SERS assisted ultra-fast peptidic screening: a new tool for drug discovery†
Rolando
Pérez-Pineiro
*ab,
Miguel A.
Correa-Duarte
c,
Veronica
Salgueirino
c and
Ramon A.
Alvarez-Puebla
*ac
aNational Institute for Nanotechnology, National Research Council, 11421 Saskatchewan Drive, Edmonton, AB, Canada T6G 2M9. E-mail: ramon.alvarez@uvigo.es
bDepartment of Chemistry, University of Alberta, 11421 Saskatchewan Drive, Edmonton, AB, Canada T6G 2M9. E-mail: perezpin@ualberta.ca
cDepartamentos de Química Física y Física Aplicada, 36310, Vigo, Spain
First published on 9th November 2011
Abstract
Herein we present a direct label-free ultra-fast method for the identification and classification of the active members of a combinatorial library directly on the solid support used for their synthesis. The method is based on the appropriate functionalization of polyethylene glycol grafted polystyrene (TentaGel®) microbeads with Au@Ag nanoparticles, the use of these materials directly as solid-phase supports for the synthesis of combinatorial libraries of peptides and the subsequent SERS analysis for identification of each peptide on each bead.
Since the 1990's combinatorial chemistry has been firmly established in academic and industrial settings as a useful strategy to speed up the drug discovery process.1 This technique entails the preparation of structurally distinct molecules in a time and resource effective manner. The subsequent generation of combinatorial libraries has been adopted in two formats: spatially resolved parallel synthesis and split synthesis of one-bead-one-compound (OBOC). Despite being more efficient in terms of cost and generation of diversity in fewer synthetic steps, split synthesis is not the preferred choice. Aspects such as poor familiarization with solid phase methodologies, on-bead biological assays and the need for fast and reliable strategies to unravel the chemical identity of active members in complex mixtures, among others, remain as technical challenges.2 Furthermore, with the rapid developments in genomics and proteomics, a huge number of genes and proteins are expected to be targeted chemically and the revival of this synthetic format, which depends on the associate single bead analysis techniques, offers the key solution.3
Deconvolution strategies such as positional scanning,1,4 sequencing of resin-bound peptides,5 and molecular tagging protocols6,7 have been extensively applied in the identification of single bead compounds (SBCs) in split libraries. On the other hand the use of tandem mass spectrometry8 and microflow NMR9 has made possible the identification of library product(s) directly released from the solid support, rather than reporting only the reaction history of a particular bead, thus offering an attractive alternative to chemical encoding/decoding strategies. Notwithstanding, the need of high loading solid supports to render the samples detectable after cleavage and the implementation of a suitable catch-released strategy are important drawbacks to the generalization of these approaches. Consequently, the development of a direct and label-free method to indentify SBCs in mixture-based libraries would have notable advantages over the existing deconvolution protocols. In this context, a recent report10 pointed out the potential of surface-enhanced Raman scattering (SERS) microspectroscopy11–14 as a reliable and ultra-sensitive analytical tool for the direct detection of label-free SBCs.
With all these considerations in mind, we report herein: (a) the fabrication, and characterization of TentaGel–NH2 bimetallic silver/gold nanoparticle composites (TG–NP); (b) their application as substrates for the simultaneous solid phase synthesis of peptides; and, (c) the ultrafast surface-selective detection and classification of SBCs using SERS. Moreover, SERS offers not only the amino acid composition but also the primary and secondary structures of the peptidic compound. The first can be achieved by exploiting the decreasing of the SERS signal in relation to the increase of the distance between the target analyte and the optical enhancer (the bimetallic nanoparticle) surface.15 The second, by taking advantage of the sensitivity of Raman to the small changes in the conformation of macromolecules.16–18 Thus, the basis for the first implementation of a SERS-based strategy for the direct deconvolution of combinatorial libraries is presented.
