate still remains unknown. Biological studies that employ NP techniques can provide novel insights into cell functions and molecular processes involving complex signaling pathways. NPs can also be used to help understand how these processes can be constructively modified and how various components of a cell work together to accomplish a task. For instance, biological processes such as apoptosis, cell division, and stem cell fate are modulated by mechanical cues and changes in their physical environment. In understanding the molecular mechanisms of mechanotransduction, NP analytical devices can be used to measure and manipulate interactions of biological molecules and cells to determine physical relationships between cellular components and understand how cells respond to mechanical forces and chemical cues.
NP monitoring and detection techniques can potentially aid in understanding the basis of biochemical pathways involved in disease and injury. NP sensors can enable the identification of biological molecules not addressable by current assays. Targeted NP systems can bind or react with these molecules with high affinity and selectivity, amplifying their signal for detection and molecular imaging. Furthermore, NPs can help overcome limitations associated with conventional delivery methods used in biological research such as insolubility and instability of hydrophobic compounds under aqueous conditions and nonspecific targeting. With significant progress in studying biological systems at the nanoscale and an integration of proteomics and genomics with nanotechnology, a deeper understanding of cell function and processes can be obtained that can in turn help in the design of novel nanoscale tools that may bring molecular systems back to normal operations after their function has been perturbed by disease. It is also important to note that as nanotechnology methods mature, there is less technical barrier for interested biologists to acquire NP techniques/platforms and adopt them into their own research. Currently, there are various commercial NPs available for use as contrast agents in imaging modalities such as fluorescence imaging and magnetic resonance imaging and for detection of low concentrations of analytes. However, as mentioned previously, NP properties such as size, shape, functional group, surface charge, and composition are all factors that may affect NP interactions with biological systems in vitro and in vivo. Thus, careful evaluation of effects such as toxicity induced by different types of NP formulations in biological systems is important in employing NPs for biological applications. The full utilization of NP technology in biological research will require chemists and nanotechnologists to standardize and simplify NP fabrication techniques. It will also involve nanotechnology researchers actively engaging and collaborating with biologists to identify new areas of application and synergy.
Notes
ECW has no potential conflicts of interest. AZW is a co-founder of Coordination Therapeutics Inc., and Capio Biosciences.Acknowledgements
This work is supported by grants from the University Cancer Research Fund from the University of North Carolina. Andrew Z. Wang is also supported by Career Development Award 5-K12-CA120780-01-05, National Institutes of Health Center for Nanotechnology Excellence Grant 1-U54-CA151652-01, and the National Academies Keck Futures Initiative Imaging Science Grant.References
- F. Allhoff, P. Lin and D. Moore, What is nanotechnology and why does it matter?: from science to ethics, Wiley-Blackwell, Chichester, UK, Malden, MA, 2010, p. 293 Search PubMed.
- A. D. Bangham, Liposomes: the Babraham connection, Chem. Phys. Lipids, 1993, 64, 275–285, DOI:10.1016/0009-3084(93)90071-A.
- V. P. Torchilin, Recent advances with liposomes as pharmaceutical carriers, Nat. Rev. Drug Discovery, 2005, 4, 145–160 CrossRef CAS PubMed.
- P. L. Felgner and G. M. Ringold, Cationic liposome-mediated transfection, Nature, 1989, 337, 387–388 CrossRef CAS PubMed.
- P. L. Felgner, T. R. Gadek, M. Holm, R. Roman, H. W. Chan, M. Wenz, J. P. Northrop, G. M. Ringold and M. Danielsen, Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 7413–7417 CrossRef CAS.
- A. D. Bangham, Liposomes-The Babraham Connection, Chem. Phys. Lipids, 1993, 64, 275–285, DOI:10.1016/0009-3084(93)90071-a.
- M. J. Hawkins, P. Soon-Shiong and N. Desai, Protein nanoparticles as drug carriers in clinical medicine, Adv. Drug Delivery Rev., 2008, 60, 876–885, DOI:10.1016/j.addr.2007.08.044.
- W. J. Gradishar, S. Tjulandin, N. Davidson, H. Shaw, N. Desai, P. Bhar, M. Hawkins and J. O'Shaughnessy, Phase III Trial of Nanoparticle Albumin-Bound Paclitaxel Compared with Polyethylated Castor Oil-Based Paclitaxel in Women with Breast Cancer, J. Clin. Oncol., 2005, 23, 7794–7803, DOI:10.1200/jco.2005.04.937.
- M. Harries, P. Ellis and P. Harper, Nanoparticle Albumin-Bound Paclitaxel for Metastatic Breast Cancer, J. Clin. Oncol., 2005, 23, 7768–7771, DOI:10.1200/jco.2005.08.002.
- W. J. Gradishar, Albumin-bound paclitaxel: a next-generation taxane, Expert Opin. Pharmacother., 2006, 7, 1041–1053, DOI:10.1517/14656566.7.8.1041.
- R. Gref, Y. Minamitake, M. Peracchia, V. Trubetskoy, V. Torchilin and R. Langer, Biodegradable long-circulating polymeric nanospheres, Science, 1994, 263, 1600–1603, DOI:10.1126/science.8128245.
- J. Chan, P. Valencia, L. Zhang, R. Langer and O. Farokhzad, in Cancer Nanotechnology, ed. S. R. Grobmyer and B. M. Moudgil, Humana Press, 2010, vol. 624, pp. 163–175 Search PubMed.
- V. P. Torchilin, Micellar Nanocarriers: Pharmaceutical Perspectives, Pharm. Res., 2007, 24, 1–16, DOI:10.1007/s11095-006-9132-0.
- A. Z. Wang, F. Gu, L. Zhang, J. M. Chan, A. Radovic-Moreno, M. R. Shaikh and O. C. Farokhzad, Biofunctionalized targeted nanoparticles for therapeutic applications, Expert Opin. Biol. Ther., 2008, 8, 1063–1070, DOI:10.1517/14712598.8.8.1063.
- S. H. Medina and M. E. H. El-Sayed, Dendrimers as Carriers for Delivery of Chemotherapeutic Agents, Chem. Rev., 2009, 109, 3141–3157, DOI:10.1021/cr900174j.
- J. M. J. Fréchet, Dendrimers and supramolecular chemistry, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4782–4787, DOI:10.1073/pnas.082013899.
- S. Svenson and D. A. Tomalia, Dendrimers in biomedical applications—reflections on the field, Adv. Drug Delivery Rev., 2005, 57, 2106–2129, DOI:10.1016/j.addr.2005.09.018.
