Task-specific membranes for the isolation of recombinant proteins with peptide tags†
Tatsuo
Maruyama
*a,
Shunsuke
Tabayashi
a,
Takafumi
Honjo
a,
Kazuki
Hoe
a,
Tsutomu
Tanaka
a,
Josui
Shimada
b,
Masahiro
Goto
b and
Hideto
Matsuyama
*a
aDepartment of Chemical Science and Engineering, Graduate School of Engineering, Kobe University, 1-1 Rokkodai, Nada-ku, Kobe 657-850. E-mail: tmarutcm@crystal.kobe-u.ac.jp, matuyama@kobe-u.ac.jp; Fax: +81-(0)78-803-6070
bDepartment of Applied Chemistry, Graduate School of Engineering and Center for Future Chemistry, Kyushu University, 744 Moto-oka, Fukuoka 819-0395, Japan. Fax: +81-(0)92-802-2810
First published on 3rd November 2011
Abstract
Task-specific membranes for recombinant proteins with peptide tags were developed by introducing surfactant-like ligands to polymeric membranes, which allowed the simple and rapid purification of target proteins from E. colicell lysates.
Progress in genetic engineering has enabled the production of a wide variety of recombinant proteins using host cells. Genetic technologies have also accelerated the discovery of protein drugs and molecularly-targeted drugs in the pharmaceutical industry. A number of efforts have discovered candidate protein drugs and chemicals that have specific affinity to disease-related proteins.1 To discover a protein drug and a particular chemical for a disease-related protein, a high-throughput technique for protein screening is required.2 To examine the pharmacological activity of a protein, the protein should be purified and isolated from other proteins coexisting in a cell. Conventionally, protein purification goes through cell disruption, centrifugation, filtration and application to at least one column purification technique. This multistep process makes protein purification time- and energy-consuming, and therefore represents a major bottleneck in isolating and exploring the functional properties of proteins. In protein screening, preliminary investigations (e.g., a simple test) are usually carried out. These preliminary tests require a small amount of the purified target protein. Consequently, high-throughput techniques for the purification of small amounts of target proteins are needed.
An affinity membrane represents a strategy for high-throughput protein purification.3 An affinity membrane is designed to selectively capture and release target proteins under given conditions. Currently, most studies employ chemical modification of a membrane surface or grafting of a membrane surface to obtain affinity membranes.4 Recently, spun nanofibers (nonwoven fabric membranes) were used for the preparation of affinity membranes.5 However, a porous membrane and a nonwoven fabric membrane have nanostructures that are too complex, which makes it difficult to achieve modification of the whole nanostructure surfaces. The degree of surface modification is also hard to control, and harsh reaction conditions required for surface modifications often damage the substrate membrane.
Porous polymeric membranes are easily prepared by phase separation of a polymer solution.6 Here we propose a novel approach using a surfactant-like ligand and phase separation for the generation of a surface-functionalized membrane. In this approach, a surfactant-like ligand was localized at an interface between the separated phases to produce a surface-functionalized membrane. In the present study, we synthesized surfactant-like ligands and introduced them to a polymeric membrane, succeeding in the rapid and selective purification of recombinant proteins with peptide tags.
We adopted cellulose diacetate as the membrane material because of its low protein binding capacity.7 As ligands for tagged recombinant proteins, nitrilotriacetic acid (NTA) and glutathione were selected, which show high affinity towards proteins with a hexahistidine tag (His-tag) and a glutathione S-transferase tag, respectively.8,9 These ligands are conjugated with hydrophobic compounds (oleic acid and 1-hexadecanol) to produce surfactant-like ligands (Scheme 1, termed NTA-surfactant (1) and glutathione-surfactant (2), respectively).
