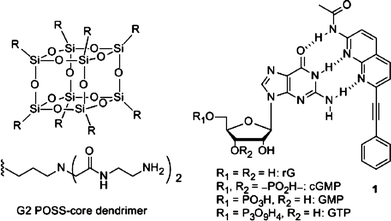
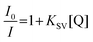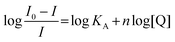Enhancement of affinity in molecular recognition via hydrogen bonds by POSS-core dendrimer and its application for selective complex formation between guanosine triphosphate and 1,8-naphthyridine derivatives†
Received
24th September 2011
, Accepted 12th October 2011
First published on 1st November 2011
Introduction
Hydrogen-bond-mediated molecular recognition in water is a topic with high relevance to biological systems. To realize stable complexation between the receptor and the target, some elaboration to maintain the binding affinity should be necessary to compensate for the inhibition of bond formation by hydration.1 A familiar examples can be seen in the RNA polymerases. In the active pocket, recognition with the nucleoside triphosphates based on hydrogen-bond patterns can be accomplished along the template sequences with high fidelity in the gene transcription.2 In contrast, complexation with the complementary nucleosides can scarcely be observed in water without enzymes.
1,8-Naphthyridine derivatives can work as a receptor for guanosine nucleotides.3 Nakatani et al. have presented a series of 1,8-naphthyridine derivatives for the highly-sensitive recognition of nucleobases.4 Cooperative interaction plays a significant role in the enhancement of affinity, leading to the specific recognition of guanine-involving mismatch sites in DNA.5 Cywinski et al. have reported cyclic GMP (cGMP)-selective recognition using modified polymer particles with naphthyridine derivatives.6 The cooperativity between the zeta potential of the particles and the recognition properties of the naphthyridine receptor can result in molecular recognition ability for cGMP with high specificity.
Hydrophilic polymeric materials which can retain guest molecules are a versatile platform as biosensors or vessels for drug delivery systems.7 In particular, because of the uniform structures, the characteristics of the dendrimers can be readily modulated by the size tuning and peripheral modification.8 These advantages of the dendrimers are feasible for site- and time-selective delivery. We have recently reported the retention ability of water-soluble dendrimers composed of polyhedral oligomeric silsesquioxane (POSS).9 In polar solvents, the POSS-core dendrimers could form globular conformations even at lower generations and could create a distinct hydrophobic space around the POSS core.10,11 Consequently, larger amounts of the hydrophobic guest molecules can be sustained inside the dendrimers than inside those of the same generation of the polyamidoamine (PAMAM, Scheme S1†) dendrimer which has similar dendrons.10 Our interest next focused on realizing selective capturing of biomolecules using the distinct hydrophobic spaces inside the POSS-core dendrimers.
Herein, we describe the influence on the binding affinity with the naphthyridine ligand and guanosine derivatives by the POSS-core dendrimer. The binding constants were evaluated from the fluorescence quenching by adding a series of guanosine nucleotides. In addition, from the investigation of the recognition mechanism with the dendrimer PAMAM and various kinds of naphthyridine derivatives (Scheme 1), it was suggested that the POSS core should offer a hydrophobic space where the strength of the hydrogen bonds should be enhanced. This is the first example, to the best of our knowledge, not only of using the amphiphilicity of the dendrimer to enhance molecular recognition via hydrogen bonding in the aqueous phase but also of capture of biomolecules by a dendrimer.
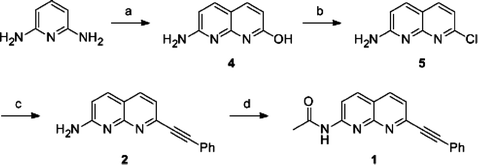 |
| | Scheme 1 Synthetic scheme for the naphthyridine derivatives. Reagents and conditions: (a) Malic acid, 95% H2SO4, 110 °C, 3 h, 92%; (b) POCl3, reflux, 4 h, 33%; (c) ethynylbenzene, PdCl2(PPh3)2, CuI, triethylamine, THF, r.t., 16 h, 30%; (d) acetic anhydride, chloroform, r.t., 3 h, 80%. | |
Experimentals
Materials
1H NMR and 13C NMR spectra were measured with a JEOL EX–400 (400 MHz for 1H and 100 MHz for 13C) spectrometer. 29Si NMR spectra were measured with a JEOL JNM–A400 (80 MHz) spectrometer. Coupling constants (J values) are reported in hertz. The chemical shifts are expressed in ppm downfield from tetramethylsilane, using residual dimethyl sulfoxide (δ = 2.50 in 1H NMR, δ = 39.5 in 13C NMR) as an internal standard. Emission from the samples was monitored using a Perkin Elmer LS50B at 25 °C using 1 cm path length cell. Mass spectra were obtained on a JEOL JMS–SX102A. The synthetic schemes for compound 312 and the G2 POSS-core dendrimer are shown in Schemes S2 and S3 in the Supporting Information.† The G2 PAMAM dendrimer was purchased from Aldrich as a methanol solution and directly used for the analyses.
