Metabonomic analysis of HIV-infected biofluids†
Lungile J. Sitole, Aurelia A. Williams and Debra Meyer*
Department of Biochemistry, University of Pretoria, Pretoria 0002, South Africa. E-mail: Debra.Meyer@up.ac.za
First published on 9th October 2012
Abstract
Monitoring the progression of HIV infection to full-blown acquired immune deficiency syndrome (AIDS) and assessing responses to treatment will benefit greatly from the identification of novel biological markers especially since existing clinical indicators of disease are not infallible. Nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry (MS) are powerful methodologies used in metabonomic analyses for an approximation of HIV-induced changes to the phenotype of an infected individual. Although early in its application to HIV/AIDS, (biofluid) metabonomics has already identified metabolic pathways influenced by both HIV and/or its treatment. To date, biofluid NMR and MS data show that the virus and highly active antiretroviral treatment (HAART) mainly influence carbohydrate and lipid metabolism, suggesting that infected individuals are susceptible to very specific metabolic complications. A number of well-defined biofluid metabonomic studies clearly distinguished HIV negative, positive and treatment experienced patient profiles from one another. While many of the virus or treatment affected metabolites have been identified, the metabonomics measurements were mostly qualitative. The identities of the molecules were not always validated neither were the statistical models used to distinguish between groups. Assigning particular metabolic changes to specific drug regimens using metabonomics also remains to be done. Studies exist where identified metabolites have been linked to various disease states suggesting great potential for the use of metabonomics in disease prognostics. This review therefore examines the field of metabonomics in the context of HIV/AIDS, comments on metabolites routinely detected as being affected by the pathogen or treatment, explains what existing data suggest and makes recommendations on future research.
Introduction
The hallmark of HIV infection is immunodeficiency. In addition, the lentivirus also causes metabolic changes in the host. The advent of a combination of three or four drugs referred to as HAART has shown success in decreasing the mortality rate and prolonging the life span and quality of life of HIV infected individuals.2,3 Long-term use of HAART is however associated with metabolic disorders such as diabetes4 atherosclerosis,5 lipodystrophy6 and cardiovascular disease.7 At present CD4 counts and viral load are the standard clinical parameters for guiding the management of HIV infection. CD4 counts are however subject to laboratory and physiological variation making it an inconsistent variable to use.8 In resource limited settings, viral load assays are either unavailable or excluded because of expense. These kinds of problems necessitate the search for additional tools for detecting and monitoring HIV and HAART-induced changes.Reliable biomarkers (indicators of normal biological processes, pathogenic processes, or responses to therapeutic interventions, fully defined in Kanekar et al.58) that can be linked to HIV infection and/or the use of HAART should provide further insight into mechanisms of HIV infection, disease progression, the response to therapy and overall management of the disease. Standard methodologies for diagnosis or prognosis of metabolic complications or for obtaining metabolic profiles include routine biochemical colorimetric assays. These are neither sensitive nor very specific, may be subject to interference and are focused on measuring one analyte at a time. Many times these methods are also really laborious to perform. Screening more than one molecule at a time using highly sensitive analytical instrumentation may be more advantageous for obtaining a holistic view of HIV and HAART induced metabolic changes. This can be achieved through the application of metabonomics, which is the study of metabolites (low molecular weight molecules detectable, primarily by NMR and MS and also Infra-Red and Raman spectroscopy) and how these molecules are altered by various stimuli. Although genomic and proteomic methods have been used to measure the responses of living systems to external factors,9 these approaches fail to provide phenotypic information which metabonomic analysis is able to do. The terminologies metabonomics and metabolomics are often used interchangeably because they differ more in practice than in definition. The goal of metabolomics is generally to characterize and quantify low molecular weight metabolites from cells, tissues and/or biofluids under specific conditions.3,10 Metabonomics on the other hand measures the global, dynamic changes in metabolic profiles of a biological system in response to disease, drugs, etc.3,10 For the purpose of this review, metabonomics is used to mean detecting metabolites or profiles affected by HIV/AIDS and/or its treatment.
This article first sets the stage by confirming metabolic changes as a consequence of HIV infection, virus induced immune deficiency and/or HAART with acknowledgement that the largest amount of existing knowledge about HIV/AIDS related metabolic abnormalities was collected by conventional biochemical methodologies. NMR and MS-based HIV/AIDS biofluid metabonomics is reviewed next, and the most prominent pathways affected by HIV (as deduced through metabonomic data) are presented along with suggestions for future research endeavours. Commentary is also provided on the link between metabolites and disease progression.
