Mechanical properties of binary DPPC/DPPS bilayers
J. J. López
Cascales
*a,
S. D. Oliveira
Costa
a,
A.
Garro
ab and
R. D.
Enriz
ab
aUniversidad Politécnica de Cartagena, Bioinformatics and Macromolecules Group (BIOMAC), Aulario II, Campus de Alfonso XIII, 30203, Cartagena, Murcia, Spain. E-mail: javier.lopez@upct.es
bUniversidad Nacional de San Luis, Facultad de Química, Bioquímica y Farmacia, IMIBIO-CONICET, Chacabuco 917, 5700, San Luis, Argentina
First published on 1st October 2012
Abstract
In this work, we studied how the lipid composition and ionic strength of an aqueous solution affect the mechanical properties of the lipid bilayer. The interfacial tension, the bending modulus, the Gaussian curvature modulus and the bilayer curvature energy of the lipid bilayer were studied by molecular dynamics simulation. For this purpose, the lipid bilayer was modelled as a binary symmetric lipid bilayer of DPPC (DiPalmitoylPhosphatidylCholine) and DPPS (DiPalmitoylPhosphatidylSerine) at different molar ratios of these two lipids in the absence of salt and in presence of 0.5 N NaCl in aqueous solution. The results of the simulations in absence of salt showed how an increase in the DPPS concentration of the lipid bilayer strongly affects most of its mechanical properties, including the lateral pressure across the membrane, interfacial tension, or the bending modulus of the lipid bilayer. However, in the presence of 0.5 N NaCl, the interfacial tension of the lipid bilayer becomes independent of the lipid composition (in the range of 15–70% DPPS), behavior that may have notable implications from a biological point of view, due to the contribution that this property may provide to the stability of biological membranes.
1 Introduction
Biological membranes play a crucial role in cells by providing a suitable environment to molecules of important biological significance, such as membrane proteins, polysaccharides or surface proteins. Furthermore, because cell membranes are not homogeneous systems because of the different lipid compositions of both leaflets of the membrane, or the existence of lipid rafts, studying how the lipid composition and ionic strength affect the physical properties of a lipid bilayer is a crucial issue to be investigated to gain insight into its biological activity.Cell membranes are mainly composed of two types of molecules, lipids and proteins. On the one hand, lipids play a crucial role by making the membrane almost impermeable to water, ions and most solutes dissolved in aqueous solutions, and, on the other, they provide a suitable environment to other types of molecule such as proteins that may be embedded in or attached to the membrane, while proteins serve as transporters and signalling devices.1
A key aspect of these cell membranes is related with their almost fluid nature, whereby dynamic lipid–protein interactions regulate their cell activity, as reflected in the fluid mosaic model.2 In this regard, membrane protein activity may well be influenced by its surrounding lipid matrix,3 and, more specifically, by the association between lipids and proteins at the lipid–protein interface.4,5 In this context, occasional patches of lipid molecules of different compositions6 (known as lipid rafts) may regulate the function of the membrane proteins by altering some physical properties of the cell membrane,1 such as its elasticity, interfacial tension or bending energy, through the redistribution, at molecular level, of the lateral pressure across the lipid bilayer.7,8
In this context, this work is devoted to studying the variation of some mechanical properties associated with the lipid composition of binary lipid bilayers formed by DPPC (DiPalmitoylPhosphatidylCholine) and DPPS (DiPalmitoylPhosphatidylSerine), in the presence and absence of salt in aqueous solution, calculating the lateral pressure profile by means of the pressure tensor decomposition proposed by Lindahl and Edholm.9 A methodical analysis of the lateral pressure profile was carried out using molecular dynamics simulation for different molecular fractions of DPPC (neutral lipid) and DPPS (charged lipid), which ranged from a pure and neutral bilayer formed only of DPPC, to a membrane exclusively formed of DPPS, in the absence and in presence of 0.5 N NaCl in aqueous solution. DPPC was chosen for our simulations because it is a common uncharged lipid found in most biological membranes of eukaryotic cells in physiological conditions, while DPPS is a charged lipid in physiological conditions, being asymmetrically distributed in the interior leaflet of biological membranes,10–13 and is involved in important biological processes, such as cell phagocytosis.14–16
Based on previous simulations involving molecular dynamics simulations of lipid bilayers in atomic detail,17–19 a methodical study was made of the mechanical properties of these lipid bilayers, depending on the DPPS/DPPC ratio that constitutes the lipid bilayers, in the absence and presence of salt in the aqueous solution. Thus, important features concerning the effect of the fraction of charged lipids conforming the bilayers and how the ionic strength of the aqueous solution affects to the bending modulus, kb, the lateral pressure profile across the bilayer, π(z), the interfacial tension γ, the Gaussian curvature modulus, KbG, and the bilayer curvature energy, Ecurve were obtained, and how the variations of these properties may present important implications on the biological activity of these lipid bilayers at a molecular level.