- C. C. Lee, J. A. MacKay, J. M. J. Frechet and F. C. Szoka, Designing dendrimers for biological applications, Nat. Biotechnol., 2005, 23, 1517–1526 CrossRef CAS PubMed.
- M. A. Mintzer and M. W. Grinstaff, Biomedical applications of dendrimers: a tutorial, Chem. Soc. Rev., 2011, 40, 173–190, 10.1039/b901839p.
- R. Weissleder, Molecular Imaging in Cancer, Science, 2006, 312, 1168–1171, DOI:10.1126/science.1125949.
- M.-C. Daniel and D. Astruc, Gold Nanoparticles: Assembly, Supramolecular Chemistry, Quantum-Size-Related Properties, and Applications toward Biology, Catalysis, and Nanotechnology, Chem. Rev., 2003, 104, 293–346, DOI:10.1021/cr030698.
- J. W. M. Bulte and D. L. Kraitchman, Iron oxide MR contrast agents for molecular and cellular imaging, NMR Biomed., 2004, 17, 484–499, DOI:10.1002/nbm.924.
- Y.-X. Wang, S. Hussain and G. Krestin, Superparamagnetic iron oxide contrast agents: physicochemical characteristics and applications in MR imaging, Eur. Radiol., 2001, 11, 2319–2331, DOI:10.1007/s003300100908.
- M. Zhao, D. A. Beauregard, L. Loizou, B. Davletov and K. M. Brindle, Non-invasive detection of apoptosis using magnetic resonance imaging and a targeted contrast agent, Nat. Med., 2001, 7, 1241–1244 CrossRef CAS PubMed.
- R. Weissleder, A. Moore, U. Mahmood, R. Bhorade, H. Benveniste, E. A. Chiocca and J. P. Basilion, In vivo magnetic resonance imaging of transgene expression, Nat. Med., 2000, 6, 351–354 CrossRef CAS PubMed.
- M. Mahmoudi, S. Sant, B. Wang, S. Laurent and T. Sen, Superparamagnetic iron oxide nanoparticles (SPIONs): development, surface modification and applications in chemotherapy, Adv. Drug Delivery Rev., 2011, 63, 24–46, DOI:10.1016/j.addr.2010.05.006.
- C. P. Collier, T. Vossmeyer and J. R. Heath, Nanocrystal Superlattices, Annu. Rev. Phys. Chem., 1998, 49, 371–404, DOI:10.1146/annurev.physchem.49.1.371.
- T. M. Jovin, Quantum dots finally come of age, Nat. Biotechnol., 2003, 21, 32–33 CrossRef CAS PubMed.
- Y.-P. Chang, F. Pinaud, J. Antelman and S. Weiss, Tracking bio-molecules in live cells using quantum dots, J. Biophotonics, 2008, 1, 287–298, DOI:10.1002/jbio.200810029.
- W. C. W. Chan, D. J. Maxwell, X. H. Gao, R. E. Bailey, M. Y. Han and S. M. Nie, Luminescent quantum dots for multiplexed biological detection and imaging, Curr. Opin. Biotechnol., 2002, 13, 40–46, DOI:10.1016/s0958-1669(02)00282-3.
- X. Michalet, F. F. Pinaud, L. A. Bentolila, J. M. Tsay, S. Doose, J. J. Li, G. Sundaresan, A. M. Wu, S. S. Gambhir and S. Weiss, Quantum Dots for Live Cells, In Vivo Imaging, and Diagnostics, Science, 2005, 307, 538–544, DOI:10.1126/science.1104274.
- X. Huang, P. K. Jain, I. H. El-Sayed and M. A. El-Sayed, Gold nanoparticles: interesting optical properties and recent applications in cancer diagnostics and therapy, Nanomedicine, 2007, 2, 681–693, DOI:10.2217/17435889.2.5.681.
- S. Nie and S. R. Emory, Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering, Science, 1997, 275, 1102–1106, DOI:10.1126/science.275.5303.1102.
- G. Peng, U. Tisch, O. Adams, M. Hakim, N. Shehada, Y. Y. Broza, S. Billan, R. Abdah-Bortnyak, A. Kuten and H. Haick, Diagnosing lung cancer in exhaled breath using gold nanoparticles, Nat. Nanotechnol., 2009, 4, 669–673 CrossRef CAS PubMed.
- X. Liu, Q. Dai, L. Austin, J. Coutts, G. Knowles, J. Zou, H. Chen and Q. Huo, A One-Step Homogeneous Immunoassay for Cancer Biomarker Detection Using Gold Nanoparticle Probes Coupled with Dynamic Light Scattering, J. Am. Chem. Soc., 2008, 130, 2780–2782, DOI:10.1021/ja711298b.
- X. Huang, I. H. El-Sayed, W. Qian and M. A. El-Sayed, Cancer Cell Imaging and Photothermal Therapy in the Near-Infrared Region by Using Gold Nanorods, J. Am. Chem. Soc., 2006, 128, 2115–2120, DOI:10.1021/ja057254a.
- V. Marx, Targeted proteomics, Nat. Methods, 2013, 10, 19–22 CrossRef CAS.
- M. R. Capecchi, Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century, Nat. Rev. Genet., 2005, 6, 507–512 CrossRef CAS PubMed.
- S. G. O'Brien, F. Guilhot, R. A. Larson, I. Gathmann, M. Baccarani, F. Cervantes, J. J. Cornelissen, T. Fischer, A. Hochhaus, T. Hughes, K. Lechner, J. L. Nielsen, P. Rousselot, J. Reiffers, G. Saglio, J. Shepherd, B. Simonsson, A. Gratwohl, J. M. Goldman, H. Kantarjian, K. Taylor, G. Verhoef, A. E. Bolton, R. Capdeville and B. J. Druker, Imatinib Compared with Interferon and Low-Dose Cytarabine for Newly Diagnosed Chronic-Phase Chronic Myeloid Leukemia, N. Engl. J. Med., 2003, 348, 994–1004, DOI:10.1056/NEJMoa022457.
- E. H. Romond, E. A. Perez, J. Bryant, V. J. Suman, C. E. Geyer, N. E. Davidson, E. Tan-Chiu, S. Martino, S. Paik, P. A. Kaufman, S. M. Swain, T. M. Pisansky, L. Fehrenbacher, L. A. Kutteh, V. G. Vogel, D. W. Visscher, G. Yothers, R. B. Jenkins, A. M. Brown, S. R. Dakhil, E. P. Mamounas, W. L. Lingle, P. M. Klein, J. N. Ingle and N. Wolmark, Trastuzumab plus Adjuvant Chemotherapy for Operable HER2-Positive Breast Cancer, N. Engl. J. Med., 2005, 353, 1673–1684, DOI:10.1056/NEJMoa052122.