HIV-and HAART-associated metabolic changes (as detected by conventional methodologies)
Owing to HIV infection, immune responses to the virus and/or the administration of HAART, metabolic modifications are an expected outcome in the infected individual. Since more than one metabolic change can be induced by the aforementioned factors, the term metabolic syndrome has been coined to refer to several of these separate but interconnected changes.11–13 HIV-induced metabolic changes were recognized during the early stages of AIDS research, before the implementation of HAART and were shown to be prevalent in asymptomatic individuals with “normal” weight and CD4 counts.1,2,11,14 A key risk factor found to be associated with the development of the metabolic syndrome during HIV infection was viral load.11 In the studies of Hommes et al.1 as well as that of Lane and Provost-Craig15 clinically stable HIV positive individuals were shown, through calorimetric experiments, to have higher rates of resting energy expenditure. This is in keeping with the high energy demands of infected cells. These individuals also had high fat oxidation rates which led the authors to speculate that the greater amount of energy lost versus that taken in would make the infected individual prone to catabolic processes. Similar studies are referenced by Salas-Salvadó and García-Lorda16 and also reflect high resting energy expenditure. Subsequent to the work of Hommes et al.;1 Pascal et al.17 through the use of positron emission tomography and magnetic resonance imaging showed increased cerebral metabolic rates for glucose in the brains of asymptomatic HIV positive patients. By doing this study, the authors demonstrated metabolic alterations in the brain before structural changes became visible. Detections such as these provide metabolic information linked to patient health and could serve as a guide for the implementation of corrective therapy prior to the development of clinical symptoms. Key metabolic defects caused by HIV also include its ability to induce malnutrition,18 disrupt the functioning of the mitochondria,19,20 induce changes in body composition, fat distribution, changes in lipid, carbohydrate and protein metabolism.14,16 Changes in body composition are largely attributed to an increase in the catabolic state of the host.13 Other metabolic changes include; changes in calcium metabolism resulting in metabolic bone disease and drug toxicity resulting in liver disease. An article by Safrin and Grunfeld21 summarises and compares some basic HIV-induced metabolic changes which are a direct consequence of chronic HIV infection and the associated immune responses.11,17HAART comprises a combination of two nucleoside reverse transcriptase inhibitors (NRTIs) combined with either a non-nucleoside reverse transcriptase inhibitor (NNRTI), or a protease inhibitor (PI).22 Since the introduction of antiretroviral treatment (ART), many patients have reported adverse metabolic and anthropometric abnormalities; of which subcutaneous fat wasting, insulin resistance, hyperlipidemia and abdominal obesity have caused the most distress.4 The publications of several case reports over the years have described a clinical entity under the name lipodystrophy syndrome. Lipodystrophy is associated with an increase in free fatty acid content, triglyceride metabolism and low density lipoprotein (LDL) cholesterol and is subdivided/characterized under the terms lipoatrophy (fat wasting) and hyperlipidaemia (visceral fat accumulation). Lipoatrophy in particular is said to be caused by the apoptosis of peripheral adipocytes, which occurs secondary to mitochondrial toxicity and may also be due to the interference of two NRTIs [namely stavudine and azidothymidine] with mitochondrial deoxyribonucleic acid (mtDNA) polymerase-γ.23,24 Interestingly, an ACTG A5142 study found that a combination of stavudine or zidovudine with efavirenz presented the highest risk of lipoatrophy.25 These findings, amongst others, led to the removal of stavudine as a first-line therapy drug in developed countries.26
Mitochondrial disorders when induced by HIV infection are associated with insulin resistance (IR) and dyslipidemia. IR is among the first metabolic complications reported in HIV/HAART patients22,27 and is associated with inflammation and the development of type-2 diabetes mellitus.22,27 Several PI drugs have also been associated with IR, lipoatrophy as well as fat accumulation. In results produced by Carr et al.28 and Silva et al.,29 increased abdominal girth was observed in patients on PI therapy, while Pei-Ying Wu et al.30 showed that long-term use of PI-inhibitors was significantly associated with the presence of metabolic syndrome in a group of infected Taiwanese patients. The development of insulin resistance in healthy volunteers exposed to PIs (indinavir and amprenavir) has also been reported.31 The relationship between exact drug regimens and specific metabolic problems has been shown using conventional biochemical assays and has yet to be investigated and confirmed using metabonomic approaches.
