Engineering of Brome mosaic virus for biomedical applications
Ibrahim
Yildiz
a,
Irina
Tsvetkova
b,
Amy M.
Wen
a,
Sourabh
Shukla
a,
M. Hema
Masarapu†
b,
Bogdan
Dragnea
b and
Nicole F.
Steinmetz
*c
aDepartment of Biomedical Engineering, Case Western Reserve University, 10900 Euclid Avenue, Cleveland, OH 44106, USA. E-mail: nicole.steinmetz@case.edu; Tel: +1 216-368-5590
bDepartment of Chemistry, Indiana University, 800 East Kirkwood Avenue, Bloomington, Indiana 47405, USA
cDepartments of Biomedical Engineering, Radiology, Materials Science and Engineering, Case Western Reserve University, 10900 Euclid Avenue, Cleveland, OH 44106, USA. E-mail: nicole.steinmetz@case.edu; Tel: +1 216-368-5590
First published on 23rd January 2012
Abstract
Viral nanoparticles (VNPs) are becoming versatile tools in platform technology development. Their well-defined structures as well as their programmability through chemical and genetic modification allow VNPs to be engineered for potential imaging and therapeutic applications. In this article, we report the application of a variety of bioconjugation chemistries to the plant VNP Brome mosaic virus (BMV). Functional BMV nanoparticles displaying multiple copies of fluorescent dyes, PEG molecules, chemotherapeutic drug moieties, targeting proteins and cell penetrating peptides were formulated. This opens the door for the application of BMV in nanomedicine.
Introduction
Nanomaterials have favorable properties for potential applications in nanomedicine as they can carry large payloads of imaging reagents and/or drugs and they can be engineered to direct the payload specifically to target cells. Many different nanomaterials have been developed, including various synthetic nanomaterials as well as naturally occurring bionanomaterials. Each platform has advantages and limitations in terms of pharmacokinetics, toxicity, immunogenicity and targeting specificity. Our research focuses on the development of bionanoparticles derived from plant viruses, also termed viral nanoparticles (VNPs).The field of VNP platform technology development is a rapidly evolving discipline. VNPs have been recognized as promising tools in biomedical research owing to their inherent biocompatibility, diversity of shapes and sizes, and having protein subunits that can be modified using molecular and chemical engineering approaches.1 The potential of VNPs for medical applications has clearly been recognized; for example, mammalian virus-based vectors for gene therapy and oncolytic virotherapy are undergoing clinical trials.2,3 There are many novel VNP platforms in the development pipeline, with bacteriophages and plant viruses favored because they are considered safer in humans than mammalian viruses (even replication-deficient strains of mammalian viruses are potentially pathogenic and/or cytotoxic).4 Plant VNPs are less likely to trigger negative downstream effects in mammals due to an inability to proliferate. Indeed, preclinical studies have shown that the plant VNPs can be administered at doses of up to 100 mg (1016 VNPs) per kg body weight without clinical toxicity.5,6 VNPs are bioavailable following oral or intravenous administration in mice and have wide biodistribution. Their highly repetitive proteinaceous and particulate structures are immunogenic, but this can be reduced significantly by PEGylation,7 a common strategy used in the development of biopharmaceuticals. The PEGylation of VNPs also reduces undesirable non-specific cell interactions, prolongs plasma circulation and increases stability.7–12 Applications of VNPs in nanomedicine such as gene and drug delivery, magnetic and fluorescence imaging, as well as vaccine development have been employed and offer promising results.8,13–17
A number of conventional and novel bioconjugation protocols have been successfully adapted to modify VNPs with a large collection of tumor-homing proteins and peptides, fluorescent dyes and imaging contrast agents, as well as therapeutic cargos to generate formulations with enhanced specificity and efficacy.18
Genetic engineering can also be used to manipulate and fine-tune the structural properties of VNPs. For example, genetic engineering can be applied to introduce novel amino acids to serve as targets for bioconjugation,19,20 to introduce affinity tags,21–23 or to alter the surface charge.24
With growing interest in the field of plant VNP platform technology, it is important to test and develop novel VNP platforms with unique properties. In this work we turned toward the plant virus Brome mosaic virus (BMV). The structure of BMV is known to atomic resolution.25 The 30 nm-sized capsid is composed of 180 identical copies of a single coat protein arranged in T = 3 symmetry. The genome is positive-sense, single-stranded, and tripartite, meaning that the genetic information is encoded on three different RNA molecules that are encapsidated into separate but morphologically identical particles.26,27
Our interest to develop BMV for potential applications in medicine is derived from its highly dynamic structure. BMV nanoparticles can be disassembled into protein dimers and re-assembled into empty virus-like particles (VLPs) or hybrid nanoparticles containing synthetic cores. A variety of synthetic materials such as gold nanoparticles, iron oxide cores, and quantum dots (QDs) have been encapsulated into BMV VLPs.28–32
To date, applications of BMV nanoparticles have been focused on its self-assembly processes. Its utility in bioconjugation chemistry has not been explored yet. The availability of bioconjugate chemistries, however, is essential for the development of BMV for medical applications. Bioconjugation will facilitate the introduction of targeting ligands as well as therapeutic and/or imaging moieties (in addition to the encapsulated cargos). In this paper we describe such bioconjugation procedures. We utilized a cysteine-containing mutant of BMV (cBMV) that displays 180 cysteine residues (Fig. 1). We report the application of thiol–maleimide chemistry and hydrazone ligation for the development of surface-modified functional cBMV nanoparticles carrying (i) fluorescent dyes, (ii) hydrophilic PEG polymers, (iii) tumor-homing proteins, (iv) chemotherapeutic moieties, and (v) cell penetrating peptides (Fig. 2).