Scheme 1 shows the experimental strategy followed for the material fabrication and spectroscopic classification processes. First, commercial TentaGel beads, functionalized with –NH2 coupling groups as the appropriate linkage for peptidic solid phase synthesis, were put in contact with a gold and silver ions solution. NaBH4 was then used as the reducing agent (Step i, Scheme 1) to yield a hybrid material composed of the common polyethylene glycol grafted polystyrene (PS-PEG) beads decorated with bimetallic Ag/Au clusters on the surface (Fig. 1A). A cross-sectional TEM image of the beads (Fig. 1B) shows a homogeneous distribution of these bimetallic nanoparticles of about 20 nm in size, into the PEG surrounding shell, but with no evidence of their presence in the PS matrix. The reason for using bimetallic nanoparticles stems from the fact that despite the negligible Ag affinity for PEG (when compared with gold, see Fig. S1, ESI†),19,20 its overall enhancing factor for SERS is three orders of magnitude larger than that for gold.21 Both features (PEG-gold affinity and silver enhancing factor for SERS) led us to use gold as the required bridge for further epitaxial deposition of silver22 onto the PS-PEG beads. High resolution X-ray photoelectron spectroscopy (HR-XPS, Fig. 1C) clearly shows peaks at 368.3 (Ag-3d5/2), 374.2 (Ag-3d3/2), 83.8 (Au-4f7/2) and 87.7 eV (Au-4f5/2) corresponding to the metallic Ag and Au, respectively. Quantitative analysis carried out on the XPS survey spectra (see Fig. S1, ESI†) shows an average elemental composition for the metallic nanoparticles of 82.5 and 17.5 atomic percentages in silver and gold, respectively. The Auger emission spectroscopy (AES) spectrum (Fig. 1D) and elemental mapping of silver and gold bands, Ag-MNN (351 eV) and Au-MNN (2015 eV), show a homogeneous distribution of the bimetallic clusters over the entire surface (Fig. 1E). Their crystalline nature was confirmed by X-ray diffraction (XRD, Fig. 1F) showing diffraction peaks for crystalline PEG (2q = 19.1, 23.2 and 26.4°)23 together with the (111), (200), (220), (311) and (420) characteristic Bragg reflections for a Ag/Au face-centered cubic lattice.24 The optical enhancing properties of the obtained hybrid material were demonstrated by using benzenethiol (BT) as SERS probe. The vibrational spectrum of the optically functionalized TG–NP beads which, as obtained, showed no Raman signals, was clearly revealed (Fig. 1G) after the addition of trace amounts of BT, offering therefore a clean substrate for SERS applications.
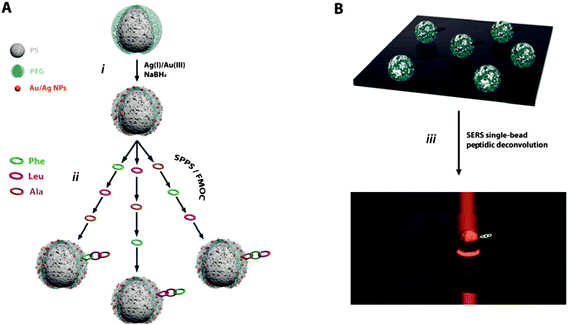 | ||
| Scheme 1 (A) Experimental strategy for the fabrication of the hybrid TG–NP bead material and its use as a support for solid-phase peptide synthesis. (B) Spectroscopy strategy for the on-bead peptidic deconvolution and classification using SERS. | ||
 | ||
| Fig. 1 (A) Optical image of the TentaGel–nanoparticle hybrid beads. (B) Cross-sectional TEM micrograph of the distribution of the bimetallic nanoparticles into the PEG shell of the TentaGel beads. (C) XPS and (D) AES spectra of the TG–NP. (E) SEM image and AES mapping for Au and Ag of the TG–NP bead surfaces. (F) XRD diffractogram of the beads. (G) Raman spectra of the hybrid beads as prepared and SERS spectra of benzenethiol after adding this molecule to the beads in minute amounts. | ||
To test the potential of SERS as a deconvolutive tool in combinatorial libraries, three different, randomly chosen, amino acids (Phe, Leu and Ala) were employed as building blocks in a set of tripeptides. In order to simplify the deconvolution protocol, only three of them, Phe-Ala-Leu, Leu-Phe-Ala, Ala-Leu-Phe, out of the 27 possible combinations, were considered for this study. Peptides were successfully synthesized on TG–NP using standard Fmoc-SPPS protocols (Step ii, Scheme 1A). Once prepared, aliquots of the three suspensions were combined and stirred to ensure a perfect blend and 20 mL of the resulting mixture were cast on a glass slide and air-dried. After that, the sample was mapped with a 633 nm laser line in a Raman microscope (Scheme 1B) avoiding photocombustion resulting from excitation with a green laser. Fig. 2A shows the SERS spectra for each of the three peptides. It is noteworthy that the data obtained by collecting only one spectrum for each bead are almost