HIV induced immune system disorders are mirrored in metabolic changes
Many diseases characterized by disturbances in metabolism (e.g. obesity) are associated with immune alterations (e.g. inflammation). On the other hand; toll like receptors, known to be the key regulators of the immune system (adaptive and innate) have been implicated in the regulation of bodily energy metabolism through acting on adipose tissue, and the energy needs of immune system cells are addressed by glucose and lipids as primary fuel sources.73 These are examples of an established link between the immune and metabolic systems, suggesting that a malfunction in one should be visible in the other.HIV infection is known for inducing immunodeficiency (CD4 cell depletion) and metabolic alterations (changes in nutrient requirements). Cytokines (immune system messengers) mediate several metabolic changes1,15,16 and induces a hypermetabolic state in the host. In the work of Cassol et al.,55 associations were made between several metabolites and inflammatory cytokines. Tumor necrosis factor (TNF) alpha is linked to changes in lipid metabolism. Williams 201260 investigated metabonomic and immune profiles of HIV positive individuals and cytokines detected in this study (IL-6, IL-10, and TNF-α) suggested increased catabolism and weight loss in HIV-infected patients. High IL-6 levels complemented the increase in measured triglycerides and glucose levels. Because there is an established link between the immune and metabolic systems73 the concurrent analysis of metabolic and immune changes as induced by the virus and/or its treatment has gained interest and should allow for a greater understanding of disease pathogenesis.
The majority of the body's energy is obtained from oxidation/catabolic processes. This occurs mainly through the breakdown of fats, carbohydrates and amino acids in mitochondria and peroxisomes, with organic acids forming as intermediates. The mitochondria serve as the metabolic hub providing an energy/adenosine triphosphate (ATP) source to cells. HIV-infected cells have been shown to have high resting energy expenditure and thus an increased demand for energy. During infection, components of the respiratory chain are impaired.63,64 Intermediates such as fumarate (salt/ester of fumaric acid) in the Krebs cycle are generally used by cells to produce ATP from food sources. When the metabolic hub, is affected (β)-oxidation of fatty acids cannot occur this then causes metabolites such as adipic acid, suberic acid and ethylmalonate to accumulate through alternative routes such as the omega (ω)-oxidation pathway. Elevated adipic and suberic acids also imply disrupted mitochondrial function. Impaired β-fatty acid oxidation contributes to ATP depletion and is compensated for by the alternative oxidative (ω) pathways and an increase in Krebs cycle intermediates such as fumarates. Reduced ATP production also triggers glycolysis,65 where glucose is converted to pyruvate and the released energy is used to form ATP. HIV infection as well as the therapeutics used to treat it disrupt glucose/carbohydrate metabolism.37 There exists an association between elevated urinary adipic and suberic acids as well as diabetes66 and glutaric aciduria type 1. Metabolic stress (induced by HIV) is also associated with a decrease in glucose levels.67 Elevated levels of adipic acid could be an indication of the development of a sugar disorder in infected patients.
Oxidative phosphorylation is a process which primarily takes place in mitochondria. During this process, carbon fuels are oxidized to yield energy with a subsequent transfer of electrons from nicotinamide adenine dinucleotide dehydrogenase (NADH) or flavin adenine dinucleotide dehydrogenase (FADH2) to oxygen (O2) which ultimately causes a proton gradient to develop and the phosphorylation of adenosine diphosphate (ADP) to ATP. When there is defective oxidative phosphorylation, as a compensatory mechanism, ATP is instead produced by the action of neurotransmitter molecules (such as tyramine),68,69 this coincides with the increased energy requirement of the infected cells. Tyramine is derived from the amino acid tyrosine and acts as a neurotransmitter being associated with a sudden rise in blood pressure.
Quinolinic acid is a tryptophan metabolite and was increased during SIV infection.54 This molecule is generally raised during chronic inflammation and neurodegeneration. It has been shown to be involved in neurodegenerative processes of the brain during AIDS, to induce lipid peroxidation, free radical production and cell death.70,71
HIV infection causes activation of the immune system and induces apoptosis. During the apoptotic process there is a subsequent rise in reactive oxygen species (ROS) which places the host under oxidative stress. ROS are involved in metabolic regulation72 and are of relevance since HIV makes use of the host biosynthetic machinery to survive. Oxidative stress is therefore involved in the pathogenesis of HIV infection. Pyroglutamic acid, a known marker of oxidative stress impairs brain energy production and contributes to the development of neuro-pathologies.29 HIV-associated neuro-complications usually present late during HIV infection. The detection of this molecule signals progression of HIV to AIDS.