2 Model and methods
2.1 Model
Five different lipid bilayers were generated with the goal of analysing the full range of lipid compositions of a binary bilayer composed of DPPC and DPPS. The starting system was formed by a bilayer composed of 288 DPPC molecules (144 DPPC per leaflet) and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 068 water molecules of the single point charge (SPC) water model.20 A precise description of how the three-dimensional periodical box was generated is given elsewhere.17,18,21,22 Once this bilayer of DPPC was generated, four additional bilayers were constructed by substituting 48, 96, 192 and 288 DPPC molecules by 48, 96, 192 and 288 DPPS molecules. To balance the negative charge associated to each DPPS molecule under physiological conditions, 48, 96, 192 and 288 sodium ions (Na+) were introduced into the system by substituting water molecules by sodium ions. In summary, five different binary bilayers of DPPC
068 water molecules of the single point charge (SPC) water model.20 A precise description of how the three-dimensional periodical box was generated is given elsewhere.17,18,21,22 Once this bilayer of DPPC was generated, four additional bilayers were constructed by substituting 48, 96, 192 and 288 DPPC molecules by 48, 96, 192 and 288 DPPS molecules. To balance the negative charge associated to each DPPS molecule under physiological conditions, 48, 96, 192 and 288 sodium ions (Na+) were introduced into the system by substituting water molecules by sodium ions. In summary, five different binary bilayers of DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPS were generated with the following compositions 288
DPPS were generated with the following compositions 288![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 240
0, 240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48, 192
48, 192![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 96, 96
96, 96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 192 and 0
192 and 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 288.
288.
The molecular fraction of DPPS that forms the lipid bilayer, χ, was defined as follows:
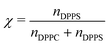 | (1) |
Finally, to simulate a 0.5 N NaCl concentration in the aqueous solution, two out of every 28 water molecules in each of the systems generated above were randomly substituted by one sodium and one chloride ion, respectively.
2.2 Local pressure profile: theory
To compute the local pressure from the simulated trajectory, the instantaneous pressure must be calculated across the phospholipid bilayer every time step. Thus, considering that the pressure tensor P is expressed as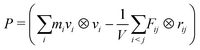 | (2) |
Since the electrostatic contribution using a long-range electrostatic interaction by the Particle Mesh Ewald method23,24 can not be decomposed in Fij terms, the contribution of the electrostatic interactions must be calculated using a cut-off method. In this regard, to calculate the pressure profile along the Z-axis perpendicular to the lipid bilayer, the P tensor must be calculated for thin slices parallel to the membrane surface across the lipid bilayer; in our case, a thickness of roughly 0.1 nm was chosen for all our simulations. However, in certain circumstances (or even in most cases), the particles i and j may lie in different slices. In these cases, the Lindahl and Edholm approximation9 can be used, and eqn (2) was accordingly modified as follows:
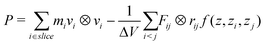 | (3) |
1. If both particles fall in the same slice, i, f = 1.
2. If both particles are outside slice i and on opposite sides,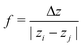 .
.
3. If one particle is in slice i and the other outside, 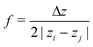
2.3 Simulation parameters
The GROMACS 3.3.3 package25,26 was used to perform the molecular dynamics simulations, with a constant integration time step of 2 fs. The electrostatic contribution was calculated using a long-range electrostatic interaction by the Particle Mesh Ewald method,23,24 in which all the coordinates of the simulated trajectories were recorded every 5 ps of simulation time. Bond lengths were constrained using the LINCS algorithm.27 All the simulation boxes were coupled to an external pressure and temperature bath, using the Berendsen algorithm,28 with temperature and pressure coupling constants of 0.1 and 1 ps, respectively. Due to the anisotropy of the membranes along the Z-axis, all the simulations were carried out using a semi-isotropic pressure algorithm coupling bath. The simulated trajectory lengths were of 100 ns, where the first 10 ns of each simulation were discarded for analysis because this was the time required by the systems to achieve an equilibrated state. All the simulations were carried out at 350 K, a temperature that was chosen because it is above the transition temperature of 314 K29 and 326 K30 for pure bilayers of DPPC and DPPS, and also because this temperature is above the transition temperature of all the binary bilayers formed by DPPC/DPPS, as deduced from its experimental phase diagram.31 In short, the temperature of 350 K ensures that all these binary bilayers are in liquid crystalline state, regardless of the fraction of DPPS in the lipid bilayer. The molecules of DPPC and DPPS were simulated using the force field employed in previous simulations of lipid bilayers of DPPC32 and DPPS,17 and the SPC,20 the water model used in our simulations.On the other hand, since calculation of the lateral pressure profile needs much computing power, the trajectories generated using the particle Mesh Ewald method were employed to recalculate the