 | ||
| Fig. 1 Structure of BMV. The 180 identical coat proteins are arranged in a T = 3 symmetry. Val 168 is highlighted in yellow. The particles described in this paper are cBMV chimeras displaying genetically engineered Cys side chains at position 168 in the BMV coat protein. The structural information was obtained from http://viperdb.scripps.edu/. The image was created using Pymol software. | ||
 | ||
| Fig. 2 Design and reaction scheme of surface modifications of cBMV with different ligands. (A) cBMV particles were reacted with fluorescent OregonGreen 488 and PEG (MW 2000 Da, n = 44) molecules via maleimide–thiol chemistry. (B) Oxidized transferrin (structure was obtained from http://pdb.com/) was introduced to particles through a two-step reaction by employing first thiol–maleimide then aldehyde–hydrazide coupling using the bivalent linker KMUH (n = 7). (C) Doxorubicin molecules were attached to cBMV particles in a two-step reaction utilizing thiol–maleimide and ketone–hydrazide coupling using the linker MHPH. (D) R5-peptide was linked to particles in a two-step reaction, first MTFB was introduced using maleimide–thiol chemistry, second R5 was conjugated via aldehyde–hydrazide linkage. | ||
Results & discussion
Production of cBMV nanoparticles
Genetic engineering of BMV to introduce unique Cys side chains is described elsewhere.33 In brief, Val 168 of the BMV coat protein was mutated to a Cys residue using site-directed mutagenesis. Infectious cDNA clones containing the full-length genome of BMV were transformed into Agrobacterium tumefaciens and then infiltrated into four-week old Nicotiana benthamiana plants to launch cBMV production. Ten days post-inoculation leaves were harvested and purified using established procedures.33 Typical yields obtained were 0.5 mg cBMV per gram infected leaf material, comparable to that of wild-type BMV. The integrity of the particles was verified by transmission electron microcopy (TEM) and size exclusion chromatography (SEC) using a Superose6 column and Äkta Explorer FPLC (GE Healthcare). cBMV chimeras were stored in buffer containing the reducing agent dithiothreitol (DTT). Aggregation was not observed even after storage for six months. The fact that aggregation did not occur reflects the design principles used for the generation of the Cys-containing particles: the Cys residue was inserted at amino acid position 168, which although being solvent-exposed, is somewhat buried on the particle surface (Fig. 1). This prevents inter-particle aggregation while allowing for accessibility in bioconjugation reactions (see below).cBMV–dye complexes for potential imaging applications
Fluorescent nanoparticles find applications as tags for in vitro and in vivo molecular imaging. Fluorescently-labeled VNPs have been shown to be versatile tools for intravital vascular and prostate cancer imaging using preclinical animal models.8,11,17 Here we established the formulation of dye-labeled cBMV conjugates using a maleimide-activated OregonGreen 488 (O488; Invitrogen) fluorophore and thiol–maleimide coupling.cBMV in SAMA buffer containing 1% DTT (SAMA buffer: 0.05 M NaOAc, 0.008 M, Mg(OAc)2; pH 4.6) was dialyzed against TNKM buffer containing 1% DTT overnight at 4 °C (TNKM buffer: 0.05 M Tris–HCl, 0.01 M KCl, 0.05 M NaCl, 0.005 M MgCl2; pH 7.4). Prior to introducing maleimide-O488, DTT was removed by four rounds of centrifuge filtration using spin columns with a 10 kDa cut-off (Millipore). cBMV (1 mg mL−1 in TNKM pH 7.4) was incubated in the dark for 2 h at 4 °C with varying molar excess of maleimide-O488: 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 100
1, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1000
1, 1000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2000
1, 2000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 5000
1, and 5000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dyes per cBMV particle. The organic dye was dissolved in DMSO, and the final DMSO concentration was adjusted to be 10% of the final volume. cBMV–O488 conjugates were purified and characterized using a combination of SEC, UV/Visible spectroscopy, and denaturing gel electrophoresis.
1 dyes per cBMV particle. The organic dye was dissolved in DMSO, and the final DMSO concentration was adjusted to be 10% of the final volume. cBMV–O488 conjugates were purified and characterized using a combination of SEC, UV/Visible spectroscopy, and denaturing gel electrophoresis.
Using the above methods covalent decoration of cBMV with O488 via a thioether bond was confirmed and the resulting cBMV–O488 particles were found to remain structurally sound (Fig. 3). The degree of conjugation was quantified. The number of dye moieties per cBMV particle was calculated based on the UV/Vis spectrum using the concentration ratio of O488 (Abs at 496, ε = 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cm−1 M−1) to the cBMV particles (Abs at 260, ε = 5.15 mg−1 mL cm−1, MW of cBMV = 4.6 × 106 g mol−1). It was found that the degree of labeling saturates at a molar excess of 1000 O488
000 cm−1 M−1) to the cBMV particles (Abs at 260, ε = 5.15 mg−1 mL cm−1, MW of cBMV = 4.6 × 106 g mol−1). It was found that the degree of labeling saturates at a molar excess of 1000 O488![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 cBMV with 110 ± 10 O488 attached per cBMV (Fig. 3A). Since cBMV display 180 Cys residues, it appears that only 50% of the Cys side chains are reactive. Further studies are ongoing to determine whether labels are randomly distributed on the cBMV surface or whether there is any spatial preference for attachment; e.g. are Cys in close proximity to the 5-fold axis more reactive than those at the 2- and 3-fold axis (Fig. 1). The reactivity of the engineered Cys residues shows high reproducibility using a variety of chemical modifiers and chemistries (see below).