identical to those obtained by mapping the whole surface of the solid support using an identical excitation line and under the same conditions (Fig. 2B). These results demonstrate that the optical enhancement of the given peptide is very homogeneous and repetitive through the entire surface of the bead. Thus, a single point is enough for the identification and classification of each peptide, elucidated by taking advantage of the unique vibrational signature for each amino acid.25 A detailed assignment for each amino acid is shown in the ESI† (Table S1) and remarkably, most of the characteristic vibrations for three of them are present in the SERS spectra of the tripeptides considered (see the markers in Fig. 2A). The relative intensity of each vibration mode changes depending on the sequence of a given tripeptide, consequence of the SERS intensity dependence with the distance of the molecular group under consideration to the optical enhancer (the bimetallic nanoparticle).15 Thus, this dependence renders the elucidation of the amino acid sequence in the peptidic chain possible. Let us consider the characteristic ring breathing of Phe (1001 cm−1). Fig. 2A shows how its intensity decreases as a function of the position of this amino acid in the chain. When Phe is directly bonded to the bead, the ring breathing mode dominates the vibrational spectrum, but its intensity decreases as positioning the Phe far away from the bead (i.e. far away from the optical enhancer). The same effect was observed in all the characteristic vibrations for the three amino acids. In the case of Ala, its characteristic vibrations at 457 cm−1 (CC stretching and amide bending), 707 cm−1 (NH deformation) or 1393 cm−1 (Cα2H2 def./C–N stretch) are clearly observed if directly attached to the bead but decrease in intensity when attached in the second position, and even disappear if located at the end of the chain.
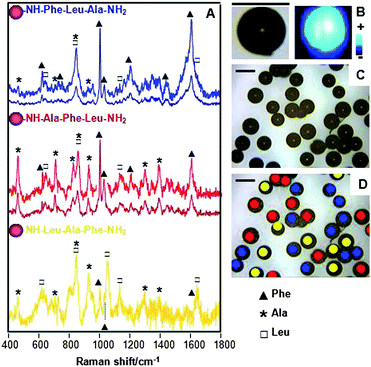 | ||
| Fig. 2 (A) SERS spectra of the prepared tripeptides on the solid phase support that acts, as well, as optical enhancer. Spectra of Phe-Leu-Ala and Ala-Phe-Leu have been as well enlarged to better observe the weaker vibrational modes. (B) Optical and SERS images of one single bead supporting the Phe-Leu-Ala peptide. (C) Optical image and (D) SERS mapping and label-free classification of a complex mixture of different tripeptides, all of them with the same composition but different sequence. SERS was collected by using a 633 nm laser line with acquisition times of 10 s per bead. | ||
Notably, the method presented here, and illustrated with a tripeptide, can be extrapolated to longer chains. Notwithstanding, and due to the dependence of the SERS intensity with the distance to the surface, the characterization of the amino acids in the chain is restricted to 7 to 10.
Conclusions
In summary, herein we present a direct label-free ultra-fast method for the identification and classification of the active members of a combinatorial library directly on their solid-phase support. We anticipate our results to be a starting point for the development of new strategies for direct high-throughput screening of large libraries with applications in drug discovery, pathogen detection and diagnosis, using the attached active molecule as a solid-support encoder.Acknowledgements
This work was conducted in part while R.A.A.-P. and R.P.-P. were at the National Institute for Nanotechnology and employed by the National Research Council and University of Alberta, respectively. Funding was provided in part by grants from the Genome Health Initiative and the National Institutes of Health held by Dr Hicham Fenniri. This work was funded by the Spanish Ministerio de Ciencia e Innovacion (MAT2008-05755) and the Xunta de Galicia (PGIDIT08TMT008314PR, INCITE09209101PR and 2010/78 Modalidade Emerxentes) and European Union's Seventh Framework Programme ([FP7/2008]) Metachem Project under grant agreement no. 228762. V. S. acknowledges the financial support from the Ramón y Cajal Program (Ministerio de Ciencia e Innovación, Spain).Notes and references
- C. Pinilla, J. R. Appel, E. Borras and R. A. Houghten, Nat. Med., 2003, 9, 118 CrossRef CAS.
- B. Yan, Curr. Opin. Chem. Biol., 2002, 6, 328 CrossRef CAS.
- S. V. Ley and I. R. Baxendale, Nat. Rev. Drug Discovery, 2002, 1, 573 CrossRef CAS.