Compound 412
A reaction mixture containing 9.07 g of malic acid (67.6 mmol) and 6.67 g of 2,6-diaminopyridine (61.1 mmol) was slowly added to 40 mL of 95% H2SO4 at 0 °C. After stirring at 110 °C for 3 h, the solution was neutralized with NH4OH, and the product was obtained as a precipitate. Compound 4 was separated by filtration as a white powder (9.05 g, 92%). 1H NMR (DMSO-d6) δ 11.85 (br, 2H), 7.64 (d, 1H, J = 9.6 Hz), 7.63 (d, 1H, J = 9.2 Hz), 6.32 (d, 1H, J = 9.2 Hz), 6.01 (d, 1H, J = 8.9 Hz). LRMS (NBA) [(M+H)+] calcd. 162, found 162.
Compound 512
The hydroxyl compound 4 (1.0 g, 6.21 mmol) was refluxed with 50 mL of POCl3 for 4 h. After cooling, the reaction solution was neutralized with NH4OH, and extracted with chloroform. Evaporation of the chloroform extract gave the crude product which was recrystallized from toluene giving 370 mg of the pure product 5 as a yellow powder (33%). 1H NMR (DMSO-d6) δ 7.86 (d, 1H, J = 8.0 Hz), 7.82 (d, 1H, J = 8.8 Hz), 7.17 (d, 1H, J = 8.0 Hz), 6.75 (d, 1H, J = 6.8 Hz), 5.33 (bs, 2H). LRMS (NBA) [(M+H)+] calcd. 180, found 180.
Compound 213
In a 25 mL round-bottom flask, 100 mg of 2-amino-7-chloro-1,8-naphthyridine (5) (0.557 mmol), 10 mg of PdCl2(PPh3)2 (cat.) and 3 mg of CuI (cat.) were stirred in 5 mL of dry THF under argon. To the solution, 0.6 mL of triethylamine was slowly added through a syringe. The mixture was stirred at room temperature for 30 min and then 0.1 mL of ethynylbenzene (0.980 mmol) was added. The reaction was stirred at room temperature overnight. Upon removal of the volatiles under reduced pressure, the residue was dissolved in dichloromethane, passed through a plug of Celite and then washed with ethyl acetate. The product was dried under vacuum. Column chromatography in silica gel with an increasing gradient of methanol in chloroform (chloroform/methanol from 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 10
1 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) yielded 41 mg of pure compound 2 (30%) as a white powder. 1H NMR (CDCl3) δ 7.92 (d, 1H, J = 8.0 Hz), 7.85 (d, 1H, J = 8.8 Hz), 7.62 (m, 2H), 7.41 (d, 1H, J = 8.4 Hz), 7.37 (m, 3H), 6.77 (d, 1H, J = 8.8 Hz), 5.05 (br,1H). 13C NMR (CDCl3) δ 159.97, 155.80, 145.35, 137.59, 136.26, 132.18, 129.07, 128.34, 122.13, 121.68, 116.77, 133.70, 90.57, 89.32. LRMS (NBA) [(M+H)+] calcd. 246, found 246: LRMS (NBA) [(M+H)+] calcd. 246.1031, found 246.1021.
1) yielded 41 mg of pure compound 2 (30%) as a white powder. 1H NMR (CDCl3) δ 7.92 (d, 1H, J = 8.0 Hz), 7.85 (d, 1H, J = 8.8 Hz), 7.62 (m, 2H), 7.41 (d, 1H, J = 8.4 Hz), 7.37 (m, 3H), 6.77 (d, 1H, J = 8.8 Hz), 5.05 (br,1H). 13C NMR (CDCl3) δ 159.97, 155.80, 145.35, 137.59, 136.26, 132.18, 129.07, 128.34, 122.13, 121.68, 116.77, 133.70, 90.57, 89.32. LRMS (NBA) [(M+H)+] calcd. 246, found 246: LRMS (NBA) [(M+H)+] calcd. 246.1031, found 246.1021.