- J. C. Yang, L. Haworth, R. M. Sherry, P. Hwu, D. J. Schwartzentruber, S. L. Topalian, S. M. Steinberg, H. X. Chen and S. A. Rosenberg, A Randomized Trial of Bevacizumab, an Anti-Vascular Endothelial Growth Factor Antibody, for Metastatic Renal Cancer, N. Engl. J. Med., 2003, 349, 427–434, DOI:10.1056/NEJMoa021491.
- B. Coiffier, E. Lepage, J. Brière, R. Herbrecht, H. Tilly, R. Bouabdallah, P. Morel, E. Van Den Neste, G. Salles, P. Gaulard, F. Reyes, P. Lederlin and C. Gisselbrecht, CHOP Chemotherapy plus Rituximab Compared with CHOP Alone in Elderly Patients with Diffuse Large-B-Cell Lymphoma, N. Engl. J. Med., 2002, 346, 235–242, DOI:10.1056/NEJMoa011795.
- R. K. Jain and T. Stylianopoulos, Delivering nanomedicine to solid tumors, Nat. Rev. Clin. Oncol., 2010, 7, 653–664 CrossRef CAS PubMed.
- H. Maeda, J. Wu, T. Sawa, Y. Matsumura and K. Hori, Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review, J. Controlled Release, 2000, 65, 271–284, DOI:10.1016/s0168-3659(99)00248-5.
- A. Verma, O. Uzun, Y. Hu, Y. Hu, H.-S. Han, N. Watson, S. Chen, D. J. Irvine and F. Stellacci, Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles, Nat. Mater., 2008, 7, 588–595 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes, Angew. Chem., Int. Ed., 2002, 41, 2596–2599, DOI:10.1002/1521-3773(20020715)41:14<2596::aid-anie2596>3.0.co;2-4.
- O. C. Farokhzad, J. Cheng, B. A. Teply, I. Sherifi, S. Jon, P. W. Kantoff, J. P. Richie and R. Langer, Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 6315–6320, DOI:10.1073/pnas.0601755103.
- M. E. Werner, J. A. Copp, S. Karve, N. D. Cummings, R. Sukumar, C. Li, M. E. Napier, R. C. Chen, A. D. Cox and A. Z. Wang, Folate-Targeted Polymeric Nanoparticle Formulation of Docetaxel is an Effective Molecularly Targeted Radiosensitizer with Efficacy Dependent on the Timing of Radiotherapy, ACS Nano, 2011, 5, 8990–8998, DOI:10.1021/nn203165z.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Gold Nanoparticles in Chemical and Biological Sensing, Chem. Rev., 2012, 112, 2739–2779, DOI:10.1021/cr2001178.
- J.-M. Nam, C. S. Thaxton and C. A. Mirkin, Nanoparticle-Based Bio-Bar Codes for the Ultrasensitive Detection of Proteins, Science, 2003, 301, 1884–1886, DOI:10.1126/science.1088755.
- H. D. Hill and C. A. Mirkin, The bio-barcode assay for the detection of protein and nucleic acid targets using DTT-induced ligand exchange, Nat. Protocols, 2006, 1, 324–336 CAS.
- J.-M. Nam, A. R. Wise and J. T. Groves, Colorimetric Bio-Barcode Amplification Assay for Cytokines, Anal. Chem., 2005, 77, 6985–6988, DOI:10.1021/ac0513764.
- D. G. Georganopoulou, L. Chang, J.-M. Nam, C. S. Thaxton, E. J. Mufson, W. L. Klein and C. A. Mirkin, Nanoparticle-based detection in cerebral spinal fluid of a soluble pathogenic biomarker for Alzheimer's disease, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 2273–2276, DOI:10.1073/pnas.0409336102.
- H. Lilja, D. Ulmert and A. J. Vickers, Prostate-specific antigen and prostate cancer: prediction, detection and monitoring, Nat. Rev. Cancer, 2008, 8, 268–278 CrossRef CAS PubMed.
- C. S. Thaxton, R. Elghanian, A. D. Thomas, S. I. Stoeva, J.-S. Lee, N. D. Smith, A. J. Schaeffer, H. Klocker, W. Horninger, G. Bartsch and C. A. Mirkin, Nanoparticle-based bio-barcode assay redefines “undetectable” PSA and biochemical recurrence after radical prostatectomy, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 18437–18442, DOI:10.1073/pnas.0904719106.
- M. Swierczewska, G. Liu, S. Lee and X. Chen, High-sensitivity nanosensors for biomarker detection, Chem. Soc. Rev., 2012, 41, 2641–2655, 10.1039/c1cs15238f.
- C.-C. You, O. R. Miranda, B. Gider, P. S. Ghosh, I.-B. Kim, B. Erdogan, S. A. Krovi, U. H. F. Bunz and V. M. Rotello, Detection and identification of proteins using nanoparticle-fluorescent polymer chemical nose sensors, Nat. Nanotechnol., 2007, 2, 318–323 CrossRef CAS PubMed.
- M. De, S. Rana, H. Akpinar, O. R. Miranda, R. R. Arvizo, U. H. F. Bunz and V. M. Rotello, Sensing of proteins in human serum using conjugates of nanoparticles and green fluorescent protein, Nat. Chem., 2009, 1, 461–465 CrossRef CAS PubMed.
- R. L. Phillips, O. R. Miranda, C.-C. You, V. M. Rotello and U. H. F. Bunz, Rapid and Efficient Identification of Bacteria using Gold-Nanoparticle–Poly(para-phenyleneethynylene) Constructs, Angew. Chem., Int. Ed., 2008, 47, 2590–2594, DOI:10.1002/anie.200703369.
- A. Bajaj, O. R. Miranda, I.-B. Kim, R. L. Phillips, D. J. Jerry, U. H. F. Bunz and V. M. Rotello, Detection and differentiation of normal, cancerous, and metastatic cells using nanoparticle-polymer sensor arrays, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10912–10916, DOI:10.1073/pnas.0900975106.
- A. Bajaj, S. Rana, O. R. Miranda, J. C. Yawe, D. J. Jerry, U. H. F. Bunz and V. M. Rotello, Cell surface-based differentiation of cell types and cancer states using a gold nanoparticle-GFP based sensing array, Chem. Sci., 2010, 1, 134–138, 10.1039/c0sc00165a.
- K. E. Shafer-Peltier, C. L. Haynes, M. R. Glucksberg and R. P. Van Duyne, Toward a Glucose Biosensor Based on Surface-Enhanced Raman Scattering, J. Am. Chem. Soc., 2002, 125, 588–593, DOI:10.1021/ja028255v.
