DNA-catalyzed reactivity of a phosphoramidate functional group and formation of an unusual pyrophosphoramidate linkage†
Amit
Sachdeva
and
Scott K.
Silverman
*
Department of Chemistry, University of Illinois at Urbana-Champaign, 600 South Mathews Avenue, Urbana, IL 61801, USA. E-mail: scott@scs.illinois.edu
First published on 14th September 2011
Abstract
During in vitro selection for DNA-catalyzed lysine reactivity, we identified a deoxyribozyme that instead catalyzes nucleophilic attack of a phosphoramidate functional group at a 5′-triphosphate-RNA, forming an unusual pyrophosphoramidate (N–PV–O–PV) linkage. This finding highlights the relatively poor nucleophilicity of nitrogen using nucleic acid catalysts, indicating a major challenge for future experimental investigation.
Introduction
Deoxyribozymes are specific DNA sequences that have catalytic activity.1–5 We have recently focused on expanding deoxyribozyme catalysis to include reactions of amino acid side chains,6–8 with the long-term goal of DNA-catalyzed modification of large proteins. Our initial effort in this direction demonstrated robust DNA catalysis (>70% yield in 2 h) of nucleopeptide linkage formation between the nucleophilic tyrosine (Tyr) phenolic OH side chain and an electrophilic 5′-triphosphate-RNA.6 In parallel, however, catalysis involving the serine (Ser) aliphatic hydroxyl side chain was extremely poor (only ∼0.2% yield), and reactivity of the lysine (Lys) amine side chain was not observed. Lys side chain reactivity has never previously been observed with either DNA or RNA enzymes, suggesting that this reactivity is a particular challenge. One report found RNA-catalyzed reaction of a peptide N-terminal α-amino group even when the Lys ε-amino side chain was available as a competing nucleophile.9Our initial study on DNA-catalyzed side chain reactivity6 used a substrate that presented a single amino acid residue in a highly preorganized three-helix-junction (3HJ) architecture (Fig. 1, Z = single amino acid).10,11 Subsequently, we found that expanding the substrate to contain a tripeptide (Z = Ala-Ser-Ala) enabled robust Ser reactivity by newly identified deoxyribozymes.7 Separately, by discarding the 3HJ preorganization altogether, we identified deoxyribozymes that function with free peptide substrates.8 In the present report, we sought DNA-catalyzed Lys reactivity using the 3HJ preorganized architecture (Z = Ala-Lys-Ala). However, we instead identified a DNA-catalyzed reaction between a phosphoramidate functional group in the substrate and 5′-triphosphate-RNA, forming an unusual pyrophosphoramidate linkage. The results provide a clear calibration point on the relative (un)reactivity of amines using nucleic acid catalysts, which is important information for our ongoing efforts to expand DNA catalysts to include the Lys side chain.
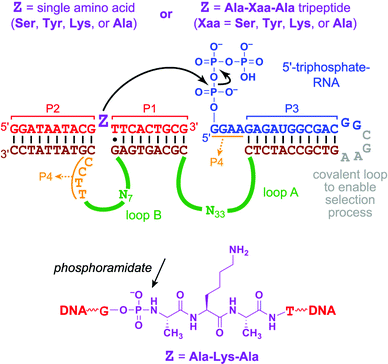 | ||
| Fig. 1 Substrates and DNA pool for in vitro selection. As established in many previous reports,6,7,11 formation of paired regions P1 through P4 creates a three-helix junction (3HJ) that juxtaposes the intended nucleophile (Z) and electrophile (5′-triphosphate). The CGAA nucleotides shown in grey are replaced with 5′-CC for the in trans (intermolecular) assays of individual deoxyribozymes. The chemical structure is drawn for the substrate with Z = Ala-Lys-Ala. Note the phosphoramidate (P–N) linkage that connects the 5′-portion of DNA to the tripeptide moiety. | ||
Results and discussion
In vitro selection of new DNA catalysts that covalently modify the Lys-containing substrate
We began our efforts to seek DNA-catalyzed Lys reactivity by retaining the 3HJ architecture of Fig. 1 while expanding the substrate to include an Ala-Lys-Ala tripeptide rather than the single Lys amino acid used previously,6 noting that analogous expansion successfully enabled Ser reactivity.7In vitro selection12,13 was used to identify individual DNA sequences with catalytic ability, following our established approach.6,7,14,15 Briefly, the 5′-triphosphate-RNA electrophile was covalently attached at its 3′-terminus to the DNA pool, which initially incorporated a total of 40 fully random nucleotide positions partitioned between two loops. Specific DNA sequences that can join the Lys-containing substrate to the 5′-triphosphate-RNA were separated by PAGE from the vast majority of nonfunctional DNA sequences and amplified by PCR. Attachment of the resulting DNA pool, now enriched in functional sequences, to a fresh sample of 5′-triphosphate-RNA enabled the next selection round to be performed. By iteration of this process for many rounds, catalytically active deoxyribozymes dominate the pool and can be cloned and characterized in detail.In each selection round's key step, during which the Lys-containing substrate can become attached to the 5′-triphosphate-RNA, we used incubation conditions of 50 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM KCl, 20 mM MnCl2, and 40 mM MgCl2 at 37 °C for 2 h. Mn2+ was included because of its previous utility in supporting both Tyr and Ser reactivity,6,7 and Mg2+ was used because of its value as a cofactor for a wide range of deoxyribozymes.15 The selections were iterated for seven rounds using an Ala-Lys-Ala substrate that included a 3′-phosphate that was intended to block nucleophilic reactivity of the 3′-terminus. However, this tactic proved ineffective, leading the majority of the pool to catalyze unwanted nucleophilic reaction of the 3′-phosphate with the 5′-triphosphate-RNA (Fig. S1 in the ESI†).