Compound 113
In a 25 mL round-bottom flask, 30 mg of 2 (0.122 mmol) and 5 mL of acetic anhydride were stirred in 10 mL of chloroform at room temperature for 3 h. After evaporation, compound 1 (28 mg, 80%) was purified as a white powder from column chromatography in silica gel with ethyl acetate. 1H NMR (CDCl3) δ 9.04 (br,1H), 8.55 (d, 1H, J = 8.8 Hz), 8.18 (d, 1H, J = 8.8 Hz), 8.11 (d, 1H, J = 8.4 Hz), 7.65 (m, 2H), 7.60 (d, 1H, J = 8.4 Hz), 7.41 (m, 3H), 2.31 (s, 3H). 13C NMR (CDCl3) δ 170.27, 154.74, 154.64, 146.78, 138.94, 136.58, 132.27, 129.42, 128.47, 124.02, 121.90, 119.70, 115.97, 91.39, 89.13, 25.08. LRMS (NBA) [(M+H)+] calcd. 288, found 288: LRMS (NBA) [(M+H)+] calcd. 288.1137, found 288.1125.
Complexation with the dendrimers
The general procedure for retention of the guest molecules by the dendrimers is described here. Stock solutions (×10) of the guest molecules and G2 POSS-core dendrimer were mixed at room temperature, and then 500 μL of the samples were prepared by adding solvents.
Evaluation of the amount of retained ligand in the G2 POSS-core dendrimer
The sample solutions were stored under ambient conditions for 2 days and filtered through Nanosep 3 K centrifugal devices (Pall Life Sciences) by centrifugation (2000g, 30 min, 25 °C). The concentration of ligand 1 was determined from the fluorescence emission of the filtrates. The amount of retention was evaluated by comparing to the intensities of the filtrates at 0 day (I0) and 2 days (I2d). The results are shown in Fig. 2.
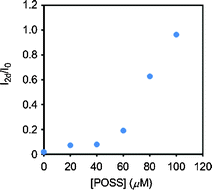 |
| | Fig. 2 The emission changes of the filtrates after 2 days. Solutions containing 10 μM 1 and variable concentrations of the G2 POSS-core dendrimer were stored under ambient conditions for 2 days and then passed through a size-exclusion filter membrane by centrifugation. Emission spectra were obtained with excitation light at 330 nm at 25 °C. | |
Fluorescence measurements of the complexes
The fluorescence emission of the ligand solution (10 μM) in the presence and absence of the dendrimers (100 μM, pH = 5.0) with or without guanosine derivatives under excitation at 330 nm was monitored using a Perkin Elmer LS50B at 25 °C with a 1 cm path length cell. The excitation bandwidth was 1 nm. The emission bandwidth was 1 nm. The quantum yields were determined as an absolute value with an integral sphere.
Stern–Volmer plots14
The data were analyzed in terms of the Stern–Volmer equation [eqn (1)]: | | 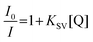 | (1) |
The emission intensity was plotted according to a Stern–Volmer equation, reporting I0/I versus the concentrations of guanosine derivatives [Q], where I0 is the intensity in the absence of quencher and I is the intensity in the presence of quencher. KSV is the Stern–Volmer quenching constant. A plot of I0/I versus [Q] yields an intercept of one and a slope equal to KSV.
Binding constants calculation14
If non-emissive complex formation between the naphthyridine ligand and guanosine nucleotides occurs, the binding constant (KA) can be calculated with the number of the binding molecules (n) from eqn (2): | | 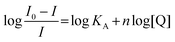 | (2) |
Results and discussion
We designed the ligand 1 as shown in Fig. 1. We expected that the POSS core could provide hydrophobic spaces inside the dendrimer, leading to the enhancement of hydrogen bonding. The 1,8-naphthyridine moiety can form a complex with the guanine skeleton via hydrogen bonds.3 In particular, as applied for fluorescence sensors for guanosine nucleotides in the previous work,3 the fluorescence emission can be quenched via electron transfer through the hydrogen bonds.6 In other words, a decrease in emission represents complex formation of the naphthyridine ligand with guanosine nucleotides. Thereby, the binding constants can be evaluated with the Stern–Volmer equation in the case of the non-emissive complex model according to eqn (2).14 These optical properties of the naphthyridine can make it possible to quantitatively assess the stability of the complexes of the naphthyridine moiety with guanosine nucleotides. The benzene ring is expected to play a role in the adsorption of the ligand on the surface of the POSS core via the hydrophobic interaction and to be engaged into the internal space of the POSS-core dendrimers. The terminal amino groups are expected to bind the guanosine nucleotides by the electrostatic interaction with the phosphate groups.