- H. Xu, E. J. Bjerneld, M. Käll and L. Börjesson, Spectroscopy of Single Hemoglobin Molecules by Surface Enhanced Raman Scattering, Phys. Rev. Lett., 1999, 83, 4357–4360 CrossRef CAS.
- B. Yan, A. Thubagere, W. R. Premasiri, L. D. Ziegler, L. Dal Negro and B. r. M. Reinhard, Engineered SERS Substrates with Multiscale Signal Enhancement: Nanoparticle Cluster Arrays, ACS Nano, 2009, 3, 1190–1202, DOI:10.1021/nn800836f.
- S. Shanmukh, L. Jones, J. Driskell, Y. Zhao, R. Dluhy and R. A. Tripp, Rapid and Sensitive Detection of Respiratory Virus Molecular Signatures Using a Silver Nanorod Array SERS Substrate, Nano Lett., 2006, 6, 2630–2636, DOI:10.1021/nl061666f.
- X. Qian, X.-H. Peng, D. O. Ansari, Q. Yin-Goen, G. Z. Chen, D. M. Shin, L. Yang, A. N. Young, M. D. Wang and S. Nie, In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags, Nat. Biotechnol., 2008, 26, 83–90 CrossRef CAS PubMed.
- X. M. Qian and S. M. Nie, Single-molecule and single-nanoparticle SERS: from fundamental mechanisms to biomedical applications, Chem. Soc. Rev., 2008, 37, 912–920, 10.1039/b708839f.
- M. P. Cecchini, V. A. Turek, J. Paget, A. A. Kornyshev and J. B. Edel, Self-assembled nanoparticle arrays for multiphase trace analyte detection, Nat. Mater., 2013, 12, 165–171 CrossRef CAS PubMed.
- J. B. Haun, T.-J. Yoon, H. Lee and R. Weissleder, Magnetic nanoparticle biosensors, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2010, 2, 291–304, DOI:10.1002/wnan.84.
- S. X. Wang and G. Li, Advances in Giant Magnetoresistance Biosensors With Magnetic Nanoparticle Tags: Review and Outlook, IEEE Trans. Magn., 2008, 44, 1687–1702, DOI:10.1109/tmag.2008.920962.
- R. S. Gaster, D. A. Hall, C. H. Nielsen, S. J. Osterfeld, H. Yu, K. E. Mach, R. J. Wilson, B. Murmann, J. C. Liao, S. S. Gambhir and S. X. Wang, Matrix-insensitive protein assays push the limits of biosensors in medicine, Nat. Med., 2009, 15, 1327–1332 CrossRef CAS PubMed.
- S. J. Osterfeld, H. Yu, R. S. Gaster, S. Caramuta, L. Xu, S.-J. Han, D. A. Hall, R. J. Wilson, S. Sun, R. L. White, R. W. Davis, N. Pourmand and S. X. Wang, Multiplex protein assays based on real-time magnetic nanotag sensing, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 20637–20640, DOI:10.1073/pnas.0810822105.
- A. Fu, W. Hu, L. Xu, R. J. Wilson, H. Yu, S. J. Osterfeld, S. S. Gambhir and S. X. Wang, Protein-Functionalized Synthetic Antiferromagnetic Nanoparticles for Biomolecule Detection and Magnetic Manipulation, Angew. Chem., Int. Ed., 2009, 48, 1620–1624, DOI:10.1002/anie.200803994.
- T. Shen, R. Weissleder, M. Papisov, A. Bogdanov and T. J. Brady, Monocrystalline iron oxide nanocompounds (MION): physicochemical properties, Magn. Reson. Med., 1993, 29, 599–604, DOI:10.1002/mrm.1910290504.
- L. Josephson, C.-H. Tung, A. Moore and R. Weissleder, High-Efficiency Intracellular Magnetic Labeling with Novel Superparamagnetic-Tat Peptide Conjugates, Bioconjugate Chem., 1999, 10, 186–191, DOI:10.1021/bc980125h.
- J. M. Perez, L. Josephson, T. O'Loughlin, D. Hogemann and R. Weissleder, Magnetic relaxation switches capable of sensing molecular interactions, Nat. Biotechnol., 2002, 20, 816–820 CrossRef CAS PubMed.
- H. Lee, E. Sun, D. Ham and R. Weissleder, Chip-NMR biosensor for detection and molecular analysis of cells, Nat. Med., 2008, 14, 869–874 CrossRef CAS PubMed.
- J. Connolly and T. G. St Pierre, Proposed biosensors based on time-dependent properties of magnetic fluids, J. Magn. Magn. Mater., 2001, 225, 156–160, DOI:10.1016/S0304-8853(00)01245-2.
- A. Fornara, P. Johansson, K. Petersson, S. Gustafsson, J. Qin, E. Olsson, D. Ilver, A. Krozer, M. Muhammed and C. Johansson, Tailored Magnetic Nanoparticles for Direct and Sensitive Detection of Biomolecules in Biological Samples, Nano Lett., 2008, 8, 3423–3428, DOI:10.1021/nl8022498.
- M. Varshney and Y. Li, Interdigitated array microelectrode based impedance biosensor coupled with magnetic nanoparticle–antibody conjugates for detection of Escherichia coli O157:H7 in food samples, Biosens. Bioelectron., 2007, 22, 2408–2414, DOI:10.1016/j.bios.2006.08.030.
- N. Xia, T. Hunt, B. Mayers, E. Alsberg, G. Whitesides, R. Westervelt and D. Ingber, Combined microfluidic-micromagnetic separation of living cells in continuous flow, Biomed. Microdevices, 2006, 8, 299–308, DOI:10.1007/s10544-006-0033-0.
- H. Lee, T.-J. Yoon and R. Weissleder, Ultrasensitive Detection of Bacteria Using Core–Shell Nanoparticles and an NMR-Filter System, Angew. Chem., Int. Ed., 2009, 48, 5657–5660, DOI:10.1002/anie.200901791.
- M. Liong, A. N. Hoang, J. Chung, N. Gural, C. B. Ford, C. Min, R. R. Shah, R. Ahmad, M. Fernandez-Suarez, S. M. Fortune, M. Toner, H. Lee and R. Weissleder, Magnetic barcode assay for genetic detection of pathogens, Nat. Commun., 2013, 4, 1752, DOI:10.1038/ncomms2745.
- H. J. Chung, C. M. Castro, H. Im, H. Lee and R. Weissleder, A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria, Nat. Nanotechnol., 2013, 5, 369–375, DOI:10.1038/nnano.2013.70.