pressure tensor of eqn (3) using the cut-off method rather than the Particle Mesh Ewald method mentioned above. In the process of calculating the lateral pressure profile, short and long spherical cut-offs of 1.4 and 1.8 nm were used for re-calculating the non-bonded forces, according to previous simulations carried out by other authors.33–35 Thus, while the list of atoms inside the short cut-off was updated and their interactions calculated every time step, the list of neighbouring atoms between the short and long cut-offs was updated and their interactions calculated every ten time steps. In an attempt to avoid any mismatch between these two ways of calculating the electrostatic interactions, several simulations were carried out at different εr, almost the same results being obtained for the surface area per lipid, using the particle Mesh Ewald or the cut-off method, when εr = 1.1. To a certain extent, this result was expected from previous simulations, involving cut-off method rather than Particle Mesh Ewald method to calculate the electrostatic interactions, because the electrostatic interaction was overestimated due to the absence of polarizability in the atomic models used in our simulations, as mentioned, too, by Berendsen et al. in several of their papers.32
3. Results and discussion
3.1 The bilayer bending modulus, kb
The bilayer bending modulus, kb, in a lipid bilayer can be calculated as follows:36| kb = KAξ2/24 | (4) |
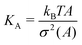 | (5) |
Table 1 shows the kb values obtained for all the bilayers studied, in the absence and presence of salt. For a lipid bilayer of pure DPPC (without DPPS), corresponding to χ = 0, values of 16.4 ± 0.2 kBT and 15.84 ± 0.08 kBT were obtained for kb in the absence and the presence of salt, respectively. These values are in good agreement with the experimental data of 16.7 kBT obtained from X-ray measurements37 of DMPC lipid bilayers in their liquid-crystalline phase and from pipette aspiration measurements, which were in a range of 11–30 kBT, depending on the length of the lipid hydrocarbon tails of PC-phospholipids38,39 and 15.8 kBT for a DOPC bilayer with unsaturation in its lipid chains.40 They also agree with the measurements from Neutron Spin Echo experiments (NSE), where a value of 25.5 kBT41 was obtained, where the difference from our results may be attributed to the different temperatures at which the experiments and simulations were performed due to the temperature dependence of the bending modulus.41
| Without NaCl | ||||
|---|---|---|---|---|
| χ | Area/nm2 | d HH/nm | K A/N m−1 | k b/(kBT) |
| 0.0 | 0.683 ± 0.007 | 1.5 | 0.468 ± 0.005 | 16.4 ± 0.18 |
| 0.17 | 0.626 ± 0.007 | 1.59 | 0.429 ± 0.005 | 17.4 ± 0.2 |
| 0.33 | 0.586 ± 0.007 | 1.83 | 0.401 ± 0.008 | 17.2 ± 0.2 |
| 0.67 | 0.531 ± 0.006 | 2.08 | 0.424 ± 0.005 | 24.9 ± 0.3 |
| 1.0 | 0.533 ± 0.006 | 1.76 | 0.426 ± 0.005 | 31.8 ± 0.4 |
| With 0.5 N NaCl | ||||
|---|---|---|---|---|
| 0.0 | 0.676 ± 0.008 | 1.52 | 0.406 ± 0.005 | 15.84 ± 0.08 |
| 0.17 | 0.620 ± 0.007 | 1.64 | 0.425 ± 0.005 | 18.5 ± 0.2 |
| 0.33 | 0.577 ± 0.006 | 1.87 | 0.461 ± 0.006 | 22.1 ± 0.2 |
| 0.67 | 0.520 ± 0.006 | 2.05 | 0.416 ± 0.005 | 26.5 ± 0.3 |
| 1.0 | 0.526 ± 0.005 | 1.79 | 0.504 ± 0.005 | 36.3 ± 0.4 |
Concerning the kb of a bilayer of DPPS, values of 31.8 ± 0.4 and 36.3 ± 0.4 kBT were estimated from simulation in the absence and presence of salt. These values are in reasonably good agreement with the value of kb provided by Petrache et al.42 from high-resolution X-ray measurements, where a value of 26 kBT was estimated for a DOPS bilayer. The slight difference between simulation and experimental data may be associated with the increases of elasticity of lipid bilayer with the unsaturation of the lipid chains, such as proposed by Rawicz et al39 from pipette aspiration measurements.
From the values of Table 1, we observe how the bending modulus increases with the fraction of DPPS in the bilayer, χ, in both the absence and presence of salt. Thus, Fig. 1 shows how kb fits a straight line with slopes of 16.51 and 20.10 (kBT), in the absence and presence of salt, respectively. From this behavior it is deduced that the presence of salt in the solution enhances the bending modulus of the lipid bilayer, in which this difference in kb increases with the molar fraction of DPPS in the lipid bilayer.
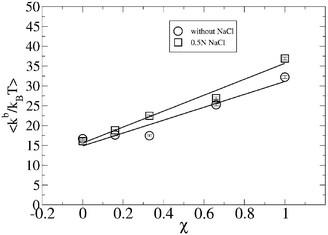 | ||
| Fig. 1 Bending modulus, kb for different binary compositions, χ, in the absence and presence of 0.5 N NaCl solution. | ||
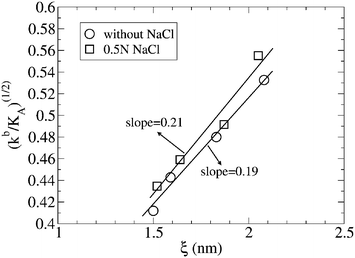 | ||
| Fig. 2 Linear representation of (kb/KA)1/2vs. ξ, where ξ was defined as the effective thickness of the lipid bilayer, with ξ = dHH − 1 in nm and dHH is the distance between the maximum of the head density distribution in both leaflets of the lipid bilayers for different composition of the lipid bilayer χ, in the absence and presence of a 0.5 N NaCl solution. For further information, see the text. | ||
From the behavior of kb with χ, and given that KA remains almost constant for the whole range of molecular fractions of DPPS that form the bilayer (see Table 1), the influence of the thickness of the lipid bilayer on its elastic properties should be noted. From a linear representation of (kb/KA)1/2vs. ξ (Fig. 2), a straight line is obtained (discarding the points corresponding to χ = 1, which deviate from the lineal regression), with slopes of 0.19 and 0.21 (dimensionless units) in the absence and presence of salt, results that are in perfect agreement with the slope of 0.20 obtained by Rawicz et al.39 for the fit of lipid bilayers of different thicknesses formed by PC of different hydrocarbon tail lengths.