1 cBMV with 110 ± 10 O488 attached per cBMV (Fig. 3A). Since cBMV display 180 Cys residues, it appears that only 50% of the Cys side chains are reactive. Further studies are ongoing to determine whether labels are randomly distributed on the cBMV surface or whether there is any spatial preference for attachment; e.g. are Cys in close proximity to the 5-fold axis more reactive than those at the 2- and 3-fold axis (Fig. 1). The reactivity of the engineered Cys residues shows high reproducibility using a variety of chemical modifiers and chemistries (see below).
 | ||
Fig. 3 Biochemical characterization of cBMV–O488. (A) UV/Visible spectra of purified cBMV–O488 nanoparticles after conjugation using varying excess of O488 dyes per particle. The inset shows the number of excess O488 per cBMV used for conjugation. (B) Transmission electron micrograph of UAc-stained cBMV–O488 particles. (C) Size exclusion chromatogram of cBMV–O488 monitored at 260 nm (red), 280 nm (blue) (Abs 260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 nm = 1.7) and 496 nm (green, O488 absorbance). Intact cBMV–O488 fractions were collected and further characterized. (D) SDS-PAGE of cBMV and cBMV–O488 visualized under UV light and white light after Coomassie staining. 280 nm = 1.7) and 496 nm (green, O488 absorbance). Intact cBMV–O488 fractions were collected and further characterized. (D) SDS-PAGE of cBMV and cBMV–O488 visualized under UV light and white light after Coomassie staining. | ||
The integrity of cBMV–O488 was confirmed using SEC and TEM. For TEM studies 0.05 mg mL−1 cBMV–O488 were applied on carbon coated copper grids (400 mesh, Electron Microscopy Sciences) and negative-stained using 2% (w/v) uranyl acetate. Images were collected at 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 magnification using a LIBRA 200FE (Zeiss). TEM confirmed that cBMV–O488 remained structurally sound (Fig. 3B). This was consistent with FPLC data. Samples were analyzed in 0.1 M potassium phosphate buffer pH 7.4 at a flow rate of 0.5 mL min−1. The elution profile and absorbance ratio of A260
000 magnification using a LIBRA 200FE (Zeiss). TEM confirmed that cBMV–O488 remained structurally sound (Fig. 3B). This was consistent with FPLC data. Samples were analyzed in 0.1 M potassium phosphate buffer pH 7.4 at a flow rate of 0.5 mL min−1. The elution profile and absorbance ratio of A260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 = 1.7 was consistent with intact cBMV particles (Fig. 3C). Further, SEC of cBMV–O488 showed that the eluted cBMV–O488 fraction also absorbed at 496 nm validating covalent attachment of the O488 dye molecules. The latter was further tested using denaturing gel electrophoresis. Protein subunits (10 μg sample, added SDS loading buffer) were analyzed on denaturing (4–12% NuPAGE gel, Invitrogen). After separation, the gel was photographed under UV light and after staining with Coomassie Blue using AlphaImager (Biosciences) imaging system. The appearance of a fluorescent band under UV light corresponding to the cBMV coat protein band after Coomassie staining confirms covalent dye-labeling (Fig. 3D). Overall, structurally sound dye-labeled cBMV conjugates were synthesized.
280 = 1.7 was consistent with intact cBMV particles (Fig. 3C). Further, SEC of cBMV–O488 showed that the eluted cBMV–O488 fraction also absorbed at 496 nm validating covalent attachment of the O488 dye molecules. The latter was further tested using denaturing gel electrophoresis. Protein subunits (10 μg sample, added SDS loading buffer) were analyzed on denaturing (4–12% NuPAGE gel, Invitrogen). After separation, the gel was photographed under UV light and after staining with Coomassie Blue using AlphaImager (Biosciences) imaging system. The appearance of a fluorescent band under UV light corresponding to the cBMV coat protein band after Coomassie staining confirms covalent dye-labeling (Fig. 3D). Overall, structurally sound dye-labeled cBMV conjugates were synthesized.
cBMV–PEG complexes for improved stability and enhanced in vivo applications
Due to their highly repetitive proteinaceous and particulate structures VNPs are immunogenic, but this can be reduced significantly by PEGylation. The PEGylation of VNPs also reduces undesirable non-specific cell interactions, prolongs plasma circulation and increases stability.7,9–12,34 PEGylation is thus an essential modification to be examined.Here we demonstrated conjugation of cBMV with PEG (MW 2000 Da). cBMV (1 mg mL−1 in TNKM pH 7.4) was incubated with a 1000-fold molar excess of maleimide-PEG2000 (NanoCS) for 2 h at 4 °C. cBMV–PEG conjugates were purified and analyzed using SEC, TEM, UV/Visible spectroscopy and denaturing gel electrophoresis. cBMV–PEG was found structurally sound as confirmed by TEM and SEC (Fig. 4A+B, the A260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 ratio was 1.7). Labeling efficiency was determined based on density measurements of the native versus PEGylated coat proteins on Coomassie-stained gels after denaturing gel electrophoresis (Fig. 4C). In agreement with the O488-labeling reaction, about 50% of the coat proteins were found to be PEGylated. This is expected to be sufficient coverage for biomedical applications. Indeed we have previously reported that coverage of as little as 10% of the coat proteins of Cowpea mosaic virus (CPMV) and Potato virus X (PVX) was sufficient to eliminate non-desired cell interactions and stabilize the particles in solution.12,35
280 ratio was 1.7). Labeling efficiency was determined based on density measurements of the native versus PEGylated coat proteins on Coomassie-stained gels after denaturing gel electrophoresis (Fig. 4C). In agreement with the O488-labeling reaction, about 50% of the coat proteins were found to be PEGylated. This is expected to be sufficient coverage for biomedical applications. Indeed we have previously reported that coverage of as little as 10% of the coat proteins of Cowpea mosaic virus (CPMV) and Potato virus X (PVX) was sufficient to eliminate non-desired cell interactions and stabilize the particles in solution.12,35