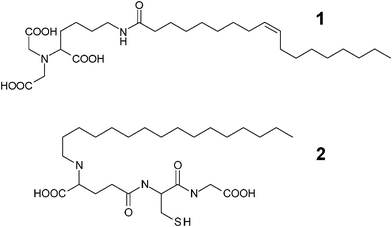 | ||
| Scheme 1 Molecular structures of surfactant-like ligands. NTA-surfactant (1) and glutathione-surfactant (2). | ||
An NTA-functionalized membrane (approx. 400 μm thick) was prepared using cellulose diacetate and an NTA-surfactant by thermally-induced phase separation.10Fig. 1 shows the field-emission SEM (FE-SEM) images of the cross-section and surface of the NTA-functionalized membrane. The microscopic observations revealed a uniformly microporous structure. The filtration of nanospheres (100 nm) through this membrane resulted in 91% rejection of the nanospheres, which indicates that the effective pore size of the membrane was approx. 100 nm.
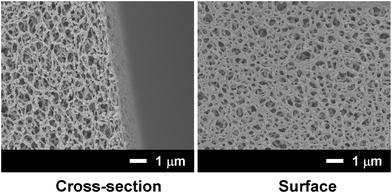 | ||
| Fig. 1 FE-SEM images of a cellulose acetate membrane containing the AB-NTA-surfactant. | ||
We adopted His-tag enhanced green fluorescent protein (His-tag GFP) as a model His-tag protein and the GFP solution was applied to an NTA-functionalized membrane. The effect of the NTA-surfactant content on the GFP-binding capacity to the functionalized membrane was investigated (Fig. 2). The binding capacity was defined as the protein amount that was eluted after loading and washing processes (not the adsorbed protein amount). The addition of the NTA-surfactant increased the binding capacity and the binding capacity reached a plateau above a content ratio of 0.05. ICP-atomic emission spectrometry analysis revealed that the Ni2+-binding capacity of the membrane was 0.24 × 10−4 mmol cm−2. These results suggest that the addition of the NTA-surfactant to the phase separation of the polymer displayed NTA moieties on a membrane surface.
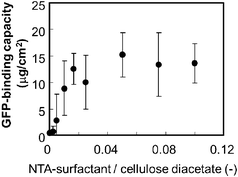 | ||
| Fig. 2 Effect of the NTA-surfactant content on the protein binding capacity of the NTA-functionalized membrane. A GFP solution (0.04 mg, 2 ml) was applied to the membrane (apparent membrane area 4.9 cm2). | ||
A mixture of His-tag GFP (90 μg) and FITC-labeled BSA (2 mg) in a 2 ml solution was applied to the NTA-functionalized membrane (Fig. 3a). The non-adsorption fraction represents the permeate fraction when a sample solution was filtrated through the membrane. The passing fraction represents the washing fraction when the protein-loaded membrane was washed using phosphate buffer. The elution fraction was the fraction eluted from the protein-loaded membrane using an imidazole solution. BSA, which had no specific affinity to an NTA moiety, did not bind to the membrane. Most of the BSA passed through the membrane, and the BSA that was trapped on the membrane was washed away by the phosphate buffer washes. There was a negligible amount of BSA in the elution fraction. In contrast, only a negligible amount of His-tag GFP was in the non-adsorption fraction. More than half of the His-tag GFP loaded was found in an elution fraction; although the passing fraction contained an amount of non-adsorbed His-tag GFP. Under the present conditions, the releasable binding capacity of the membrane was 9.2 μg cm−2 of His-tag GFP. These results indicate the selective separation of His-tag GFP from a protein mixture using an NTA-functionalized membrane.