- K. El-Boubbou, C. Gruden and X. Huang, Magnetic Glyco-nanoparticles: A Unique Tool for Rapid Pathogen Detection, Decontamination, and Strain Differentiation, J. Am. Chem. Soc., 2007, 129, 13392–13393, DOI:10.1021/ja076086e.
- Y. Pan, X. Du, F. Zhao and B. Xu, Magnetic nanoparticles for the manipulation of proteins and cells, Chem. Soc. Rev., 2012, 41, 2912–2942, 10.1039/c2cs15315g.
- A. J. Kell, G. Stewart, S. Ryan, R. Peytavi, M. Boissinot, A. Huletsky, M. G. Bergeron and B. Simard, Vancomycin-Modified Nanoparticles for Efficient Targeting and Preconcentration of Gram-Positive and Gram-Negative Bacteria, ACS Nano, 2008, 2, 1777–1788, DOI:10.1021/nn700183g.
- H. D. Hill, R. A. Vega and C. A. Mirkin, Nonenzymatic Detection of Bacterial Genomic DNA Using the Bio Bar Code Assay, Anal. Chem., 2007, 79, 9218–9223, DOI:10.1021/ac701626y.
- D. Zhang, D. J. Carr and E. C. Alocilja, Fluorescent bio-barcode DNA assay for the detection of Salmonella enterica serovar Enteritidis, Biosens. Bioelectron., 2009, 24, 1377–1381, DOI:10.1016/j.bios.2008.07.081.
- R. Edgar, M. McKinstry, J. Hwang, A. B. Oppenheim, R. A. Fekete, G. Giulian, C. Merril, K. Nagashima and S. Adhya, High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4841–4845, DOI:10.1073/pnas.0601211103.
- M. Cristofanilli, D. F. Hayes, G. T. Budd, M. J. Ellis, A. Stopeck, J. M. Reuben, G. V. Doyle, J. Matera, W. J. Allard, M. C. Miller, H. A. Fritsche, G. N. Hortobagyi and L. W. M. M. Terstappen, Circulating Tumor Cells: A Novel Prognostic Factor for Newly Diagnosed Metastatic Breast Cancer, J. Clin. Oncol., 2005, 23, 1420–1430, DOI:10.1200/jco.2005.08.140.
- M. C. Miller, G. V. Doyle and L. W. M. M. Terstappen, Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer, J. Oncol., 2010, 2010, 617421, DOI:10.1155/2010/617421.
- S. J. Cohen, C. J. A. Punt, N. Iannotti, B. H. Saidman, K. D. Sabbath, N. Y. Gabrail, J. Picus, M. Morse, E. Mitchell, M. C. Miller, G. V. Doyle, H. Tissing, L. W. M. M. Terstappen and N. J. Meropol, Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients With Metastatic Colorectal Cancer, J. Clin. Oncol., 2008, 26, 3213–3221, DOI:10.1200/jco.2007.15.8923.
- H. Xu, Z. P. Aguilar, L. Yang, M. Kuang, H. Duan, Y. Xiong, H. Wei and A. Wang, Antibody conjugated magnetic iron oxide nanoparticles for cancer cell separation in fresh whole blood, Biomaterials, 2011, 32, 9758–9765, DOI:10.1016/j.biomaterials.2011.08.076.
- E. I. Galanzha, E. V. Shashkov, T. Kelly, J.-W. Kim, L. Yang and V. P. Zharov, In vivo magnetic enrichment and multiplex photoacoustic detection of circulating tumour cells, Nat. Nanotechnol., 2009, 4, 855–860 CrossRef CAS PubMed.
- J. H. Myung, K. A. Gajjar, Y. E. Han and S. Hong, The role of polymers in detection and isolation of circulating tumor cells, Polym. Chem., 2012, 3, 2336–2341, 10.1039/c2py20420g.
- X. Wang, X. Qian, J. J. Beitler, Z. G. Chen, F. R. Khuri, M. M. Lewis, H. J. C. Shin, S. Nie and D. M. Shin, Detection of Circulating Tumor Cells in Human Peripheral Blood Using Surface-Enhanced Raman Scattering Nanoparticles, Cancer Res., 2011, 71, 1526–1532, DOI:10.1158/0008-5472.can-10-3069.
- E. C. Dreaden, A. M. Alkilany, X. Huang, C. J. Murphy and M. A. El-Sayed, The golden age: gold nanoparticles for biomedicine, Chem. Soc. Rev., 2012, 41, 2740–2779, 10.1039/c1cs15237h.
- R. Karnik, S. Hong, H. Zhang, Y. Mei, D. G. Anderson, J. M. Karp and R. Langer, Nanomechanical Control of Cell Rolling in Two Dimensions through Surface Patterning of Receptors, Nano Lett., 2008, 8, 1153–1158, DOI:10.1021/nl073322a.
- J. H. Myung, K. A. Gajjar, J. Saric, D. T. Eddington and S. Hong, Dendrimer-Mediated Multivalent Binding for the Enhanced Capture of Tumor Cells, Angew. Chem., Int. Ed., 2011, 50, 11769–11772, DOI:10.1002/anie.201105508.
- J.-H. Lee, K. Lee, S. H. Moon, Y. Lee, T. G. Park and J. Cheon, All-in-One Target-Cell-Specific Magnetic Nanoparticles for Simultaneous Molecular Imaging and siRNA Delivery, Angew. Chem., Int. Ed., 2009, 48, 4174–4179, DOI:10.1002/anie.200805998.
- A. Albanese, P. S. Tang and W. C. W. Chan, The Effect of Nanoparticle Size, Shape, and Surface Chemistry on Biological Systems, Annu. Rev. Biomed. Eng., 2012, 14, 1–16, DOI:10.1146/annurev-bioeng-071811-150124.
- F. Alexis, E. Pridgen, L. K. Molnar and O. C. Farokhzad, Factors Affecting the Clearance and Biodistribution of Polymeric Nanoparticles, Mol. Pharmaceutics, 2008, 5, 505–515, DOI:10.1021/mp800051m.
- J. A. Kim, C. Aberg, A. Salvati and K. A. Dawson, Role of cell cycle on the cellular uptake and dilution of nanoparticles in a cell population, Nat. Nanotechnol., 2012, 7, 62–68 CrossRef CAS PubMed.
- S. W. Jones, R. A. Roberts, G. R. Robbins, J. L. Perry, M. P. Kai, K. Chen, T. Bo, M. E. Napier, J. P. Y. Ting, J. M. DeSimone and J. E. Bear, Nanoparticle clearance is governed by Th1/Th2 immunity and strain background, J. Clin. Invest., 2013, 123, 3061–3073, DOI:10.1172/jci66895.