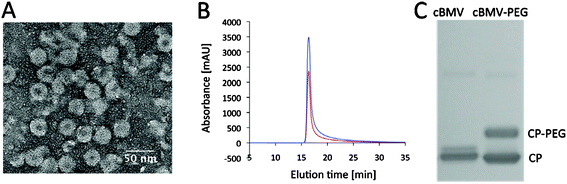 | ||
Fig. 4 Biochemical characterization of PEG modified cBMV nanoparticles. (A) Transmission electron micrograph of UAc-stained cBMV–PEG particles. (B) Size exclusion chromatogram of cBMV–PEG monitored at 260 nm (blue), 280 nm (red) (Abs 260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 nm = 1.7). (C) SDS-PAGE of cBMV and cBMV–PEG. PEGylated coat proteins (CPs) show lower mobility, ratio of CP-PEG versus (total) CP is (1 280 nm = 1.7). (C) SDS-PAGE of cBMV and cBMV–PEG. PEGylated coat proteins (CPs) show lower mobility, ratio of CP-PEG versus (total) CP is (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 50%. 1) 50%. | ||
cBMV–transferrin conjugates—toward tissue-specific targeting applications
One of the advantages of (bio)nanoparticles lies in the fact that they can be engineered with targeting ligands to confer tissue-specificity. Multifunctional nanoparticles displaying imaging/therapeutic cargos and targeting ligands have the potential to deliver these cargos to specific tissues with high specificity and efficacy. A number of targeting ligands are currently under exploration; one such ligand is the iron storage protein human holo-transferrin (Tf). Rapidly dividing cancer cells overexpress Tf receptors; transferrin thus offers an attractive ligand to target nanoparticle conjugates to tumors and metastatic sites.36cBMV–Tf conjugates were generated using a two-step ligation reaction (Fig. 2B). First, a maleimide bifunctional linker carrying a hydrazide end group was introduced using N-[κ-maleimidoundecanoic acid] hydrazide, trifluoroacetic acid salt (KMUH; Pierce Biosciences). cBMV (1 mg mL−1 in TNKM pH 7.4) and 1000 equivalent KMUH dissolved in DMSO were incubated for 2 h at 4 °C, the final DMSO concentration adjusted to be 10% of the final volume. The cBMV–KMUH conjugate was purified and analyzed using SEC, UV/Visible spectroscopy, and denaturing gel electrophoresis. Subsequently, oxidized transferrin with exposed and reactive aldehyde groups was introduced: Tf was oxidized using 0.01 M sodium periodate in 0.1 M sodium acetate buffer (pH 5.5) for 30 min at 0 °C in the dark. The buffer was exchanged to 0.1 M potassium phosphate buffer using 10 kDa centrifuge filters (Amicon). The oxidized Tf was then added to cBMV–KMUH using an 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar excess of Tf
1 molar excess of Tf![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cBMV–KMUH. The reaction mix was incubated in 0.1 M potassium phosphate buffer pH 7.4 for 12 h at 4 °C. cBMV–transferrin conjugates were purified and analyzed using SEC, UV/Visible spectroscopy, denaturing gel electrophoresis, and dot blotting (Fig. 5).
cBMV–KMUH. The reaction mix was incubated in 0.1 M potassium phosphate buffer pH 7.4 for 12 h at 4 °C. cBMV–transferrin conjugates were purified and analyzed using SEC, UV/Visible spectroscopy, denaturing gel electrophoresis, and dot blotting (Fig. 5).
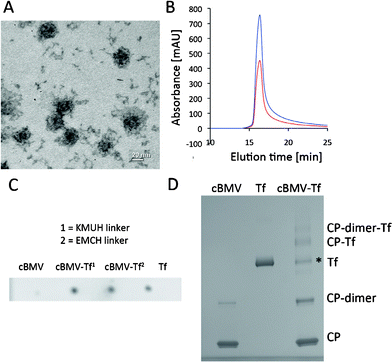 | ||
Fig. 5 Biochemical characterization of cBMV–Tf conjugates. (A) Transmission electron micrograph of cBMV–Tf. (B) Size exclusion chromatogram of cBMV–Tf monitored at 260 nm (blue) and 280 nm (red) (A260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 = 1.7). The cBMV–Tf fractions were collected and further characterized. (C) Dot blot of cBMV, cBMV–Tf generated using KMUH and EMCH linker, and free Tf (positive control). The blot was probed with an anti-transferrin and alkaline phosphatase-conjugated secondary antibody. (D) SDS-PAGE of cBMV, Tf, and cBMV–Tf (KMUH reaction). Free coat proteins (CPs), CP dimer, Tf, CP–Tf, and CP-dimer–Tf are detected after Coomassie Blue staining. * Tf is a dimer which dissociates in the denaturing conditions of the gel. The ratio of “free” Tf in the BMV–Tf samples equates to the amount of conjugated Tf. It is thus concluded the Tf* is not derived from impurities of the preparation but Tf released from the cBMV–Tf conjugate in the gel. This is as previously reported using HK97.38 280 = 1.7). The cBMV–Tf fractions were collected and further characterized. (C) Dot blot of cBMV, cBMV–Tf generated using KMUH and EMCH linker, and free Tf (positive control). The blot was probed with an anti-transferrin and alkaline phosphatase-conjugated secondary antibody. (D) SDS-PAGE of cBMV, Tf, and cBMV–Tf (KMUH reaction). Free coat proteins (CPs), CP dimer, Tf, CP–Tf, and CP-dimer–Tf are detected after Coomassie Blue staining. * Tf is a dimer which dissociates in the denaturing conditions of the gel. The ratio of “free” Tf in the BMV–Tf samples equates to the amount of conjugated Tf. It is thus concluded the Tf* is not derived from impurities of the preparation but Tf released from the cBMV–Tf conjugate in the gel. This is as previously reported using HK97.38 | ||
Tf labeling was also attempted using the bifunctional linker 3,3′-N-[ε-maleimidocaproic acid] hydrazide, trifluoroacetic acid salt (EMCH, Pierce Biosciences). Although labeling with EMCH and Tf was confirmed (Fig. 5C), labeling efficiency was low and could not be quantified. The difference between EMCH and KMUH lies in the alkyl chain linker. KMUH includes a longer alkyl chain (n = 7) (Fig. 2B). The low Tf loading efficiency using cBMV–EMCH is likely explained by steric hindrance. Only the longer KMUH linker enables the large protein Tf to engage efficiently in chemical conjugation.
The cBMV–Tf (with KMUH linker) conjugate was purified using SEC to remove excess Tf. TEM imaging and the SEC elution profile as well as the A260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 ratio indicated intact cBMV–Tf particles (Fig. 5A+B).
280 ratio indicated intact cBMV–Tf particles (Fig. 5A+B).