Initially, 1-amino-7-chloronaphthyridine was prepared according to the previous reports.12 The ligand 1 was obtained via Sonogashira–Hagihara cross coupling reaction, following acetylation at the amino group on the 7 position. The second generation of the POSS-core dendrimer (G2 POSS-core dendrimer) was prepared according to the literature.9,15 Complexation of the G2 POSS-core dendrimer with the naphthyridine ligand was performed by mixing. To confirm the retention in the dendrimer, we investigated the fluorescence emission of the filtrate passed through the size-exclusion filter.11 If the naphthyridine ligand forms aggregates or adsorbs onto the vessel walls, the filtrate shows significant low emission intensity. On the other hand, if the naphthyridine ligand can be retained in the dendrimers, the adsorption should be inhibited. Thereby, the fluorescence can be observed from the filtrate. In addition, the emission intensity of the filtrate can represent the amount of retention. We prepared solutions with various concentrations of G2 POSS-core dendrimer and investigated the intensity changes of the filtrate after 2 days from the ligand 1 (Fig. 2). The filtrate in the absence of G2 POSS-core dendrimer provided subtle emission from the ligand 1. In contrast, fluorescence emission from the filtrate was mostly maintained in the presence of G2 POSS-core dendrimer. These results suggest that 10 μM of the ligand 1 should be retained in the dendrimer (100 μM). Moreover, it was found that the naphthyridine ligand retained in the G2 POSS-core dendrimer can maintain a good dispersion state without non-specific aggregation or adsorption on the vessel wall for at least two days. This is long enough to do further experiments.
We investigated the changes in fluorescence intensity from the ligand 1 by adding a series of guanosine nucleotides (Table 1). The emission spectra were obtained from the aqueous solutions containing 10 μM ligand 1 and 100 μM G2 POSS-core dendrimer with excitation light at 330 nm (Fig. 3). The strong fluorescence emission with the peak around 380 nm was observed from the ligand 1 in the aqueous solution. The addition of guanosine triphosphate (GTP) caused the significant quenching of the emission from the ligand 1 (Fig. 3a). Complex formation with GTP was suggested. In the presence of cGMP, the emission was also quenched (Fig. 3b). Corresponding to the results reported by Cywinski et al., complex formation via hydrogen bonds should occur between the ligand 1 and cGMP.6 A slight effect on emission intensity was observed by adding rG and GMP to the solution. These results suggest that the interaction particularly between GTP and the ligand 1 can be enhanced by the G2 POSS-core dendrimer.
Table 1 Optical properties and the binding constants of the ligand 1a
| Nucleotidesb |
Φ
|
K
SV [×103 mol L−1]d |
n
|
K
A [M−1]e |
|
Procedures and conditions are described in the Experimental section in the Supporting Information.†
100 μM solutions.
Determined as an absolute value.
Quenching constants were determined from the slopes of the fitting line in the Stern–Volmer plots.
Calculated according to ref. 14
n.d. = not determined because of too weak an interaction.