The effect of the ionic strength on the bending rigidity agrees well with the experimental data provided by Claessens et al.43 for a DOPG and DOPC bilayer at different ionic strengths of aqueous solution. Thus, these authors determined how the bending modulus of charged bilayers formed by DOPG is higher than the corresponding one formed only by DOPC. Furthermore, the bending modulus of charged bilayers of DOPG increases as well with the ionic strength of the aqueous solution, behavior that is in full agreement with the results from our simulations.
3.2 Lateral pressure profiles, π(z)
Phospholipid bilayers in their liquid crystalline state are fluid systems subjected to different types of molecular interactions, some parallel and others perpendicular to the bilayer plane, due to the anisotropic nature of these molecular systems. In this regard, the lateral pressure π(z) along the Z-axis can be defined as follows:| π(z) = plat(z) − pN | (6) |
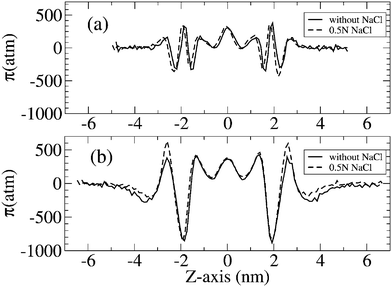 | ||
| Fig. 3 Lateral pressure profile of DPPC and DPPS bilayers corresponding to χ = 0 (a) and χ = 1 (b), in the absence (solid line) and presence (dashed line) of 0.5 N NaCl in solution. | ||
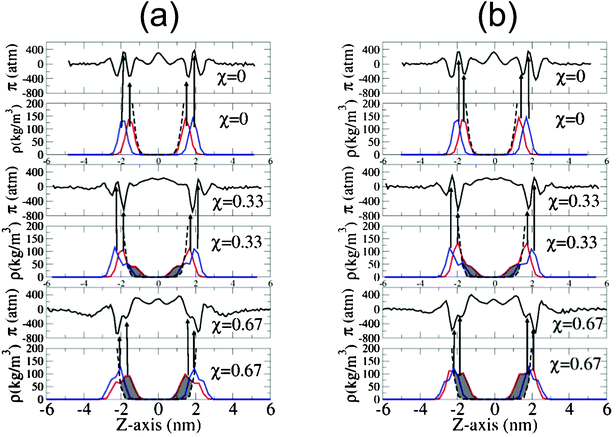 | ||
| Fig. 4 Lateral pressure profile of bilayers of different molecular fractions, χ, in the absence of salt (a) and with 0.5 N NaCl in solution (b). Red lines correspond to the atomic density distribution of oxygen carbonyl, and the blue lines to the phosphorus atomic density distribution across the lipid bilayer. The dashed line corresponds to the water density distribution and the arrows associate the maximum of the carbonyl and phosphorus distribution with their respective positions in the lateral pressure profiles at that point. | ||
Unfortunately, the lateral pressure profiles obtained from numerical calculations can not easily be contrasted with experimental data, because, to the best of our knowledge, there are very few experimental works in which lateral pressure has been estimated at a molecular level. One of such works was the study performed by Kamo et al.,44 in which they estimated the lateral pressure in a lipid bilayer based on fluorescence measurements using dipyrenylphosphatidylcholines (DipyPCs) as a fluorescent probe.
Thus, for a bilayer of DPPC without DPPS (corresponding to the case in which χ = 0), a maximum in the lateral pressure profile π(z) of = 320 atm is identified for a Z-axis that matches the peak in the atomic distribution of the phosphorus atom of the head of the DPPC. This positive value of π(z) means that, at this depth in the bilayer plane, repulsive interactions are taking place, which contribute to the expansion of the lipid bilayer. However, in the plane of the bilayer mainly occupied by the carbonyl oxygens of the lipid tails, the attractive interactions between the adjacent lipids leads to a negative value in the lateral pressure π(z) = −342 atm to compensate the positive values in the region occupied by the phosphate groups, as mentioned above. Hence, in the molecular region between the phosphate and the oxygen carbonyl groups of the lipid tails, a pressure gradient of around 660 atm is estimated, a value that is of the same order of magnitude as the value estimated by Gullinsgsrud and Schulten,33 who predicted a pressure gradient of 1000 atm for the lateral pressure at the boundary between the hydrophilic and the hydrophobic part of the lipid bilayer. On the other hand, in the core of the hydrocarbon region of the lipid bilayer, a value of 350 atm was predicted by these authors, a value in close agreement with the 308 atm value obtained in our simulations. Compensating these positive values in the hydrocarbon region in the interior of the lipid bilayer, a minimum value of π = −350 atm was observed at the lipid/water interface, in the vicinity of the head distribution of the lipid bilayer. Furthermore, from Fig. 4, we observe how the bilayers of DPPC are highly hydrated, regardless of the absence or presence of salt, as can be deduced from the atomic density distribution of phosphorus, carbonyl oxygens and water across the lipid bilayer.