Dot blot analysis confirmed the presence of conjugated Tf: 1 μL Tf, cBMV, and cBMV–Tf (1 mg mL−1) were spotted onto a methanol-activated nitrocellulose membrane and allowed to dry for 30 min. The membrane was blocked at room temperature for 60 min using 0.1 M Tris-buffered saline (TBS) pH 7.6 containing 5% w/v skimmed milk powder and 0.05% w/v Tween 20 (blocking buffer). The detection was carried out using rabbit anti-transferrin antibody and a secondary anti-rabbit alkaline phosphatase-labeled antibody (Pierce Biosciences). Each antibody was used at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution and incubated with the membranes in blocking buffer. Alkaline phosphatase activity was detected using the BCIP/NBT liquid substrate system (Sigma Aldrich) (Fig. 5C).
1000 dilution and incubated with the membranes in blocking buffer. Alkaline phosphatase activity was detected using the BCIP/NBT liquid substrate system (Sigma Aldrich) (Fig. 5C).
Denaturing gel electrophoresis confirmed the covalent decoration of cBMV with Tf targeting proteins. Band density analysis indicated 35 ± 5 Tf moieties being attached per cBMV nanoparticle (Fig. 5D). Based on the dimensions of the cBMV particles (30 nm in diameter) and the size of Tf (5 × 8 nm) (the structural data of Tf were obtained at http://pdb.com/), it is indicated that maximum surface coverage was achieved. This is in good agreement with previous studies using the phages Qβ and HK97.37,38 In each study Tf-conjugated VNPs were generated, maximum surface coverage with Tf labels was achieved, and in vitro target-specificity was confirmed using various Tf-receptor overexpressing cell lines. These data indicate that the cBMV–Tf has potential for use in tissue-specific imaging or targeted drug delivery applications.
cBMV–DOX formulations—toward potential cancer medicines
Doxorubicin (DOX) is a state-of-the-art approved cancer drug that intercalates into nucleic acids and interferes with DNA synthesis, thus inhibiting cell proliferation and inducing cell death. DOX–nanoparticle conjugates include DOXIL, an approved PEGylated liposomal formulation (Ben Venue Laboratories), and a variety of conjugates are in development and clinical testing. Here we tested the conjugation of DOX to cBMV.DOX was conjugated to cBMV using a two-step chemical reaction (Fig. 2C). First, 5-maleimido-2-hydraziniumpyridine hydrochloride (MHPH; Solulink), a maleimide bifunctional linker carrying a hydrazine end group was conjugated to surface thiols displayed on cBMV. cBMV (1 mg mL−1 in TNKM pH 7.4) and 1000 equivalents of MHPH dissolved in DMSO (the final DMSO was adjusted to be 10% of the final volume) were reacted for 2 h at 4 °C. The cBMV–MHPH conjugate was purified by SEC and analyzed using UV/Visible spectroscopy.
To quantify MHPH modification per cBMV particle the Solulink molar substitution ratio protocol (MSR) assay was used. In brief, 4-nitrobenzaldehyde was reacted with cBMV–MHPH resulting in the formation of a UV traceable hydrazone bond (Abs at 350 nm, ε = 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1) between 4-nitrobenzaldehyde and the hydraziniumpyridine group. We found that 100 ± 5 MHPH groups were introduced per cBMV. This is consistent with the above described results. Maximum coverage using small chemical modifiers such as fluorophores, PEG, or chemical linkers results in labeling of 50% of the cBMV coat proteins.
000 M−1 cm−1) between 4-nitrobenzaldehyde and the hydraziniumpyridine group. We found that 100 ± 5 MHPH groups were introduced per cBMV. This is consistent with the above described results. Maximum coverage using small chemical modifiers such as fluorophores, PEG, or chemical linkers results in labeling of 50% of the cBMV coat proteins.
Next, DOX (doxorubicin hydrochloride, Fisher Bioreagents) was introduced. The ketone moiety of DOX was coupled with the hydrazine functionality of cBMV–MHPH as follows: 600 molar equivalents of doxorubicin was mixed with cBMV–MHPH in 0.1 M potassium phosphate buffer pH 7.4 containing 100 mM aniline for 2 h at 4 °C. Aniline was used as a catalyst in the reaction.39 The cBMV–DOX conjugate was purified and analyzed using SEC, UV/Visible spectroscopy, and denaturing gel electrophoresis (Fig. 6).
 | ||
Fig. 6 Biochemical characterization of cBMV–DOX. (A) Transmission electron micrograph of cBMV–DOX. (B) Size exclusion chromatogram of cBMV–DOX monitored at 260 nm (blue), 280 nm (red) (Abs 260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 nm = 1.7) and 496 nm (green, DOX absorbance). Intact cBMV–DOX fractions were collected and further characterized. (C) SDS-PAGE of cBMV and cBMV–DOX visualized under UV light and white light after Coomassie staining. 280 nm = 1.7) and 496 nm (green, DOX absorbance). Intact cBMV–DOX fractions were collected and further characterized. (C) SDS-PAGE of cBMV and cBMV–DOX visualized under UV light and white light after Coomassie staining. | ||
TEM and SEC confirmed that intact cBMV–DOX was recovered after purification using SEC. It should be noted that the negative stain of cBMV–DOX using 2% UAc was difficult to achieve. It is possible that the hydrophobic DOX moieties interfere with the negative stain (Fig. 6A). The elution profile in SEC indicated the particles remained structurally sound. Further co-elution of DOX was indicated (Abs at 496 nm) and thus confirmed covalent attachment (Fig. 6B). UV/Visible spectroscopy using the concentration ratio of DOX (Abs at 496 ε = 8000 M−1 cm−1) to the cBMV particles (Abs at 260, ε = 5.15 mg−1 mL cm−1, MW of cBMV = 4.6 × 106 g mol−1) indicated labeling with 90 ± 5 DOX moieties per cBMV. Covalent attachment was further confirmed by denaturing gel electrophoresis; the DOX-labeled coat proteins are fluorescent under UV light (Fig. 6C).