We therefore redirected the selection effort16 by replacing the 3′-phosphate group with a 2′,3′-dideoxycytidine nucleotide (3′-ddC), which lacks any strong nucleophile. The selection process was resumed from round 8, and by round 13, 12% ligation activity of the pool was observed. On the basis of PAGE migration rates, the uncloned products have a linkage to the RNA substrate that is either within or very near to the tripeptide region (Fig. S2A in the ESI†). However, an Ala-Ala-Ala substrate was comparable in reactivity to Ala-Lys-Ala, demonstrating that the linkage to the RNA was created not directly at the Lys side chain but instead at a nearby functional group. Upon cloning, a single deoxyribozyme sequence named 13LS3 was identified and found to form a product in modest 6% yield after 21 h in the presence of 20 mM Mn2+ either with or without 40 mM Mg2+. (This modest yield for 13LS3 is consistent with the relatively low activity observed for the corresponding uncloned pool.) The 13LS3 product appears to have the same connectivity as the product from the uncloned Lys selection pool (Fig. S2B in the ESI†). The tripeptide region of the substrate was required for 13LS3 activity; i.e., an all-DNA substrate that lacks the tripeptide region was unreactive (data not shown). In addition, as observed for the uncloned pool, the central amino acid of the substrate could be either Lys or Ala, indicating that the Lys side chain does not provide the nucleophile (Fig. S2B in the ESI†). Indeed, the 13LS3 product that was synthesized with the Ala-Lys-Ala substrate was shown to retain an aliphatic amine that can undergo reductive amination with a periodate-oxidized 3′-rA oligonucleotide17–19 (Fig. S3 and Fig. S4 in the ESI†), confirming that reaction does not occur directly at the Lys side chain (which is therefore available for reductive amination). Independent selection experiments with the Ala-Lys-Ala substrate where the 3′-ddC was present from the outset of selection led to the same overall observations (data not shown).
Assignment of 13LS3 deoxyribozyme product structure
The 13LS3 deoxyribozyme catalyzes an in trans (intermolecular) reaction between the tripeptide-containing substrate and 5′-triphosphate-RNA (see Fig. 1; note that the covalent loop at the right side is dispensable for activity). MALDI mass spectrometry of the PAGE-purified 13LS3 product was consistent with a reaction between a functional group of the Ala-Lys-Ala (or Ala-Ala-Ala) substrate and the α-phosphorus of the 5′-triphosphate-RNA, with pyrophosphate as the leaving group (see Experimental section and Fig. S5 in the ESI†). Biochemical cleavage assays of the product allowed its assignment as 1 (Fig. 2), which contains an unusual pyrophosphoramidate linkage that arises from nucleophilic attack at the 5′-triphosphate-RNA of the O atom at the DNA-tripeptide phosphoramidate junction. This product assignment was made by excluding all other reasonable candidates. Treatment with 80% acetic acid is known to hydrolyze phosphoramidate (P–N) bonds,20 and the tripeptide-containing substrate has a phosphoramidate linkage directly to the N-terminal side of the tripeptide moiety (Fig. 1). Therefore, treatment of the 13LS3 ligation product with 80% acetic acid should lead to a readily distinguishable PAGE pattern that depends sensitively on the site of nucleophilic reactivity within the tripeptide-containing substrate. Overall, the observations in Fig. 2 compel the conclusion that the product has the structure of pyrophosphoramidate 1. The data exclude the alternative, isomeric diphosphoramidate 2, which would have been formed by nucleophilic attack of the phosphoramidate N atom at the 5′-triphosphate. Product 2 would be susceptible to cleavage by 80% acetic acid, considering that a simple phosphoramidate is itself highly reactive under such conditions,20 and 2 should be even more reactive.