When the fraction of DPPS in the lipid bilayer increases, the minimum in the π(z) distribution associated with the region occupied by oxygen carbonyl of the lipid tails also increases considerably, reflecting the enhancement of the lipid–lipid interaction between neighbouring lipids, these favourable interactions being associated with the molecular interactions between the NH3+ of DPPS and the carbonyl oxygens of the neighbouring lipids (DPPC or DPPS), as discussed elsewhere.17,19 To gain further insight, the radial distribution function, g(r) between the NH3+ belonging to the DPPS and the oxygen carbonyl group of the lipid tails of the DPPC is shown in Fig. 5, where the radial distribution function, g(r), is defined as follows:
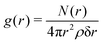 | (7) |
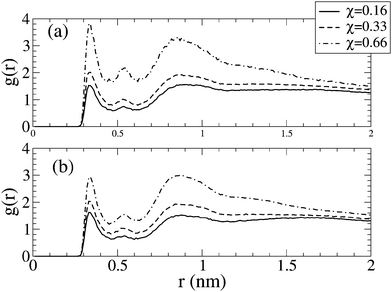 | ||
| Fig. 5 Radial distribution function, g(r), between nitrogen of DPPS and oxygen carbonyl group of DPPC for bilayers of different molecular fractions, χ, in the absence of salt (a) and with 0.5 N NaCl in solution (b). | ||
Parallel to this increase in molecular interactions between neighbouring lipids, lipid dehydration also increased. The extent of this dehydration is shown in Fig. 4 by an increase in the shaded area, where the increase in the molecular fraction in DPPS, is clearly matched by the dehydration of the lipid heads. Similarly, Fig. 6 shows how the reduction in the hydration of the carbonyl groups of DPPC and DPPS decreases with an increasing molecular fraction in DPPS, in part compensated by interactions between neighbouring lipids, as mentioned above.
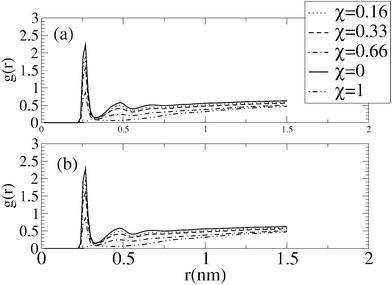 | ||
| Fig. 6 Radial distribution function g(r) of water around the oxygen carbonyl group of the lipid tails for bilayers of different molecular fraction, χ, in the absence of salt (a) and with 0.5 N NaCl in solution (b). | ||
From Fig. 6, no great variation in the hydration of the oxygen carbonyl group of the lipid tails in the absence and presence of salt is evident. However, in the presence of salt, Fig. 5 points to a decrease in the interactions between neighbouring lipids as seen from the reduction in peaks associated to the radial distribution function between neighbouring lipids, although this decreasing is compensated by the coordination of these oxygen carbonyl groups by sodium ions.
Finally the peak associated with this minimum of π(z) reflected a value of −700 and −840 atm when χ = 0.67 and 1 in the absence of salt, and −680 and −869 atm, when χ = 0.67 and 1 in a 0.5 N NaCl solution.
3.3 Lipid/water interfacial tension, (γb)
The interfacial tension measured experimentally is related with the interfacial tension calculated from simulation by eqn (8):| γb = γr + γs | (8) |
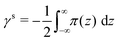 | (9) |
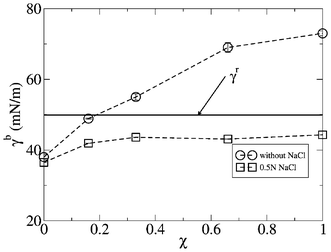 | ||
| Fig. 7 Interfacial tension vs. the molecular fraction χ of the lipid bilayer. Error bars were calculated from three sub-trajectories of 30 ns each, where the first 10 ns of simulation trajectory were discarded as being the equilibration time. | ||
In the case of a bilayer formed only by DPPC, χ = 0, an interfacial tension of γb = 38.1 ± 0.8 and 36.50 ± 0.07 mN m−1 was estimated from simulation, in the absence of salt and with 0.5 N NaCl in the aqueous solution, values that are of the same order of magnitude as the value measured for the oil–water interface tension, where values of ≈40 mN m−1 have been reported.45,47 In addition, these values agree with the simulation data calculated by Gullingsrud et al,33 who reported values in a range from 27.6 ± 10 to 39.6 ± 1.6 mN m−1 for different surface areas of DOPC.