|
| None |
0.39 |
n.d.f |
n.d.f |
n.d.f |
| rG |
0.39 |
n.d.f |
n.d.f |
n.d.f |
| GMP |
0.39 |
n.d.f |
n.d.f |
n.d.f |
| cGMP |
0.34 |
1.14 |
0.95 |
670 |
| GTP |
0.30 |
2.83 |
1.10 |
7500 |
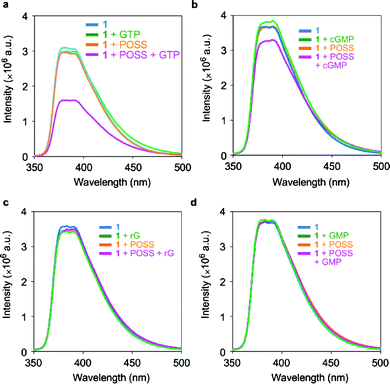 |
| | Fig. 3 Emission changes of 10 μM 1 in the presence and absence of 100 μM G2 POSS-core dendrimers by adding 100 μM guanosine nucleotides in water at 25 °C. Excitation wavelength was 330 nm. | |
To evaluate the role of the POSS core, we performed fluorescence measurements using the same procedure using the G2 PAMAM dendrimer which possesses an ethylene diamine core instead of POSS. On addition of guanosine nucleotides including GTP, the emission from the ligand 1 was less affected (Fig. S1†). These results clearly indicate that the POSS core is responsible for enhancing the complex formation between ligand 1 and GTP. The hydrophobic space around the POSS core could reduce the hydration of the naphthyridine ligand, resulting in enhancement of the affinity of the complex via hydrogen bonds. Moreover, fluorescence measurements were also carried out with adenosine triphosphate (ATP), and it was found that the emission was hardly changed (Fig. S2). This result suggests that the triphosphate group has less influence on the emission properties of the naphthyridine derivatives.
We prepared Stern–Volmer plots with guanosine nucleotides as quencher (Fig. 4). Based on the formation of a non-emissive complex model, the binding constants were calculated (Fig. 5). All plots obtained from the titration of GTP and cGMP were fitted on the line, and the binding constants could be determined. It was found that POSS can enhance the binding affinity with GTP approximately 10 times more than that with cGMP. These results suggest that the ligand 1 can selectively recognize GTP in the G2 POSS-core dendrimer.
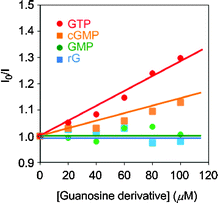 |
| | Fig. 4 Stern–Volmer plots with a solution containing 10 μM 1 by adding various kinds of guanosine derivatives. | |
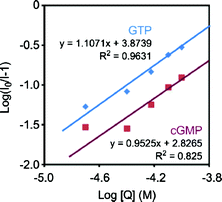 |
| | Fig. 5 Plots for determining binding constants between the ligand 1 and guanosine nucleotides according to eqn (2). The slopes represent the number of binding guanosine nucleotides. The y-intercepts represent the logKA values. The lines are prepared with the least-squares method. R2 is the determination coefficient. | |
In order to investigate the influence of the substituents at the naphthyridine ligand on the recognition, the emission spectra were measured with the various kinds of naphthyridine derivatives12 listed in Fig. 6. The retention of these derivatives in the G2 POSS-core dendrimer in the same manner as above, and the emission changes, were examined. Significant quenching was hardly observed on addition of guanosine nucleotides to the series of naphthyridine ligands. These results mean that all substituents on the ligand 1 are necessary to maintain the recognition ability. The hydrogen bonds and the hydrophobic interaction with the surface of the POSS core might cooperatively contribute to the selective recognition of ligand 1 with GTP in the G2 POSS-core dendrimer.
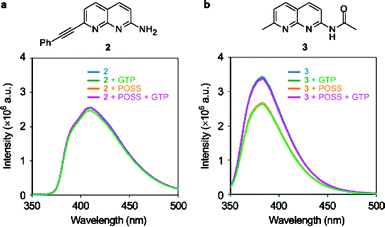 |
| | Fig. 6 Chemical structures and emission spectra of 10 μM (a) 2 and (b) 3 in the presence and absence of 100 μM G2 POSS-core dendrimers by adding 100 μM GTP in water at 25 °C. Excitation wavelength was 330 nm. | |
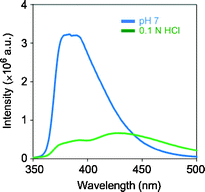
To elaborate the influence of the charge on the surface of the dendrimer, the emission intensity of the complex was compared in solutions of various pH (Fig. 7). Under alkaline conditions, the degree of fluorescence quenching was reduced, while the quenching ability can be maintained under acidic conditions. These results indicate that the terminal-ammonium groups should contribute to binding GTP via electrostatic interaction. Under the extremely acidic conditions in 0.1 N hydrochloric acid, the fluorescence emission was largely suppressed (Fig. 8, Φ = 0.13), and a new peak appeared in the longer wavelength region. Therefore, the protonation of the naphthyridine moiety induced by the triphosphate moiety of GTP should hardly be responsible for the quenching. From the series of data, a recognition model for guanosine nucleotides by the dendrimer complex can be proposed. The naphthyridine ligand and G2 POSS-core dendrimer form a stable complex in which guanosine nucleotides can enter and leave. GTP should be retained in the complex for a relatively longer time because of the cooperative interaction via hydrogen bonds with the naphthyridine ligand and the stronger electrostatic attraction. Thus, selective capturing of GTP by the dendrimer complex could be observed.