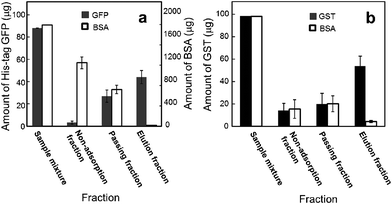 | ||
| Fig. 3 Protein amounts, in fractions, eluted from the NTA-functionalized membrane (a) and glutathione-functionalized membrane (b). Open circles represent BSA. Filled circles represent His-tag GFP (a) and GST (b). The protein amount represents the total amount of protein in a fraction. The weight ratio of ligand-surfactant/cellulose diacetate was 0.05. Apparent membrane area was 4.9 cm2. | ||
A glutathione-functionalized membrane was also prepared in a similar manner using the glutathione-surfactant. A mixture of GST and TRITC-labeled BSA was applied to a glutathione-functionalized membrane. Fig. 3b shows that GST (56 μg cm−2) was selectively collected in the elution fraction from the protein mixture, indicating that the glutathione-functionalized membrane can separate GST from BSA. This result also indicates the validity of our strategy using a surfactant-like ligand in an affinity membrane preparation.
Finally we tried the purification of tagged recombinant proteins from a host cell (E. coli). E. coli overexpressing a target protein was disrupted by ultrasonication. The cell lysates were applied to the NTA- and glutathione-functionalized membranes. SDS-PAGE analyses (Fig. 4) showed that the elution fractions from the membranes contained single bands corresponding to the molecular weights of His-tag GFP (42.7 kDa) and GST (26.6 kDa), whereas the cell lysate contained many bands of various proteins.11,12 These results demonstrate that the NTA-functionalized and glutathione-functionalized membranes effectively purified His-tag GFP and GST from the cell lysate.
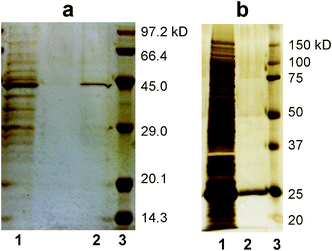 | ||
| Fig. 4 SDS-PAGE analyses (silver staining) of proteins purified by the task-specific membranes. a) His-tagged GFP and b) GST. Lanes 1, 2 and 3 represent the cell lysate, elution from the membrane and molecular weight standards, respectively. | ||
These investigations demonstrate that our present membranes allow the selective and facile purification of tagged proteins. The separation behavior is very similar to the commercial systems and also to the literatures reported, probably due to the same ligand–protein interactions. The yield of the target protein in the present system is based on the surface area of the membrane, while bead amounts determine the capacity of protein purification in the conventional column system.
In conclusion, we have synthesized surfactant-like ligands that target recombinant proteins, and have introduced these ligands onto polymeric membranes using a thermally-induced phase separation procedure. The ligands were successfully displayed on the membrane surfaces and the ligand-functionalized membranes enabled highly-selective purification of target proteins (GFP and GST) from cell lysates. The task-specific membranes proposed in this study will aid high-throughput screening or high-throughput analysis of genetically designed proteins.
The authors thank Prof. A. Mori for his help of ESI-MS measurements. This work was supported financially by the Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), MEXT, Japan and also by Grants for Research and Technology Development on Waste Management (K2336) from the Ministry of Environment, Japan.
References
- (a) V. P. Torchilin and A. N. Lukyanov, Drug Discovery Today, 2003, 8, 259 CrossRef CAS; (b) D. Schneidman-Duhovny, R. Nussinov and H. J. Wolfson, Curr. Med. Chem., 2004, 11, 91 CrossRef CAS; (c) A. B. Weinglass, J. P. Whitelegge and H. R. Kaback, Curr. Opin. Drug Discov. Dev., 2004, 7, 589 CAS; (d) J. Campbell Larry and J. C. Yves Le Blanc, Bioanalysis, 2011, 3, 645 CrossRef CAS.
- (a) J. K. Buolamwini, Curr. Pharm. Des., 2000, 6, 379 CrossRef CAS; (b) J. P. Goddard and J. L. Reymond, Curr. Opin. Biotechnol., 2004, 15, 314 CrossRef CAS; (c) D. H. Thompson, M. Zhou, J. Grey and H.-K. Kim, Chem. Lett., 2007, 36, 956 CrossRef CAS; (d) A. Das, J. Zhao, G. C. Schatz, S. G. Sligar and R. P. Van Duyne, Anal. Chem., 2009, 81, 3754 CrossRef CAS.