Fig. 7 shows how the interface tension increases with the fraction of DPPS in the lipid bilayer. Thus, up to a fraction of χ = 0.18, the interfacial tension has values below the reference interfacial tension, γb < γr, and from that point on, the interfacial tension becomes higher than the reference interfacial tension. This enhancement of the interfacial tension with the fraction of DPPS in the lipid bilayer is directly related with the increase in the dehydration of the lipid bilayer associated with the increasing the lipid–lipid molecular interactions, as was described in the previous section.
On the other hand, the presence of salt in aqueous solution substantially changes the trend of the interface tension related with the fraction of DPPS in the lipid bilayer. Thus, in general, for the full range of fractions of DPPS in the lipid bilayer, the interfacial tension adopts values below the interfacial tension of the reference, γb < γr. Hence, the lipid–lipid interactions associated with the presence of DPPS in the lipid bilayer are partially screened by the ionic strength of the aqueous solution, so that γb reaches a plateau for a range of molecular fractions of DPPS between 0.2 and 1, γb taking values of ≈43 mN m−1 in this range of DPPS molecular fractions.
This behavior of the interfacial tension associated with the presence of salt in the aqueous solution and the molecular fraction of charged lipids in the lipid bilayer may be of a great biological relevance, since under a certain ionic strength of the aqueous solution, the interfacial tension of a cellular membrane becomes independent of the fraction of the charged lipids present in the membrane. This fact is of a crucial importance considering the asymmetry in the molecular fraction of charged lipids between both leaflets of the membrane of an eukaryotic cell, whereby the ionic strength could equal the interfacial tension between both sides of the membrane, even when they have different lipidic compositions.
3.4 Gaussian curvature modulus, KbG, and the bilayer curvature energy, Ecurve
From simulation, the bilayer Gaussian Curvature Modulus, KbG, can be estimated as follows:36| KbG = 2(KmG − kmCm0ξ) | (10) |
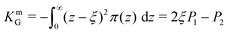 | (11) |
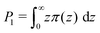 | (12) |
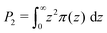 | (13) |
Thus, KbG can adopt positive or negative values. In films that prefer isotropic shapes such as spheres or planes, the Gaussian modulus adopts negative values, KbG < 0, while in films that prefer saddle-splay shapes the modulus adopts positive values, KbG > 0, in accordance with Safran's theoretical studies.49Fig. 8 depicts the variation of the Gaussian modulus, KbG, in the absence and presence of salt, for different molecular fractions of charged lipids in the bilayer. Thus, in the absence of salt, KbG fits a straight line for χ values between 0 and 0.7. Note how, in the absence of salt, for lipid compositions below χ = 0.1, the bilayer adopts an isotropic shape, corresponding to its negative KbG value. In this regard, for χ = 0 a value of KbG = −6 ± 2 kBT was estimated from our simulations, a value that is in a reasonably good agreement with the value of −10 ± 2.3 kBT estimated by Orsi et al.36 for a DPPC bilayer without salt in aqueous solution. However, for values of χ > 0.1, the bilayer adopts a saddle-splay shape, according with Safran's theoretical studies. On the other hand, in the presence of salt, a KbG value of −9.9 ± 1.5 kBT is obtained, which is also in a good agreement with the value estimated by Orsi et al.36 In this regard, we observed a similar qualitative behavior to that obtained in absence of salt for all the bilayers with molecular fractions below χ = 0.7, where KbG also fits a straight line, although with a lower slope than in the absence of salt.
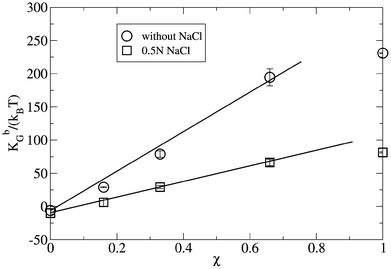 | ||
| Fig. 8 Gaussian constant, KbG, for different bilayer compositions, χ, in the absence of salt and in the presence of 0.5 N NaCl aqueous solution. Error bars were estimated from the last three sub-trajectories of 30 ns each, where the first 10 ns were discarded as being the equilibration time. | ||
With the goal of providing additional insight related with the bilayer stability, Claessens et al43 proposed that bilayer stability depends on the values of the bilayer bending modulus, kb, and the Gaussian curvature module, KbG. Thus, mechanically stable bilayers are only found if the total free energy of bending the bilayer is positive with respect to the original shape.43 Hence, in a first approximation and considering that the system passes from a planar to a spherical geometry, the energy needed to curve a flat bilayer into a closed vesicle, Ecurve, is obtained from kb and KbG, as follows: Ecurve = 4π(2kb + KbG).43 In this respect, Fig. 9 shows the curve energy, Ecurve, of the lipid bilayers for different molecular fractions of DPPS, χ, in the absence and presence of salt obtained from our simulations.