- J. Fang, H. Nakamura and H. Maeda, The EPR effect: unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect, Adv. Drug Delivery Rev., 2011, 63, 136–151, DOI:10.1016/j.addr.2010.04.009.
- V. P. Chauhan, T. Stylianopoulos, J. D. Martin, Z. Popovic, O. Chen, W. S. Kamoun, M. G. Bawendi, D. Fukumura and R. K. Jain, Normalization of tumour blood vessels improves the delivery of nanomedicines in a size-dependent manner, Nat. Nanotechnol., 2012, 7, 383–388 CrossRef CAS PubMed , http://www.nature.com/nnano/journal/v7/n6/abs/nnano.2012.45.html#supplementary-information.
- J. Liu, S. Liao, B. Diop-Frimpong, W. Chen, S. Goel, K. Naxerova, M. Ancukiewicz, Y. Boucher, R. K. Jain and L. Xu, TGF-β blockade improves the distribution and efficacy of therapeutics in breast carcinoma by normalizing the tumor stroma, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16618–16623, DOI:10.1073/pnas.1117610109.
- M. J. Welch, C. J. Hawker and K. L. Wooley, The Advantages of Nanoparticles for PET, J. Nucl. Med., 2009, 50, 1743–1746, DOI:10.2967/jnumed.109.061846.
- W. Cai, K. Chen, Z.-B. Li, S. S. Gambhir and X. Chen, Dual-Function Probe for PET and Near-Infrared Fluorescence Imaging of Tumor Vasculature, J. Nucl. Med., 2007, 48, 1862–1870, DOI:10.2967/jnumed.107.043216.
- S. Clarke, F. Pinaud, O. Beutel, C. You, J. Piehler and M. Dahan, Covalent Monofunctionalization of Peptide-Coated Quantum Dots for Single-Molecule Assays, Nano Lett., 2010, 10, 2147–2154, DOI:10.1021/nl100825n.
- M. Howarth, W. Liu, S. Puthenveetil, Y. Zheng, L. F. Marshall, M. M. Schmidt, K. D. Wittrup, M. G. Bawendi and A. Y. Ting, Monovalent, reduced-size quantum dots for imaging receptors on living cells, Nat. Methods, 2008, 5, 397–399 CrossRef CAS PubMed.
- W. Cai, D.-W. Shin, K. Chen, O. Gheysens, Q. Cao, S. X. Wang, S. S. Gambhir and X. Chen, Peptide-Labeled Near-Infrared Quantum Dots for Imaging Tumor Vasculature in Living Subjects, Nano Lett., 2006, 6, 669–676, DOI:10.1021/nl052405t.
- B. Ballou, L. A. Ernst, S. Andreko, T. Harper, J. A. J. Fitzpatrick, A. S. Waggoner and M. P. Bruchez, Sentinel Lymph Node Imaging Using Quantum Dots in Mouse Tumor Models, Bioconjugate Chem., 2007, 18, 389–396, DOI:10.1021/bc060261j.
- S. Lin, X. Xie, M. Patel, Y.-H. Yang, Z. Li, F. Cao, O. Gheysens, Y. Zhang, S. Gambhir, J. Rao and J. Wu, Quantum dot imaging for embryonic stem cells, BMC Biotechnol., 2007, 7, 67 CrossRef PubMed.
- W. J. Kang, J. R. Chae, Y. L. Cho, J. D. Lee and S. Kim, Multiplex Imaging of Single Tumor Cells Using Quantum-Dot-Conjugated Aptamers, Small, 2009, 5, 2519–2522, DOI:10.1002/smll.200900848.
- (a) X. Gao, Y. Cui, R. M. Levenson, L. W. K. Chung and S. Nie, In vivo cancer targeting and imaging with semiconductor quantum dots, Nat. Biotechnol., 2004, 22, 969–976 CrossRef CAS PubMed; (b) J. Gao, K. Chen, R. Luong, D. M. Bouley, H. Mao, T. Qiao, S. S. Gambhir and Z. Cheng, A Novel Clinically Translatable Fluorescent Nanoparticle for Targeted Molecular Imaging of Tumors in Living Subjects, Nano Lett., 2011, 12, 281–286, DOI:10.1021/nl203526f.
- A. Samanta, K. K. Maiti, K.-S. Soh, X. Liao, M. Vendrell, U. S. Dinish, S.-W. Yun, R. Bhuvaneswari, H. Kim, S. Rautela, J. Chung, M. Olivo and Y.-T. Chang, Ultrasensitive Near-Infrared Raman Reporters for SERS-Based In Vivo Cancer Detection, Angew. Chem., Int. Ed., 2011, 50, 6089–6092, DOI:10.1002/anie.201007841.
- C. L. Zavaleta, B. R. Smith, I. Walton, W. Doering, G. Davis, B. Shojaei, M. J. Natan and S. S. Gambhir, Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13511–13516, DOI:10.1073/pnas.0813327106.
- K. K. Maiti, U. S. Dinish, A. Samanta, M. Vendrell, K.-S. Soh, S.-J. Park, M. Olivo and Y.-T. Chang, Multiplex targeted in vivo cancer detection using sensitive near-infrared SERS nanotags, Nano Today, 2012, 7, 85–93, DOI:10.1016/j.nantod.2012.02.008.
- M. Mahmoudi, H. Hosseinkhani, M. Hosseinkhani, S. Boutry, A. Simchi, W. S. Journeay, K. Subramani and S. Laurent, Magnetic Resonance Imaging Tracking of Stem Cells In Vivo using Iron Oxide Nanoparticles as a Tool for the Advancement of Clinical Regenerative Medicine, Chem. Rev., 2010, 111, 253–280, DOI:10.1021/cr1001832.
- L.-Y. Chien, J.-K. Hsiao, S.-C. Hsu, M. Yao, C.-W. Lu, H.-M. Liu, Y.-C. Chen, C.-S. Yang and D.-M. Huang, In vivo magnetic resonance imaging of cell tropsim, trafficking mechanism, and therapeutic impact of human mesenchymal stem cells in a murine glioma model, Biomaterials, 2011, 32, 3275–3284, DOI:10.1016/j.biomaterials.2011.01.042.
- J. W. M. Bulte, T. Douglas, B. Witwer, S.-C. Zhang, E. Strable, B. K. Lewis, H. Zywicke, B. Miller, P. van Gelderen, B. M. Moskowitz, I. D. Duncan and J. A. Frank, Magnetodendrimers allow endosomal magnetic labeling and in vivo tracking of stem cells, Nat. Biotechnol., 2001, 19, 1141–1147 CrossRef CAS PubMed.