- (a) R. Ghosh, J. Chromatogr., A, 2002, 952, 13–27 CrossRef CAS; (b) C. Boi, J. Chromatogr., B: Anal. Technol. Biomed. Life Sci., 2007, 848, 19 CrossRef CAS; (c) A. Saxena, B. P. Tripathi, M. Kumar and V. K. Shahi, Adv. Colloid Interface Sci., 2009, 145, 1 CrossRef CAS.
- (a) H. Iwata, K. Saito, S. Furusaki, T. Sugo and J. Okamoto, Biotechnol. Prog., 1991, 7, 412 CrossRef CAS; (b) M. Kim, K. Saito, S. Furusaki, T. Sugo and I. Ishigaki, J. Chromatogr., A, 1991, 585, 45 CrossRef CAS; (c) C. Dietrich, O. Boscheinen, K.-D. Scharf, L. Schmitt and R. Tampe, Biochemistry, 1996, 35, 1100 CrossRef CAS; (d) X. Zeng and E. Ruckenstein, Ind. Eng. Chem. Res., 1998, 37, 159 CrossRef CAS; (e) J. A. Hestekin, L. G. Bachas and D. Bhattacharyya, Ind. Eng. Chem. Res., 2001, 40, 2668 CrossRef CAS; (f) Q. Luo, H. Zou, Q. Zhang, X. Xiao, Z. Guo, L. Kong and X. Mao, J. Chromatogr., A, 2001, 926, 255 CrossRef CAS; (g) A. M. Hollman, D. A. Christian, P. D. Ray, D. Galey, J. Turchan, A. Nath and D. Bhattacharyya, Biotechnol. Prog., 2005, 21, 451 CrossRef CAS; (h) P. Jain, L. Sun, J. Dai, G. L. Baker and M. L. Bruening, Biomacromolecules, 2007, 8, 3102 CrossRef CAS; (i) P. Jain, M. K. Vyas, J. H. Geiger, G. L. Baker and M. L. Bruening, Biomacromolecules, 2010, 11, 1019 CrossRef CAS.
- (a) Z. W. Ma, M. Kotaki and S. Ramakrishna, J. Membr. Sci., 2005, 265, 115 CrossRef CAS; (b) K. Yoshimatsua, L. Yea, J. Lindbergb and I. S. Chronakis, Biosens. Bioelectron., 2008, 23, 1208 CrossRef; (c) A.-F. Che, Z.-M. Liu, X.-J. Huang, Z.-G. Wang and Z.-K. Xu, Biomacromolecules, 2008, 9, 3397 CrossRef CAS.
- (a) P. M. Atkinson and D. R. Lloyd, J. Membr. Sci., 2000, 171, 1 CrossRef CAS.
- http://www.whatman.com/CelluloseAcetateMembranes.aspx .
- E. Hochuli, H. Doeli and A. Schacher, J. Chromatogr., A, 1987, 411, 177 CrossRef CAS.
- D. B. Smith and K. S. Johnson, Gene, 1988, 67, 31 CrossRef CAS.
- T. Shibutani, T. Kitaura, Y. Ohmukai, T. Maruyama, S. Nakatsuka, T. Watabe and H. Matsuyama, J. Membr. Sci., 2011, 376, 102 CrossRef CAS.
- T. Tanaka, N. Kamiya and T. Nagamune, FEBS Lett., 2005, 579, 2092 CrossRef CAS.
- J. Shimada, T. Maruyama, M. Kitaoka, N. Kamiya and M. Goto, Anal. Biochem., 2011, 414, 103 CrossRef CAS.
Footnote |
| † Electronic Supplementary Information (ESI) available: Experimental section. See DOI: 10.1039/c1ra00856k/ |
| This journal is © The Royal Society of Chemistry 2012 |