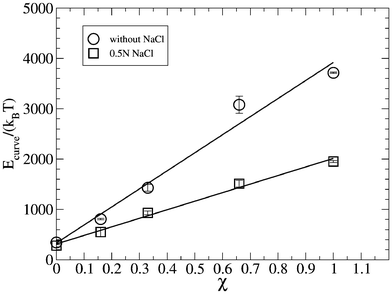 | ||
| Fig. 9 Energy curve Ecurve of lipid bilayers of different binary composition in the absence and the presence of salt. Error bars were estimated from the last three sub-trajectories of 30 ns each, where the first 10 ns were discarded as being the equilibration time. | ||
Fig. 9 also shows how, in the absence of salt, the curve energy is always positive for the whole range of molecular fractions, with Ecurve fitting a straight line for the full range of compositions of the lipid bilayer, with a positive slope. In the presence of salt, the curve energy is also positive for the full range of molecular fractions, χ, of the lipid bilayer, fitting a straight line with a positive, but lower slope than in absence of salt. From the data of Fig. 9, we conclude that the presence of salt in aqueous solution diminishes the energy needed to curve a planar bilayer of analogous composition in pure water. Due to the positive values of the curve energy as a function of molecular fraction of charged lipids in the lipid bilayer, we also conclude that the energy required to close the bilayer in a spherical shape, as in a lipid vesicle, becomes higher (and the vesicle much more unstable) with an increase in the charged lipid composition of the bilayer. This simulation result would explain the instability of POPG/POPC and POPA/POPC lipid vesicles observed with the increasing anionic lipid content (POPG or POPA) measured experimentally by Shoemaker and Vanderlik50 using the micropipette aspiration technique. These authors identified the existence of a critical tension that they could apply to the lipid vesicles, with the fraction of charged lipids in a lipid bilayer. The same authors obtained how this critical tension followed a straight line (with negative slope) with respect to the fraction of negative lipids (anionic lipid) in the bilayer of the lipid vesicle. These results can be easily related with our simulation results. Thus, due to the fact that the curve energy increases linearly with the fraction of charged lipid in the bilayer, much lower external tensions are sufficient to destroy the vesicle, i.e. there is perfect correlation between the curve energy obtained from simulation, and the critical tension measured by Shoemaker et al50 in charged vesicles.
3.5 Concluding remarks
This study has provided further insight at molecular level into the different mechanical properties of binary lipid bilayers of DPPC/DPPS, in the absence and presence of salt. In this regard, from calculation of the lateral pressure profile across a lipid bilayer, the influence of the lipid composition and salt concentration on the interfacial tension and elastic properties of the lipid bilayer were estimated. It is seen how an increase in the DPPS composition of the lipid bilayer tends to increase the interfacial tension, and lead to a noticeable enhancement of the stiffness of the lipid bilayer as well, as a consequence of the increase of the molecular interactions between neighboring lipids such as was evidenced from the radial distribution function g(r) between adjacent lipids. On the other hand, with salt in the aqueous solution, the interfacial tension reaches a constant value for a wide range of molecular fractions of the charged lipids in the lipid bilayer, or put in a different way, the interfacial tension becomes independent of the molecular fraction of the charged lipid in the membrane. These results may be of a great biological interest concerning our knowledge of the stability of the biological membranes, due to the fact that cellular membranes of eukaryotic cells are constituted by different fractions of charged lipids on both leaflets of the membrane, the charged lipids being asymmetrically located in the inner leaflet. Hence, by regulating the ionic strength, the interfacial tension can be compensated on both leaflets of the membrane, regardless of its composition, thus contributing to the stability of the membrane and to its biological activity by providing a suitable environment for other biological molecules, such as, for example, protein membranes.Acknowledgements
R. D. E. acknowledges the Fundacion Seneca de la Region de Murcia for financial support during his stay in the Universidad Politécnica de Cartagena. R. D. E. is a member of the CONICET (Argentina) staff. The authors wish to thank Centro de Calculo Cientifico of the Universidad Politecnica de Cartagena, for their support in carrying out the calculations on which this work is based.References
- G. Karp, Cell and Molecular Biology: Concepts and Experiments, John Wiley and Sons, 3rd edn, 2003 Search PubMed.
- S. Singer and G. Nicolson, Science, 1972, 175, 720–731 CAS.
- A. Spector and M. Yorek, J. Lipid Res., 1985, 26, 1015–1035 CAS.
- J. Gullingsrud and K. Schulten, Biophys. J., 2003, 85, 2087–2099 CrossRef CAS.
- O. H. S. Ollila, T. Rog, M. Karttunen and I. Vattulainen, J. Struct. Biol., 2007, 159, 311–323 CrossRef.
- J. Allen, R. Halverson-Tamboli and M. Rasenick, Nature, 2007, 8, 128–140 CAS.
- R. S. Cantor, Chem. Phys. Lipids, 1999, 101, 45–56 CrossRef CAS.
- Y. Ermakov, K. Kamaraju, K. Sengupta and S. Sukharev, Biophys. J., 2010, 98, 1018–1027 CrossRef CAS.
- E. Lindahl and O. Edholm, J. Chem. Phys., 2000, 113, 3882–3893 CrossRef CAS.
- P. Devaux, Biochemistry, 1991, 30, 1163–1173 CrossRef CAS.
- P. Williamson and R. Schlegel, Mol. Membr. Biol., 1994, 11, 199–216 CrossRef CAS.