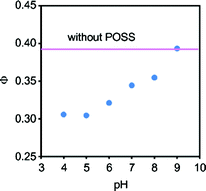 |
| | Fig. 7 The influence of pH on the quantum yield of emission from solutions containing 10 μM 1, 100 μM G2 POSS-core dendrimer, and 100 μM GTP in 50 mM sodium phosphate buffer at 25 °C. Excitation wavelength was 330 nm. | |
Conclusions
In conclusion, we present that the binding affinity of the naphthyridine ligand with GTP can be enhanced by the POSS-core dendrimer. Although the affinity is lower than those of the aptamers16 as previously reported, our system is applicable over a wide pH range and salt concentration. Our data indicate that the dendrimer complex with the naphthyridine ligand can selectively recognize and capture GTP. Furthermore, in this system, the POSS-core dendrimer and the ligand molecules interact only via non-covalent bonds. Therefore, it can be expected that the target guest molecules can be tuned by replacing the ligand units. This might be applicable not only to development of new tools for biosensing but also for modulating the concentrations of targets in the cells.
Notes and references
- T. Sawada, M. Yoshizawa, S. Sato and M. Fujita, Nat. Chem., 2009, 1, 53 CrossRef CAS.
- K. D. Westover, D. A. Bushnell and R. D. Kornberg, Cell, 2004, 119, 481 CrossRef CAS.
- J. H. Liao, C. T. Chen, H. C. Chou, C. C. Cheng, P. T. Chou, J. M. Fang, Z. Slanina and T. J. Chow, Org. Lett., 2002, 4, 3107 CrossRef CAS; J. M. Fang, S. Selvi, J. H. Liao, Z. Slanina, C. T. Chen and P. T. Chou, J. Am. Chem. Soc., 2004, 126, 3559 CrossRef; Z. Xu, K. Morita, Y. Sato, Q. Dai, S. Nishizawa and N. Teramae, Chem. Commun., 2009, 6445 RSC; Y. Sato, S. Nishizawa, K. Yoshimoto, T. Seino, T. Ichizawa, K. Morita and N. Teramae, Nucleic Acids Res., 2009, 37, 1411 CrossRef.
- K. Nakatani, S. Hagihara, Y. Goto, A. Kobori, M. Hagihara, G. Hayashi, M. Kyo, M. Nomura, M. Mishima and C. Kojima, Nat. Chem. Biol., 2005, 1, 39 CrossRef CAS.
- K. Nakatani, S. Sando and I. Saito, Nat. Biotechnol., 2001, 19, 51 CrossRef CAS; K. Nakatani, S. Sando, H. Kumasawa, J. Kikuchi and I. Saito, J. Am. Chem. Soc., 2001, 123, 12650 CrossRef; T. Peng, C. Dohno and K. Nakatani, Angew. Chem., Int. Ed., 2006, 45, 5623 CrossRef.
- P. J. Cywinski, A. J. Moro, T. Ritschel, N. Hildebrandt and H. G. Löhmannsröben, Anal. Bioanal. Chem., 2011, 399, 1215 CrossRef CAS.
- S. Matsumoto, S. Yamaguchi, S. Ueno, H. Komatsu, M. Ikeda, K. Ishizuka, Y. Iko, K. V. Tabata, H. Aoki, S. Ito, H. Noji and I. Hamachi, Chem.–Eur. J., 2008, 14, 3977 CrossRef CAS; S. Matsumoto, S. Yamaguchi, A. Wada, T. Matsui, M. Ikeda and I. Hamachi, Chem. Commun., 2008, 1545 RSC; H. Komatsu, S. Matsumoto, S. Tamaru, K. Kaneko, M. Ikeda and I. Hamachi, J. Am. Chem. Soc., 2009, 131, 5580 CrossRef; J. Boekhoven, A. M. Brizard, K. N. K. Kowlgi, G. J. M. Koper, R. Eelkema and J. H. van Esch, Angew. Chem., Int. Ed., 2010, 49, 4825 Search PubMed.