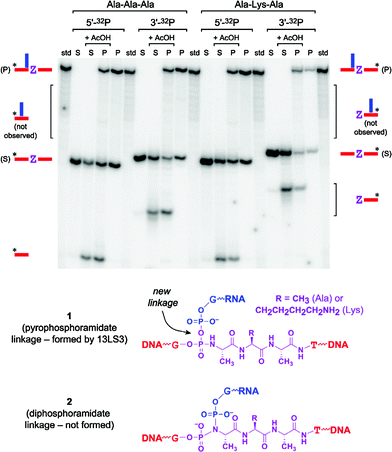 | ||
| Fig. 2 Treatment with 80% acetic acid for 5 h at 23 °C of the 5′- or 3′-32P-radiolabeled 13LS3 product, to provide evidence regarding its connectivity. S = substrate; P = product. The product was prepared using either the Ala-Ala-Ala or Ala-Lys-Ala substrate, with 5′-radiolabel from γ-32P-ATP and T4 PNK or 3′-radiolabel from α-32P-dCTP and terminal deoxytransferase. Each product led to essentially the same assay pattern. The cleavage band formed from each of the 13LS3 products (either 5′- or 3′-radiolabeled) arises solely from the regenerated substrate unavoidably present within the product sample due to hydrolysis, as discussed in the text (see also Fig. S3 in the ESI†). The upper bracketed areas denote regions of the gel where either the 5′- or 3′-32P-radiolabeled 13LS3 product would be expected to provide an acetic acid cleavage band, if 13LS3 were to attach the 5′-triphosphate-RNA to the DNA portion of the substrate anywhere either to the left or the right of the tripeptide region. The key point is that the absence of any such cleavage bands compels the conclusion that the 13LS3 product lacks the simple phosphoramidate linkage that is present in the substrate (Fig. 1). A phosphoramidate linkage readily hydrolyzes under the incubation conditions, as revealed by the cleavage observed both for the S samples and for the regenerated substrate within the P samples. | ||
The 13LS3 ligation product 1 was observed to regenerate a variable amount of the original tripeptide-containing substrate by cleavage of its pyrophosphate linkage. Note that this linkage is different from the P–N bond that is hydrolyzed by 80% AcOH within the phosphoramidate substrate (but not within 1). In particular, after separation by PAGE, the 13LS3 product 1 was partially cleaved during extraction from the polyacrylamide gel. Extraction in the conventional TEN buffer (10 mM Tris, pH 8.0, 300 mM NaCl, 1 mM EDTA) led to 40–50% substrate regeneration by pyrophosphate cleavage, whereas extraction in buffers of lower pH value (e.g., 10 mM HEPES, pH 7.0 or 10 mM NaOAc, pH 4.6) led to only 10–20% substrate regeneration. The extent of pyrophosphate cleavage during TEN extraction was time-dependent; e.g., 10–15% after 30 min extraction, but 40–50% after 2 h extraction. The regenerated substrate could itself be isolated by PAGE and was equally as competent for 13LS3-catalyzed reaction as fresh substrate (data not shown). Curiously, the substrate regeneration reaction does not occur when the gel-extracted product sample is incubated in free solution of the same composition as the extraction buffer; the ratio of product to regenerated substrate is unchanged by such incubation. Substrate regeneration appears to be a heterogeneous reaction that requires the polyacrylamide matrix and therefore occurs only during the extraction process.