Drug-loaded cBMV–DOX nanoparticles could find applications in drug delivery applications. A variety of DOX–nanoparticle conjugates, including VNP–DOX conjugates are currently under investigation. For example, the phages MS2 and Qβ have been decorated with taxol and DOX, respectively, using pH-labile hydrazone ligation strategies and efficient cell killing in tissue culture has been reported.40,41 In a different study, VLPs of rotavirus were modified with DOX; however in this case the drug was ligated using pH stable amide bonds.42 Efficient cell killing in tissue culture was also reported. It was hypothesized that the VNPs enter the cells via endocytosis and inside the acidic endolysosomes the protein structure is denatured resulting in release of the drug molecules. In our case DOX was attached using a hydrazone ligation, however the hydrazide pyridine ring may stabilize the bond even in acidic pH. Drug-release studies using cBMV–DOX are currently under investigation in our laboratory. We propose that cell killing could be achieved even if the bond is stable at low pH, and this would be based on the degradation of the protein nanoparticle carrier in the lysosomal cell compartment.
cBMV–R5 conjugates—candidates with potential cell penetrating properties
To be effective for drug delivery applications, nanoparticle formulations must be taken up into target cells. Cell penetrating peptides (CPPs) have been shown to promote nanoparticle translocation across membranes and are therefore potentially valuable in nanomedicine.43 Cationic CPPs contain clusters of primarily arginine and lysine residues, e.g. the transcription factor TAT and poly-arginine peptides such as the R5 peptide promote the uptake of nanoparticles and at least partial endosomal escape, as recently shown for TAT-labeled MS2 phage14 and poly-arginine-labeled liposomes.44Here we demonstrated the labeling of cBMV with R5 peptides using a two-step hydrazone ligation protocol. First, a maleimide bifunctional linker with a PEG3 spacer and a benzaldehyde end group (maleimido-PEG3-4-benzylamide analogue, MTFB; Solulink) was introduced to the cBMV surface as follows (Fig. 2D): cBMV (1 mg mL−1 in TNKM pH 7.4) and 1000 equivalents of MTFB (final DMSO concentration was 10% by volume) were incubated for 2 h at 4 °C. cBMV–MTFB conjugate was purified using SEC and analyzed with UV/Visible spectroscopy. Quantification of MTFB modification was determined using Solulink MSR assay and 2-hydrazinepyridine. The resulting hydrazone bond is UV traceable (Abs at 350 nm, ε = 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1); we found that 50% of the cBMV coat proteins were covalently modified with the MTFB linker (100 ± 5), consistent with the above-reported results.
000 M−1 cm−1); we found that 50% of the cBMV coat proteins were covalently modified with the MTFB linker (100 ± 5), consistent with the above-reported results.
The aldehyde moiety of cBMV–MTFB was then coupled with the hydrazide functionality of R5-peptide using a 336 molar equivalent of R5 peptide in 0.1 M potassium phosphate buffer pH 7.4 containing 100 mM aniline (catalyst) for 2 h at 4 °C. The synthesis of the R5 peptide (Fig. 2D) is described elsewhere (Wu & Steinmetz, in preparation). cBMV–R5 was purified from excess reagents using SEC and characterized using TEM, SEC, UV/Visible spectroscopy, denaturing gel electrophoresis, and western blotting (Fig. 7).
 | ||
Fig. 7 Biochemical characterization of cBMV–R5. (A) Transmission electron micrograph of cBMV–R5. (B) Size exclusion chromatogram of cBMV–R5 monitored at 260 nm (blue), 280 nm (red) (Abs 260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 nm = 1.7). Intact cBMV–R5 fractions were collected and further characterized. (C) SDS-PAGE of cBMV and cBMV–R5 visualized under white light after Coomassie staining. Modified coat proteins (CP–R5) have an increased molecular weight. The ratio between CP–R5 280 nm = 1.7). Intact cBMV–R5 fractions were collected and further characterized. (C) SDS-PAGE of cBMV and cBMV–R5 visualized under white light after Coomassie staining. Modified coat proteins (CP–R5) have an increased molecular weight. The ratio between CP–R5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CP was determined based on band intensity and found to be 1 CP was determined based on band intensity and found to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. (D) Western blot probed with streptavidin–alkaline phosphatase detecting the biotin-tag of the R5 peptide. 3. (D) Western blot probed with streptavidin–alkaline phosphatase detecting the biotin-tag of the R5 peptide. | ||
TEM and SEC indicated that cBMV–R5 remained structurally sound (Fig. 7A+B). Quantification of peptide-labeling was based on the UV-traceable hydrazone bond formed (Abs at 354 nm, ε = 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1) and band density analysis in denaturing protein gels (Fig. 7C). Quantifications from these two independent methods were in good agreement indicating 60 ± 5 R5 peptides per cBMV particle. We attempted to increase the labeling efficiency by increasing the molar excess of R5 used or by extending the incubation time. However, in both cases aggregation was observed and recovered yield was less than 5% indicating that the highly positively charged R5 peptide induces electrostatically-driven aggregation upon a threshold. cBMV–R5 displaying 60 R5 peptides were found structurally sound and dispersed.
000 M−1 cm−1) and band density analysis in denaturing protein gels (Fig. 7C). Quantifications from these two independent methods were in good agreement indicating 60 ± 5 R5 peptides per cBMV particle. We attempted to increase the labeling efficiency by increasing the molar excess of R5 used or by extending the incubation time. However, in both cases aggregation was observed and recovered yield was less than 5% indicating that the highly positively charged R5 peptide induces electrostatically-driven aggregation upon a threshold. cBMV–R5 displaying 60 R5 peptides were found structurally sound and dispersed.
The peptide used in this study displays a biotin-tag (Fig. 2D) allowing detection using enzyme-tagged streptavidin probes and western blotting. cBMV–R5 and cBMV coat proteins were separated on denaturing gels; the proteins were transferred onto methanol-activated nitrocellulose membranes. The membrane was blocked at room temperature for 60 min using 0.1 M TBS pH 7.6 containing 5% w/v skimmed milk powder and 0.05% w/v Tween 20. Detection was carried out using alkaline phosphatase conjugated streptavidin (Sigma Aldrich) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) in blocking buffer solution. Alkaline phosphatase activity was detected using the BCIP/NBT liquid substrate system (Sigma Aldrich). Western blotting confirmed the covalent decoration of cBMV with R5 (Fig. 7D).