- N. Morimoto, N. Ogino, T. Narita and K. Akiyoshi, J. Biotechnol., 2009, 140, 246 CrossRef CAS; A. Morimoto, X.-P. Qiu, F. M. Winnik and K. Akiyoshi, Macromolecules, 2008, 41, 5985 CrossRef; N. Morimoto, T. Ohki, K. Kurita and K. Akiyoshi, Macromol. Rapid Commun., 2008, 29, 672 CrossRef.
- P.-A. Jaffrés and R. E. Morris, J. Chem. Soc., Dalton Trans., 1998, 2767 RSC; F. J. Feher and K. D. Wyndham, Chem. Commun., 1998, 323 RSC; F. J. Feher, K. D. Wyndham, D. Soulivong and F. Nguyen, J. Chem. Soc., Dalton Trans., 1999, 1491 RSC; X. Zhang, K. J. Haxton, L. Ropartz, D. J. Cole-Hamilton and R. E. Morris, J. Chem. Soc., Dalton Trans., 2001, 3261 RSC.
- K. Tanaka, K. Inafuku, K. Naka and Y. Chujo, Org. Biomol. Chem., 2008, 6, 3899 Search PubMed; K. Tanaka, K. Inafuku and Y. Chujo, Chem. Commun., 2010, 46, 4378 RSC; K. Tanaka, K. Inafuku, S. Adachi and Y. Chujo, Macromolecules, 2009, 42, 3489 CrossRef CAS.
- K. Tanaka, W. Ohashi, N. Kitamura and Y. Chujo, Bull. Chem. Soc. Jpn., 2011, 84, 612 CrossRef CAS.
- R. G. Newkome, J. S. Garbis, K. V. Majestic, R. F. Fronczek and G. Chiari, J. Org. Chem., 1981, 46, 833 CrossRef.
- The 1H NMR spectrum is shown in the Supporting Information.
- Y. Sun, S. Bi, D. Song, C. Qiao, D. Mu and H. Zhang, Sens. Actuators, B, 2008, 129, 799 CrossRef.
- M. C. Gravel, C. Zhang, M. Dinderman and R. M. Laine, Appl. Organomet. Chem., 1999, 13, 329 CrossRef CAS; F. J. Feher and K. D. Wyndham, Chem. Commun., 1998, 323 RSC.
- J. H. Davis and J. W. Szostak, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11616 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details. See DOI: 10.1039/c1ob06630g |
|
| This journal is © The Royal Society of Chemistry 2012 |
Click here to see how this site uses Cookies. View our privacy policy here. 
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 10
1 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) yielded 41 mg of pure compound 2 (30%) as a white powder. 1H NMR (CDCl3) δ 7.92 (d, 1H, J = 8.0 Hz), 7.85 (d, 1H, J = 8.8 Hz), 7.62 (m, 2H), 7.41 (d, 1H, J = 8.4 Hz), 7.37 (m, 3H), 6.77 (d, 1H, J = 8.8 Hz), 5.05 (br,1H). 13C NMR (CDCl3) δ 159.97, 155.80, 145.35, 137.59, 136.26, 132.18, 129.07, 128.34, 122.13, 121.68, 116.77, 133.70, 90.57, 89.32. LRMS (NBA) [(M+H)+] calcd. 246, found 246: LRMS (NBA) [(M+H)+] calcd. 246.1031, found 246.1021.
1) yielded 41 mg of pure compound 2 (30%) as a white powder. 1H NMR (CDCl3) δ 7.92 (d, 1H, J = 8.0 Hz), 7.85 (d, 1H, J = 8.8 Hz), 7.62 (m, 2H), 7.41 (d, 1H, J = 8.4 Hz), 7.37 (m, 3H), 6.77 (d, 1H, J = 8.8 Hz), 5.05 (br,1H). 13C NMR (CDCl3) δ 159.97, 155.80, 145.35, 137.59, 136.26, 132.18, 129.07, 128.34, 122.13, 121.68, 116.77, 133.70, 90.57, 89.32. LRMS (NBA) [(M+H)+] calcd. 246, found 246: LRMS (NBA) [(M+H)+] calcd. 246.1031, found 246.1021.