Relationship of 13LS3 activity to previous observations
The newly formed linkage 1 is related to linkage 3 (Fig. 3), which was found by Moffatt and Khorana to have similar stability properties.21 These authors reported the formation of 3 by slow, uncatalyzed self-condensation of adenosine 5′-phosphoromorpholidate; i.e., attack of a phosphoramidate into an activated 5′-phosphorus (reaction A in Fig. 3). This uncatalyzed reaction is directly analogous to formation of 1 by DNA-catalyzed attack of the phosphoramidate linkage of the tripeptide-containing substrate into 5′-triphosphate-RNA (reaction B in Fig. 3), thereby providing a chemical precedent for the DNA-catalyzed reaction leading to 1. Moffatt and Khorana also reported that a simple phosphodiester is much less nucleophilic towards adenosine 5′-phosphoromorpholidate than is the phosphoromorpholidate itself. Consistent with this, we do not observe any DNA phosphodiester group within the tripeptide-containing substrate attacking the 5′-triphosphate-RNA.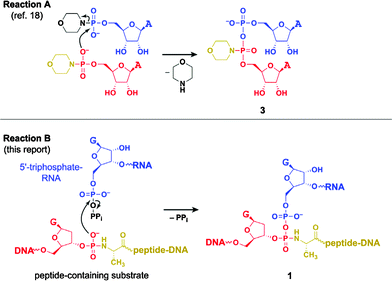 | ||
| Fig. 3 Comparison of reactions to form 3 (reaction A,21 uncatalyzed) and 1 (reaction B, this report, catalyzed by the 13LS3 deoxyribozyme). Both 3 and 1 have pyrophosphoramidate linkages formed by attack of a phosphoramidate (red/gold) into an activated 5′-phosphorus (blue, with black leaving group). The colour scheme here is modified relative to Fig. 1 in order to simplify comparison between the structural components of reactions A and B. | ||
Phosphoramidate linkages are found naturally as the lysine-adenylated intermediates of DNA ligase enzymes, among other instances.22–24 The 13LS3-catalyzed reaction of its substrate to form 1 rather than 2 indicates that the ambident phosphoramidate functional group of the substrate is more reactive at oxygen than at nitrogen, although 13LS3 necessarily offers only a single data point in this regard.
Broader implications of the results
A key implication of the finding that 13LS3 forms pyrophosphoramidate 1 is that at least under the evaluated incubation conditions, which are typical for in vitro selection experiments with ribozymes and deoxyribozymes, the phosphoramidate functional group within the Ala-Lys-Ala substrate is much more nucleophilic (at oxygen) than is the originally intended Lys amine side chain (at nitrogen). This result highlights the relative unreactivity of nitrogen as a nucleophile with the assistance of nucleic acid catalysts. Our ongoing efforts towards DNA-catalyzed Lys reactivity include use of more reactive electrophiles than 5′-triphosphate, which is intended to counterbalance the relatively poor nucleophilicity of the Lys amine. More broadly, in combination with our observation that DNA-catalyzed peptide bond cleavage is disfavored relative to phosphodiester hydrolysis,25 the results indicate that any kind of DNA-catalyzed reactivity of nitrogen-containing substrates (where the N atom is either nucleophile or leaving group) is a major experimental challenge.Experimental section
Materials were prepared and procedures were performed as described in our previous report.7 The reductive amination procedure is described in the ESI.† The 13LS3 sequence for the in trans (intermolecular) assays was 5′-CCGTCGCCATCTC![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CGCAGTGAG
CGCAGTGAG![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TTCCCGTATTATCC-3′, where the two underlined sequences are those of loops A and B derived from the N33+N7 random regions, and the three non-underlined sequences are those of the fixed P3, P1, and P4+P2 regions (in that order), all as shown in Fig. 1. Note that loop A of 13LS3 contains 34 rather than 33 nucleotides, presumably due to an insertion by Taq polymerase during one of the selection rounds. The two substrate sequences are shown in Fig. 1.
TTCCCGTATTATCC-3′, where the two underlined sequences are those of loops A and B derived from the N33+N7 random regions, and the three non-underlined sequences are those of the fixed P3, P1, and P4+P2 regions (in that order), all as shown in Fig. 1. Note that loop A of 13LS3 contains 34 rather than 33 nucleotides, presumably due to an insertion by Taq polymerase during one of the selection rounds. The two substrate sequences are shown in Fig. 1.
MALDI mass spectrometry was performed on 13LS3 reaction products that were each prepared as follows. A 560 μL sample containing 10 nmol of tripeptide-containing substrate, 10.5 nmol of 13LS3 deoxyribozyme, and 11 nmol of 5′-triphosphate-RNA was annealed in 5 mM HEPES, pH 7.5, 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. The pyrophosphoramidate linkage formation reaction was initiated by bringing the sample to 800 μL total volume containing 50 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM KCl, 20 mM MnCl2, and 40 mM MgCl2 and incubating at 37 °C for 14 h. The product was precipitated with ethanol, separated by 20% PAGE, extracted from the polyacrylamide gel in TEN buffer (10 mM Tris, pH 8.0, 300 mM NaCl, 1 mM EDTA), and precipitated with ethanol. The sample was dissolved in 20 μL of water; 10 μL was desalted by C18 ZipTip and used for mass spectrometry. All observed mass spectra were in accord with expectations (Fig. S5 in the ESI†). Product from the Ala-Lys-Ala substrate: calcd. 12038.8, found 12030.8 (Δ = −0.07%). Product from the Ala-Ala-Ala substrate: calcd. 11981.7, found 11976.5 (Δ = −0.04%). Within each sample was also observed the regenerated substrate from cleavage of the pyrophosphate linkage during extraction from the polyacrylamide gel. This regenerated substrate was fully reactive in subsequent 13LS3-catalyzed reactions and had the expected mass: calcd. 6312.2, found 6310.8 (Δ = −0.02%). The 5′-phosphorylated RNA formed by pyrophosphate cleavage during substrate regeneration (see 1 in Fig. 2) was also observed by MS: calcd. 5686.5, found 5685.4 (Δ = −0.02%).