1000) in blocking buffer solution. Alkaline phosphatase activity was detected using the BCIP/NBT liquid substrate system (Sigma Aldrich). Western blotting confirmed the covalent decoration of cBMV with R5 (Fig. 7D).
Structurally sound peptide-decorated cBMV–R5 particles were successfully synthesized. We envision the application of cBMV–R5 in cell penetration for the delivery of medically relevant moieties, in which high cellular uptake is desired. We recently showed that the multivalent display of R5 clusters on the VNP CPMV promotes highly efficient cellular uptake (Wu & Steinmetz, in preparation).
Conclusions
Although BMV has been studied and developed for potential medical applications, its chemical bioconjugation has not previously been reported. We demonstrated here that maleimide–thiol conjugation and hydrazone ligation reactions can be applied to a cBMV chimera displaying 180 cysteine side chains at amino acid position 168. 50% of these thiols were found reactive and could be labeled with several chemical modifiers: (1) fluorescent dyes, (2) PEG chains, (3) tumor-homing protein, (4) potent cancer drug, and (5) cell penetrating peptides. Structurally sound nanoparticle formulations were generated, and we envision potential applications in tissue-specific imaging and/or drug delivery.To be most useful for future medical applications it is desired to generate multifunctional nanoparticle formulations combining tissue-specificity and drug delivery and/or imaging in a single formulation. One of the advantages of BMV over other plant VNPs is that its highly dynamic self-assembly process is well-understood and can be exploited to load the interior cavity with cargos.28–32 A combination of encapsulation and bioconjugate chemistry is expected to yield multifunctional nanoparticles.
In addition, additional amino acid side chains could be targeted on the BMV surface. In this study we described the chemical labeling of genetically engineered Cys side chains. Native BMV display 1440 solvent-exposed surface Lys side chains, that is eight Lys residues per coat protein (the structural coordinates are available on http://viperdb.scripps.edu/). We have confirmed that the Lys side chains are also addressable and double labeling can be performed. For example BMV particles displaying Tf attached to Cys residues and O488 attached to Lys side chains have been successfully generated; the application of dual-labeled cBMV–Tf–O488 particles for molecular imaging will be reported elsewhere (Yildiz & Steinmetz, unpublished).
In summary, we established bioconjugate chemistries on cBMV and decorated the nanoparticles with a variety of biomedically-relevant cargos. This will contribute to the further development of important virus platform technologies.
Acknowledgements
This work was supported by NIH/NIBIB grants R00 EB009105 (PI Steinmetz) and P30 EB011317 (PI Duerk, Recruited Faculty Steinmetz) and Case Western Reserve University Interdisciplinary Alliance Investment Grant (PIs Steinmetz & Samia), and NIH/NIBIB training grant, T32 EB007509 (to AMW) and NIH GM 081029 and grant 0832651 from NSF to B.D. M. Hema Masarapu acknowledges the Department of Biotechnology, New Delhi, India for providing DBT Long-term Overseas Associateship. We would like to thank Prof. C. Cheng Kao (Indiana University) for providing the cBMV. CK acknowledges funding from R01 AI090280 from NIH/NIAID. Dr Anouk Dirksen and Prof. Phil Dawson (The Scripps Research Institute) are thanked for providing the R5 peptide. We also thank Reza Sharghi-Moshtaghin (CWRU) for assistance with TEM studies.References
- E. Strable and M. G. Finn, Curr. Top. Microbiol. Immunol., 2009, 327, 1–21 CrossRef CAS.
- L. Barzon, A. L. Stefani, M. Pacenti and G. Palu, Expert Opin. Biol. Ther., 2005, 5, 639–662 CrossRef CAS.
- T. C. Liu and D. Kirn, Gene Ther., 2008, 15, 877–884 CrossRef CAS.
- P. Singh, M. J. Gonzalez and M. Manchester, Drug Dev. Res., 2006, 67, 23–41 CrossRef CAS.
- C. R. Kaiser, M. L. Flenniken, E. Gillitzer, A. L. Harmsen, A. G. Harmsen, M. A. Jutila, T. Douglas and M. J. Young, Int. J. Nanomedicine, 2007, 2, 715–733 CAS.
- P. Singh, D. Prasuhn, R. M. Yeh, G. Destito, C. S. Rae, K. Osborn, M. G. Finn and M. Manchester, J. Controlled Release, 2007, 120, 41–50 CrossRef CAS.
- K. S. Raja, Q. Wang, M. J. Gonzalez, M. Manchester, J. E. Johnson and M. G. Finn, Biomacromolecules, 2003, 3, 472–476 CrossRef.
- N. F. Steinmetz, A. L. Ablack, J. L. Hickey, J. Ablack, B. Manocha, J. S. Mymryk, L. G. Luyt and J. D. Lewis, Small, 2011, 7, 1664–1672 CrossRef CAS.
- F. M. Brunel, J. D. Lewis, G. Destito, N. F. Steinmetz, M. Manchester, H. Stuhlmann and P. E. Dawson, Nano Lett., 2010, 10, 1093–1097 CrossRef CAS.
- G. Destito, R. Yeh, C. S. Rae, M. G. Finn and M. Manchester, Chem. Biol., 2007, 14, 1152–1162 CrossRef CAS.
- J. D. Lewis, G. Destito, A. Zijlstra, M. J. Gonzalez, J. P. Quigley, M. Manchester and H. Stuhlmann, Nat. Med., 2006, 12, 354–360 CrossRef CAS.
- N. F. Steinmetz and M. Manchester, Biomacromolecules, 2009, 10, 784–792 CrossRef CAS.
- E. M. Plummer and M. Manchester, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol., 2010 Search PubMed.
- B. Wei, Y. Wei, K. Zhang, J. Wang, R. Xu, S. Zhan, G. Lin, W. Wang, M. Liu, L. Wang, R. Zhang and J. Li, Biomed. Pharmacother., 2009, 63, 313–318 CrossRef CAS.