- C. H. Tung, U. Mahmood, S. Bredow and R. Weissleder, In vivo imaging of proteolytic enzyme activity using a novel molecular reporter, Cancer Res., 2000, 60, 4953–4958 CAS.
- W. S. Enochs, G. Harsh, F. Hochberg and R. Weissleder, Improved delineation of human brain tumors on MR images using a long-circulating, superparamagnetic iron oxide agent, J. Magn. Reson. Imaging, 1999, 9, 228–232, DOI:10.1002/(sici)1522-2586(199902)9:2<228::aid-jmri12>3.0.co;2-k.
- C. Leuschner, C. Kumar, W. Hansel, W. Soboyejo, J. Zhou and J. Hormes, LHRH-conjugated Magnetic Iron Oxide Nanoparticles for Detection of Breast Cancer Metastases, Breast Cancer Res. Treat., 2006, 99, 163–176, DOI:10.1007/s10549-006-9199-7.
- J.-H. Lee, Y.-M. Huh, Y.-w. Jun, J.-w. Seo, J.-t. Jang, H.-T. Song, S. Kim, E.-J. Cho, H.-G. Yoon, J.-S. Suh and J. Cheon, Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging, Nat. Med., 2007, 13, 95–99 CrossRef CAS PubMed.
- J.-H. Park, G. von Maltzahn, L. Zhang, M. P. Schwartz, E. Ruoslahti, S. N. Bhatia and M. J. Sailor, Magnetic Iron Oxide Nanoworms for Tumor Targeting and Imaging, Adv. Mater., 2008, 20, 1630–1635, DOI:10.1002/adma.200800004.
- D. Ghosh, Y. Lee, S. Thomas, A. G. Kohli, D. S. Yun, A. M. Belcher and K. A. Kelly, M13-templated magnetic nanoparticles for targeted in vivo imaging of prostate cancer, Nat. Nanotechnol., 2012, 7, 677–682 CrossRef CAS PubMed.
- K. Fan, C. Cao, Y. Pan, D. Lu, D. Yang, J. Feng, L. Song, M. Liang and X. Yan, Magnetoferritin nanoparticles for targeting and visualizing tumour tissues, Nat. Nanotechnol., 2012, 7, 459–464 CrossRef CAS PubMed.
- S. J. H. Soenen, M. Hodenius and M. De Cuyper, Magnetoliposomes: versatile innovative nanocolloids for use in biotechnology and biomedicine, Nanomedicine, 2009, 4, 177–191, DOI:10.2217/17435889.4.2.177.
- J. W. M. Bulte, M. de Cuyper, D. Despres and J. A. Frank, Short- vs. long-circulating magnetoliposomes as bone marrow-seeking MR contrast agents, J. Magn. Reson. Imaging, 1999, 9, 329–335, DOI:10.1002/(sici)1522-2586(199902)9:2<228::aid-jmri12>3.0.co;2-k.
- A. Khaled, S. Guo, F. Li and P. Guo, Controllable Self-Assembly of Nanoparticles for Specific Delivery of Multiple Therapeutic Molecules to Cancer Cells using RNA Nanotechnology, Nano Lett., 2005, 5, 1797–1808, DOI:10.1021/nl051264s.
- A. de Fougerolles, H.-P. Vornlocher, J. Maraganore and J. Lieberman, Interfering with disease: a progress report on siRNA-based therapeutics, Nat. Rev. Drug Discovery, 2007, 6, 443–453 CrossRef CAS PubMed.
- I. Lebedeva and C. Stein, Antisense Oligonucleotides: Promise and Reality, Annu. Rev. Pharmacol. Toxicol., 2001, 41, 403–419, DOI:10.1146/annurev.pharmtox.41.1.403.
- M. A. Behlke, Progress towards in vivo use of siRNAs, Mol. Ther., 2006, 13, 644–670 CrossRef CAS PubMed.
- G. Sahay, W. Querbes, C. Alabi, A. Eltoukhy, S. Sarkar, C. Zurenko, E. Karagiannis, K. Love, D. Chen, R. Zoncu, Y. Buganim, A. Schroeder, R. Langer and D. G. Anderson, Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling, Nat. Biotechnol., 2013, 31, 653–658, DOI:10.1038/nbt.2614.
- S. Deshayes, M. C. Morris, G. Divita and F. Heitz, Cell-penetrating peptides: tools for intracellular delivery of therapeutics, Cell. Mol. Life Sci., 2005, 62, 1839–1849, DOI:10.1007/s00018-005-5109-0.
- C. A. Alabi, G. Sahay, R. Langer and D. G. Anderson, Development of siRNA-probes for studying intracellular trafficking of siRNA nanoparticles, Integr. Biol., 2013, 5, 224–230, 10.1039/c2ib20155k.
- C. A. Alabi, K. T. Love, G. Sahay, T. Stutzman, W. T. Young, R. Langer and D. G. Anderson, FRET-Labeled siRNA Probes for Tracking Assembly and Disassembly of siRNA Nanocomplexes, ACS Nano, 2012, 6, 6133–6141, DOI:10.1021/nn3013838.
- D. Peer, E. J. Park, Y. Morishita, C. V. Carman and M. Shimaoka, Systemic Leukocyte-Directed siRNA Delivery Revealing Cyclin D1 as an Anti-Inflammatory Target, Science, 2008, 319, 627–630, DOI:10.1126/science.1149859.
- T. I. Novobrantseva, A. Borodovsky, J. Wong, B. Klebanov, M. Zafari, K. Yucius, W. Querbes, P. Ge, V. M. Ruda, S. Milstein, L. Speciner, R. Duncan, S. Barros, G. Basha, P. Cullis, A. Akinc, J. S. Donahoe, K. Narayanannair Jayaprakash, M. Jayaraman, R. L. Bogorad, K. Love, K. Whitehead, C. Levins, M. Manoharan, F. K. Swirski, R. Weissleder, R. Langer, D. G. Anderson, A. de Fougerolles, M. Nahrendorf and V. Koteliansky, Systemic RNAi-mediated Gene Silencing in Nonhuman Primate and Rodent Myeloid Cells, Mol. Ther.–Nucleic Acids, 2012, 1, e4, DOI:10.1038/mtna.2011.3.
- A. Silva, A. Nguyen, C. Ye, J. Verchot and J. Moon, Conjugated polymer nanoparticles for effective siRNA delivery to tobacco BY-2 protoplasts, BMC Plant Biol., 2010, 10, 291 CrossRef CAS PubMed.