Acknowledgements
This research was supported by grants to S.K.S. from the National Institutes of Health (GM065966) and the Defense Threat Reduction Agency (BRBAA08-L-2-0001). A.S. was partially supported by NIH T32 GM070421.Notes and references
- A. Peracchi, ChemBioChem, 2005, 6, 1316–1322 CrossRef CAS.
- S. K. Silverman, Chem. Commun., 2008, 3467–3485 RSC.
- K. Schlosser and Y. Li, Chem. Biol., 2009, 16, 311–322 CrossRef CAS.
- S. K. Silverman, Angew. Chem., Int. Ed., 2010, 49, 7180–7201 CrossRef CAS.
- M. Hollenstein, C. J. Hipolito, C. H. Lam and D. M. Perrin, ChemBioChem, 2009, 10, 1988–1992 CrossRef CAS.
- P. I. Pradeepkumar, C. Höbartner, D. A. Baum and S. K. Silverman, Angew. Chem., Int. Ed., 2008, 47, 1753–1757 CrossRef CAS.
- A. Sachdeva and S. K. Silverman, Chem. Commun., 2010, 46, 2215–2217 RSC.
- O. Y. Wong, P. I. Pradeepkumar and S. K. Silverman, Biochemistry, 2011, 50, 4741–4749 CrossRef CAS.
- S. Baskerville and D. P. Bartel, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9154–9159 CrossRef CAS.
- R. L. Coppins and S. K. Silverman, Nat. Struct. Mol. Biol., 2004, 11, 270–274 CAS.
- R. L. Coppins and S. K. Silverman, J. Am. Chem. Soc., 2005, 127, 2900–2907 CrossRef CAS.
- G. F. Joyce, Annu. Rev. Biochem., 2004, 73, 791–836 CrossRef CAS.
- G. F. Joyce, Angew. Chem., Int. Ed., 2007, 46, 6420–6436 CrossRef CAS.
- A. Flynn-Charlebois, Y. Wang, T. K. Prior, I. Rashid, K. A. Hoadley, R. L. Coppins, A. C. Wolf and S. K. Silverman, J. Am. Chem. Soc., 2003, 125, 2444–2454 CrossRef CAS.
- S. K. Silverman, Acc. Chem. Res., 2009, 42, 1521–1531 CrossRef CAS.
- Y. Wang and S. K. Silverman, Biochemistry, 2005, 44, 3017–3023 CrossRef CAS.
- P. C. Zamecnik, M. L. Stephenson and J. F. Scott, Proc. Natl. Acad. Sci. U. S. A., 1960, 46, 811–822 CrossRef CAS.
- S. A. Reines and C. R. Cantor, Nucleic Acids Res., 1974, 1, 767–786 CrossRef CAS.
- O. W. Odom, Jr., D. J. Robbins, J. Lynch, D. Dottavio-Martin, G. Kramer and B. Hardesty, Biochemistry, 1980, 19, 5947–5954 CrossRef.
- T. Wada, T. Moriguchi and M. Sekine, J. Am. Chem. Soc., 1994, 116, 9901–9911 CrossRef CAS.
- J. G. Moffatt and H. G. Khorana, J. Am. Chem. Soc., 1961, 83, 649–658 CrossRef CAS.
- B. Weiss, A. Thompson and C. C. Richardson, J. Biol. Chem., 1968, 243, 4556–4563 CAS.
- I. R. Lehman, Science, 1974, 186, 790–797 CAS.
- N. P. Higgins and N. R. Cozzarelli, Methods Enzymol., 1979, 68, 50–71 CrossRef CAS.
- M. Chandra, A. Sachdeva and S. K. Silverman, Nat. Chem. Biol., 2009, 5, 718–720 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Assays of uncloned pools and 13LS3 deoxyribozyme; reductive amination assay of 13LS3 ligation product; mass spectra for 13LS3 ligation products. See DOI: 10.1039/c1ob06088k |
| This journal is © The Royal Society of Chemistry 2012 |