- N. F. Steinmetz, V. Hong, E. D. Spoerke, P. Lu, K. Breitenkamp, M. G. Finn and M. Manchester, J. Am. Chem. Soc., 2009, 131, 17093–17095 CrossRef CAS.
- A. Datta, J. M. Hooker, M. Botta, M. B. Francis, S. Aime and K. N. Raymond, J. Am. Chem. Soc., 2008, 130, 2546–2552 CrossRef CAS.
- H. S. Leong, N. F. Steinmetz, A. Ablack, G. Destito, A. Zijlstra, H. Stuhlmann, M. Manchester and J. D. Lewis, Nat. Protoc., 2010, 5, 1406–1417 CrossRef CAS.
- J. K. Pokorski and N. F. Steinmetz, Mol. Pharm., 2011, 8, 29–43 CrossRef CAS.
- R. A. Miller, A. D. Presley and M. B. Francis, J. Am. Chem. Soc., 2007, 129, 3104–3109 CrossRef CAS.
- Q. Wang, T. Lin, J. E. Johnson and M. G. Finn, Chem. Biol., 2002, 9, 813–819 CrossRef CAS.
- A. Chatterji, W. F. Ochoa, T. Ueno, T. Lin and J. E. Johnson, Nano Lett., 2005, 5, 597–602 CrossRef CAS.
- I. L. Medintz, K. E. Sapsford, J. H. Konnert, A. Chatterji, T. Lin, J. E. Johnson and H. Mattoussi, Langmuir, 2005, 21, 5501–5510 CrossRef CAS.
- A. K. Udit, S. Brown, M. M. Baksh and M. G. Finn, J. Inorg. Biochem., 2008, 102, 2142–2146 CrossRef CAS.
- T. Douglas, E. Strable and D. Willits, Adv. Mater., 2002, 14, 415–418 CrossRef CAS.
- R. W. Lucas, S. B. Larson and A. McPherson, J. Mol. Biol., 2002, 317, 95–108 CrossRef CAS.
- P. Ahlquist, R. Dasgupta and P. Kaesberg, J. Mol. Biol., 1984, 172, 369–383 CrossRef CAS.
- P. Ahlquist, V. Luckow and P. Kaesberg, J. Mol. Biol., 1981, 153, 23–38 CrossRef CAS.
- M. C. Daniel, I. B. Tsvetkova, Z. T. Quinkert, A. Murali, M. De, V. M. Rotello, C. C. Kao and B. Dragnea, ACS Nano, 2010, 4, 3853–3860 CrossRef CAS.
- S. K. Dixit, N. L. Goicochea, M. C. Daniel, A. Murali, L. Bronstein, M. De, B. Stein, V. M. Rotello, C. C. Kao and B. Dragnea, Nano Lett., 2006, 6, 1993–1999 CrossRef CAS.
- X. Huang, L. M. Bronstein, J. Retrum, C. Dufort, I. Tsvetkova, S. Aniagyei, B. Stein, G. Stucky, B. McKenna, N. Remmes, D. Baxter, C. C. Kao and B. Dragnea, Nano Lett., 2007, 7, 2407–2416 CrossRef CAS.
- X. Huang, B. D. Stein, H. Cheng, A. Malyutin, I. B. Tsvetkova, D. V. Baxter, N. B. Remmes, J. Verchot, C. Kao, L. M. Bronstein and B. Dragnea, ACS Nano, 2011, 5, 4037–4045 CrossRef CAS.
- J. Sun, C. DuFort, M. C. Daniel, A. Murali, C. Chen, K. Gopinath, B. Stein, M. De, V. M. Rotello, A. Holzenburg, C. C. Kao and B. Dragnea, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 1354–1359 CrossRef CAS.
- M. Hema Masarapu, A. Murali, P. Ni, R. C. Vaughan, K. Fujisaki, I. Tsvetkova, B. Dragnea and C. C. Kao, Mol. Plant-Microbe Interact., 2010, 23, 1433–1447 CrossRef.
- N. F. Steinmetz, C.-F. Cho, A. Ablack, J. D. Lewis and M. Manchester, Nanomedicine, 2011, 6(2), 351–64 CrossRef CAS.
- N. F. Steinmetz, M. E. Mertens, R. E. Taurog, J. E. Johnson, U. Commandeur, R. Fischer and M. Manchester, Nano Lett., 2010, 10, 305–312 CrossRef CAS.
- K. C. Gatter, G. Brown, I. S. Trowbridge, R. E. Woolston and D. Y. Mason, J. Clin. Pathol., 1983, 36, 539–545 CrossRef CAS.
- D. Banerjee, A. P. Liu, N. R. Voss, S. L. Schmid and M. G. Finn, ChemBioChem, 2010, 11, 1273–1279 CrossRef CAS.
- R. K. Huang, N. F. Steinmetz, C. Y. Fu, M. Manchester and J. E. Johnson, Nanomedicine, 2011, 6, 55–68 CrossRef CAS.
- A. Dirksen and P. E. Dawson, Bioconjugate Chem., 2008, 19, 2543–2548 CrossRef CAS.
- E. W. Kovacs, J. M. Hooker, D. W. Romanini, P. G. Holder, K. E. Berry and M. B. Francis, Bioconjugate Chem., 2007, 18, 1140–1147 CrossRef CAS.
- J. K. Pokorski, K. Breitenkamp, L. O. Liepold, S. Qazi and M. G. Finn, J. Am. Chem. Soc., 2011, 133, 9242–9245 CrossRef CAS.
- Q. Zhao, W. Chen, Y. Chen, L. Zhang, J. Zhang and Z. Zhang, Bioconjugate Chem., 2011, 22, 346–352 CrossRef CAS.
- L. N. Patel, J. L. Zaro and W. C. Shen, Pharm. Res., 2007, 24, 1977–1992 CrossRef CAS.
- A. El-Sayed, S. Futaki and H. Harashima, AAPS J., 2009, 11, 13–22 CrossRef CAS.
Footnote |
| † Present address: Department of Virology, Sri Venkateswara University, Tirupati-517 502, A.P., India. |
| This journal is © The Royal Society of Chemistry 2012 |