- S. Karve, M. E. Werner, R. Sukumar, N. D. Cummings, J. A. Copp, E. C. Wang, C. Li, M. Sethi, R. C. Chen, M. E. Pacold and A. Z. Wang, Revival of the abandoned therapeutic wortmannin by nanoparticle drug delivery, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 8230–8235, DOI:10.1073/pnas.1120508109.
- L. Rajendran, H.-J. Knolker and K. Simons, Subcellular targeting strategies for drug design and delivery, Nat. Rev. Drug Discovery, 2010, 9, 29–42 CrossRef CAS PubMed.
- C. W. Pouton, K. M. Wagstaff, D. M. Roth, G. W. Moseley and D. A. Jans, Targeted delivery to the nucleus, Adv. Drug Delivery Rev., 2007, 59, 698–717, DOI:10.1016/j.addr.2007.06.010.
- J. K. Vasir and V. Labhasetwar, Biodegradable nanoparticles for cytosolic delivery of therapeutics, Adv. Drug Delivery Rev., 2007, 59, 718–728, DOI:10.1016/j.addr.2007.06.003.
- Y. Yamada and H. Harashima, Mitochondrial drug delivery systems for macromolecule and their therapeutic application to mitochondrial diseases, Adv. Drug Delivery Rev., 2008, 60, 1439–1462, DOI:10.1016/j.addr.2008.04.016.
- L. M. Bareford and P. W. Swaan, Endocytic mechanisms for targeted drug delivery, Adv. Drug Delivery Rev., 2007, 59, 748–758, DOI:10.1016/j.addr.2007.06.008.
- J. B. Lloyd, Lysosome membrane permeability: implications for drug delivery, Adv. Drug Delivery Rev., 2000, 41, 189–200, DOI:10.1016/S0169-409X(99)00065-4.
- Z. P. Xu, M. Niebert, K. Porazik, T. L. Walker, H. M. Cooper, A. P. J. Middelberg, P. P. Gray, P. F. Bartlett and G. Q. Lu, Subcellular compartment targeting of layered double hydroxide nanoparticles, J. Controlled Release, 2008, 130, 86–94, DOI:10.1016/j.jconrel.2008.05.021.
- B. Kang, M. A. Mackey and M. A. El-Sayed, Nuclear Targeting of Gold Nanoparticles in Cancer Cells Induces DNA Damage, Causing Cytokinesis Arrest and Apoptosis, J. Am. Chem. Soc., 2010, 132, 1517–1519, DOI:10.1021/ja9102698.
- A. G. Tkachenko, H. Xie, D. Coleman, W. Glomm, J. Ryan, M. F. Anderson, S. Franzen and D. L. Feldheim, Multifunctional Gold Nanoparticle–Peptide Complexes for Nuclear Targeting, J. Am. Chem. Soc., 2003, 125, 4700–4701, DOI:10.1021/ja0296935.
- S. A. Durazo and U. B. Kompella, Functionalized nanosystems for targeted mitochondrial delivery, Mitochondrion, 2012, 12, 190–201, DOI:10.1016/j.mito.2011.11.001.
- M. Breunig, S. Bauer and A. Goepferich, Polymers and nanoparticles: Intelligent tools for intracellular targeting?, Eur. J. Pharm. Biopharm., 2008, 68, 112–128, DOI:10.1016/j.ejpb.2007.06.010.
- S. V. Boddapati, G. G. M. D'Souza, S. Erdogan, V. P. Torchilin and V. Weissig, Organelle-Targeted Nanocarriers: Specific Delivery of Liposomal Ceramide to Mitochondria Enhances Its Cytotoxicity In Vitro and In Vivo, Nano Lett., 2008, 8, 2559–2563, DOI:10.1021/nl801908y.
- N. R. Patel, S. Hatziantoniou, A. Georgopoulos, C. Demetzos, V. P. Torchilin, V. Weissig and G. G. M. D'Souza, Mitochondria-targeted liposomes improve the apoptotic and cytotoxic action of sclareol, J. Liposome Res., 2010, 20, 244–249, DOI:10.3109/08982100903347931.
- S. Marrache and S. Dhar, Engineering of blended nanoparticle platform for delivery of mitochondria-acting therapeutics, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16288–16293, DOI:10.1073/pnas.1210096109.
- Y. Xing, Q. Chaudry, C. Shen, K. Y. Kong, H. E. Zhau, L. W. Chung, J. A. Petros, R. M. O'Regan, M. V. Yezhelyev, J. W. Simons, M. D. Wang and S. Nie, Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry, Nat. Protocols, 2007, 2, 1152–1165 CAS.
- I. Chung, R. Akita, R. Vandlen, D. Toomre, J. Schlessinger and I. Mellman, Spatial control of EGF receptor activation by reversible dimerization on living cells, Nature, 2010, 464, 783–787 CrossRef CAS PubMed.
- F. Etoc, D. Lisse, Y. Bellaiche, J. Piehler, M. Coppey and M. Dahan, Subcellular control of Rac-GTPase signalling by magnetogenetic manipulation inside living cells, Nat. Nanotechnol., 2013, 8, 193–198 CrossRef CAS PubMed.
- M. H. Cho, E. J. Lee, M. Son, J.-H. Lee, D. Yoo, J.-w. Kim, S. W. Park, J.-S. Shin and J. Cheon, A magnetic switch for the control of cell death signalling in in vitro and in vivo systems, Nat. Mater., 2012, 11, 1038–1043 CrossRef CAS PubMed.
- G. R. Souza, J. R. Molina, R. M. Raphael, M. G. Ozawa, D. J. Stark, C. S. Levin, L. F. Bronk, J. S. Ananta, J. Mandelin, M.-M. Georgescu, J. A. Bankson, J. G. Gelovani, T. C. Killian, W. Arap and R. Pasqualini, Three-dimensional tissue culture based on magnetic cell levitation, Nat. Nanotechnol., 2010, 5, 291–296 CrossRef CAS PubMed.
- S. A. Stanley, J. E. Gagner, S. Damanpour, M. Yoshida, J. S. Dordick and J. M. Friedman, Radio-Wave Heating of Iron Oxide Nanoparticles Can Regulate Plasma Glucose in Mice, Science, 2012, 336, 604–608, DOI:10.1126/science.1216753.
- S. E. Lee, G. L. Liu, F. Kim and L. P. Lee, Remote Optical Switch for Localized and Selective Control of Gene Interference, Nano Lett., 2009, 9, 562–570, DOI:10.1021/nl802689k.
| This journal is © The Royal Society of Chemistry 2014 |