- C. Pinilla, J. R. Appel, P. Blanc and R. A. Houghten, BioTechniques, 1992, 13, 901 CAS.
- K. S. Lam, S. E. Salmon, E. M. Hersh, V. J. Hruby, W. M. Kazmierski and R. J. Knapp, Nature, 1991, 354, 82 CrossRef CAS.
- M. H. J. Ohlmeyer, R. N. Swanson, L. W. Dillard, J. C. Reader, G. Asouline, R. Kobayashi, M. Wigler and W. C. Still, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 10922 CrossRef CAS.
- W. Bannwarth and B. Hinzen, Combinatorial Chemistry: from Theory to Application, Wiley-VCH, Weinheim, 2006 Search PubMed.
- C. Gibson, G. A. G. Sulyok, D. Hahn, S. L. Goodman, G. Hölzemann and H. Kessler, Angew. Chem., Int. Ed., 2001, 40, 165 CrossRef CAS.
- R. A. Simon, L. Schuresko, N. Dendukuri, E. Goers, B. Murphy and R. S. Lokey, J. Comb. Chem., 2005, 7, 697 CrossRef CAS.
- C. Schmuck, P. Wich, B. Kustner, W. Kiefer and S. Schlucker, Angew. Chem., Int. Ed., 2007, 46, 1 CrossRef.
- R. F. Aroca, Surface Enhanced Vibrational Spectroscopy, Wiley-VCH, New York, 2006 Search PubMed.
- M. Moskovits, Nature, 2010, 464, 357 CrossRef CAS.
- J. Kneipp, B. Wittig, H. Bohr and K. Kneipp, Theor. Chem. Acc., 2010, 125, 319–327 CrossRef CAS.
- R. A. Alvarez-Puebla and L. M. Liz-Marzan, Small, 2010, 6, 604–610 CrossRef CAS.
- G. J. Kovacs, R. O. Loutfy, P. S. Vincett, C. Jennings and R. Aroca, Langmuir, 1986, 2, 689 CrossRef CAS.
- R. A. Alvarez-Puebla, A. Agarwal, P. Manna, B. P. Khanal, P. Aldeanueva-Potel, E. Carbó-Argibay, N. Pazos-Pérez, L. Vigderman, E. R. Zubarev, N. A. Kotov and L. M. Liz-Marzán, Proc. Natl. Acad. Sci. U. S. A., 2011, 118, 8157–8161 CrossRef.
- M. Sanles-Sobrido, L. Rodriguez-Lorenzo, S. Lorenzo-Abalde, A. Gonzalez-Fernandez, M. A. Correa-Duarte, R. A. Alvarez-Puebla and L. M. Liz-Marzan, Nanoscale, 2009, 1, 153–158 RSC.
- L. L. He, T. Rodda, C. L. Haynes, T. Deschaines, T. Strother, F. Diez-Gonzalez and T. P. Labuza, Anal. Chem., 2011, 83, 1510–1513 CrossRef CAS.
- A. A. Farah, J. P. Bravo-Vasquez, R. A. Alvarez-Puebla, J. Y. Cho and H. Fenniri, Small, 2009, 5, 1283–1286 CrossRef CAS.
- M. Sanles-Sobrido, W. Exner, L. Rodriguez-Lorenzo, B. Rodriguez-Gonzalez, M. A. Correa-Duarte, R. A. Alvarez-Puebla and L. M. Liz-Marzan, J. Am. Chem. Soc., 2009, 131, 2699–2705 CrossRef CAS.
- F. J. García de Abajo, Rev. Mod. Phys., 2007, 79, 1267–1290 CrossRef.
- D. Tsoutsi, J. M. Montenegro, F. Dommershausen, U. Koert, L. M. Liz-Marzán, W. J. Parak and R. A. Alvarez-Puebla, ACS Nano, 2011, 5, 7539–7546 CrossRef CAS.
- X. Li, X. J. Loh, K. Wang, C. He and J. Li, Biomacromolecules, 2005, 6, 2740 CrossRef CAS.
- V. V. Agrawal, P. Mahalakshmi, G. U. Kulkarni and C. N. R. Rao, Langmuir, 2006, 22, 1846 CrossRef CAS.
- R. Tuma, J. Raman Spectrosc., 2005, 36, 307 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental materials and methods. XPS spectra and SERS assignments for Phe, Ala and Leu. See DOI: 10.1039/c1nr11293g |
| This journal is © The Royal Society of Chemistry 2012 |