- H. Hauser and G. Poupart, Lipid structure, in The Structure of Biological Membranes, ed. P. L. Yeagle, CRC Press, Boca Raton, FL, 1991 Search PubMed.
- J. Vance, J. Lipid Res., 2008, 49, 1377–1387 CrossRef CAS.
- R. Schleger and P. Williamson, Cell Death Differ., 2001, 8, 551–563 CrossRef.
- J. Weitzman, J. Biol., 2004, 3, 1–5 CrossRef.
- P. Williamson and R. Schlegel, J. Biol., 2004, 3, 14–4 CrossRef.
- J. J. López Cascales, J. García de la Torre, S. Marrink and H. Berendsen, J. Chem. Phys., 1996, 104, 2713–2720 CrossRef.
- J. López Cascales, M. Huertas and J. de la Torre, Biophys. J., 1997, 69, 1–8 CrossRef.
- R. Porasso and J. López Cascales, Colloids Surf., B, 2009, 73, 42–50 CrossRef CAS.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren and J. Hermans, Intermolecular Forces, D. Reidel Publishing Company, Dortrecht, 1981 Search PubMed.
- J. López Cascales, H. Berendsen and J. G. de la Torre, J. Phys. Chem., 1996, 100, 8621–8627 CrossRef.
- R. D. Porasso, W. F. D. Bennett, S. D. Oliveira-Costa and J. López Cascales, J. Phys. Chem. B, 2009, 113, 9988–9994 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- U. Essmann, L. Perera, M. B. T. Darden, H. Lee and L. Pedersen, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
- H. Berendsen, D. van der Spoel and R. van Drunen, Comput. Phys. Commun., 1995, 91, 43–56 CrossRef CAS.
- E. Lindahl, B. Hess and D. van der Spoel, J. Mol. Modeling, 2001, 7, 306–317 CAS.
- B. Hess, H. Bekker, H. Berendsen and H. J. C. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, A. DiNola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS.
- A. Seeling and J. Seeling, Biochemistry, 1974, 13, 4839–4845 CrossRef.
- G. Cevc, A. Watts and D. Marsh, Biochemistry, 1981, 20, 4955–4965 CrossRef CAS.
- E. Luna and H. Mcconnell, Biochim. Biophys. Acta, Biomembr., 1977, 470, 303–316 CrossRef CAS.
- E. Egberts, S. J. Marrink and H. J. C. Berendsen, Eur. Biophys. J., 1994, 22, 423–436 CrossRef CAS.
- J. Gullingsrud and K. Schulten, Biophys. J., 2004, 86, 3496–3509 CrossRef CAS.
- J. Sonne, F. Hansen and G. Peters, J. Chem. Phys., 2005, 122(124903), 1–9 Search PubMed.
- M. Patra, Eur. Biophys. J., 2005, 35, 79–88 CrossRef CAS.
- M. Orsi, D. Haubertin, W. Sanderson and J. Essex, J. Phys. Chem. B, 2008, 112, 802–815 CrossRef CAS.
- N. Chu, N. Kucerka, Y. Liu, S. Tristram-Nagle and J. Nagle, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2005, 71, 041904 CrossRef.
- E. Evans and W. Rawicz, Phys. Rev. Lett., 1990, 64, 2094–2097 CrossRef CAS.
- W. Rawicz, K. Olbrich, T. McIntosh, D. Needham and E. Evans, Biophys. J., 2000, 79, 328–339 CrossRef CAS.
- J. Pan, S. Tristram-Nagle, N. Kucerka and J. Nagle, Biophys. J., 2008, 94, 117–124 CrossRef CAS.
- H. Seto, N. Yamada, M. Nagao, M. Hishida and T. Takeda, Eur. Phys. J. E, 2008, 26, 217–223 CrossRef CAS.
- H. Petrache, S. Tristram-Nagle, K. Gawrisch, D. Harries, V. Parseguian and J. Nagle, Biophys. J., 2004, 86, 1574–1586 CrossRef CAS.
- M. Claessens, B. van Oort, F. Leermakers, F. Hoekstra and M. C. Stuart, Biophys. J., 2004, 87, 3882–3893 CrossRef CAS.
- T. Kamo, M. Nakano, Y. Kuroda and T. Handa, J. Phys. Chem. B, 2006, 110, 24987–24992 CrossRef CAS.
- S. Zeppieri, J. Rodriguez and A. Lopez-de-Ramos, J. Chem. Eng. Data, 2001, 46, 1086–1088 CrossRef CAS.
- J. Rowlinson and B. Widom, Molecular Theory of Capillarity, Oxford University Press, 1982 Search PubMed.
- E. Evans and W. Rawicz, Phys. Rev. Lett., 1997, 79, 2379–2382 CrossRef CAS.
- G. Shearman, O. Ces, R. Templer and J. Seddon, J. Phys.: Condens. Matter, 2006, 18, S1105–1124 CrossRef CAS.
- S. Safran, Adv. Phys., 1999, 48, 395–448 CrossRef CAS.
- S. Shoemaker and T. Vanderlick, Biophys. J., 2002, 83, 2007–2014 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2012 |