Based on the detection of oxidative stress (OS) markers, it would seem that the antioxidant levels of HIV positive biofluids are lowered as the disease progresses. These data were collected using standard biochemical techniques rather than metabonomics methodology. Experiments to specifically measure antioxidant capacity of infected biofluids using metabonomics methodology and/or using existing antioxidant capacity detection kits to provide complementary data for the same sample, could in addition to expanding the existing knowledge base, also serve a validation purpose. Fig. 1 is a simplified scheme of how HIV, oxidative stress, the metabolic and immune systems may be linked.
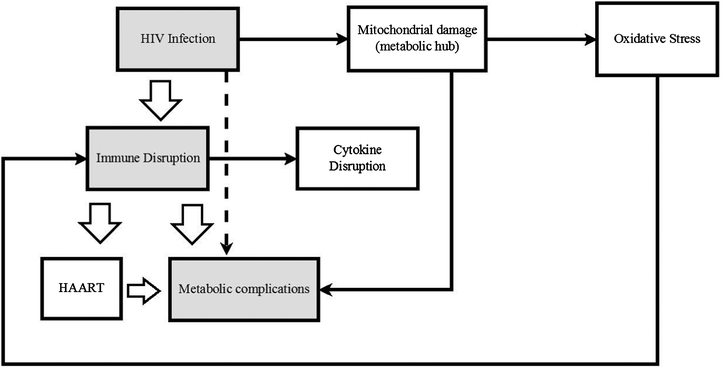 | ||
| Fig. 1 A schematic representation of how HIV infection triggers immune system disruption which then leads to metabolic complications is indicated. The associated changes triggered by the immune and metabolic systems or HIV directly are also presented. | ||
Why metabonomics for HIV infection?
HIV infection is characterized by four stages during which the infected individual presents different biochemical profiles. The changes that occur during these four stages of infection serve as an avenue for biomarker discovery. The virus also impacts on the metabolic signature of the host32 altering several metabolites which in turn is reflected in the affected biochemical pathways. Metabonomics has the ability to non-invasively detect all metabolites in HIV-infected cells, tissues (lymph) or biofluids (e.g. serum) making the unbiased identification of potential molecular markers for HIV possible. Research to date has focused on proteins as biomarkers of HIV infection (Kanekar et al.58) while the application of metabonomics has been geared towards measuring the influence of HAART in HIV infected individuals. Linking specific drug regimens to metabolic complications involving multiple metabolites and the quantification of HIV affected metabolites is largely un-investigated9 but could also be successfully explored through metabonomics because of the nature of the methodologies involved (NMR primarily detects multiple low molecular weight metabolites while MS detects all molecular weights present in a sample).The complexity of metabonomic data is interpreted through chemometrics (the statistical analysis of biological data). Pattern recognition statistics approaches are used to reduce the complex datasets obtained. This is achieved by identifying spectral differences and molecules which differ significantly between groups. Examples of statistical methods include but are not limited to; principle component analysis (PCA), linear discriminant analysis (LDA) and analysis of variance (ANOVA). Validation of these statistical approaches (in relation to HIV metabonomics) is not always done but is important for confirming the reliability and fit of a new external data set to the predicted model.
The choice of sample to use for metabonomic investigations is largely dependent on the biological question being addressed and the available instrumentation. Samples are also chosen according to their ease of access. Those that can be obtained non-invasively are usually the first choice and are most often chosen in such a way as to represent the in vivo state of the individual. Homogeneity of samples is also of paramount importance in human metabonomic studies. This is because; differences in diet, gender, lifestyle, age and genetic factors can influence the metabolome. It is therefore advisable to match (e.g. in terms of age, gender etc.) control samples to diseased samples and to analyse a large number of samples in order to detect biologically relevant sample clustering.
The biofluids mainly used for HIV metabonomics studies include; serum, whole blood, plasma, saliva, cells and urine. These samples are used because HIV infects blood-borne immune cells and the biofluids bathing these immune cells, contain metabolites which can reflect responses to the virus. To investigate the metabolic profile of these samples, various metabonomics approaches involving NMR and MS have been utilized e.g.1H-NMR, gas chromatography (GC)-MS and liquid chromatography (LC)-MS. Fig. 2 is representative of a typical metabonomics workflow and incorporates some of the common sample types, techniques, data processing and statistical methods that have been applied specifically to HIV-infected biofluids, also including relevant references. As the final step in the workflow, altered metabolites and the associated metabolic pathways that are affected are considered.
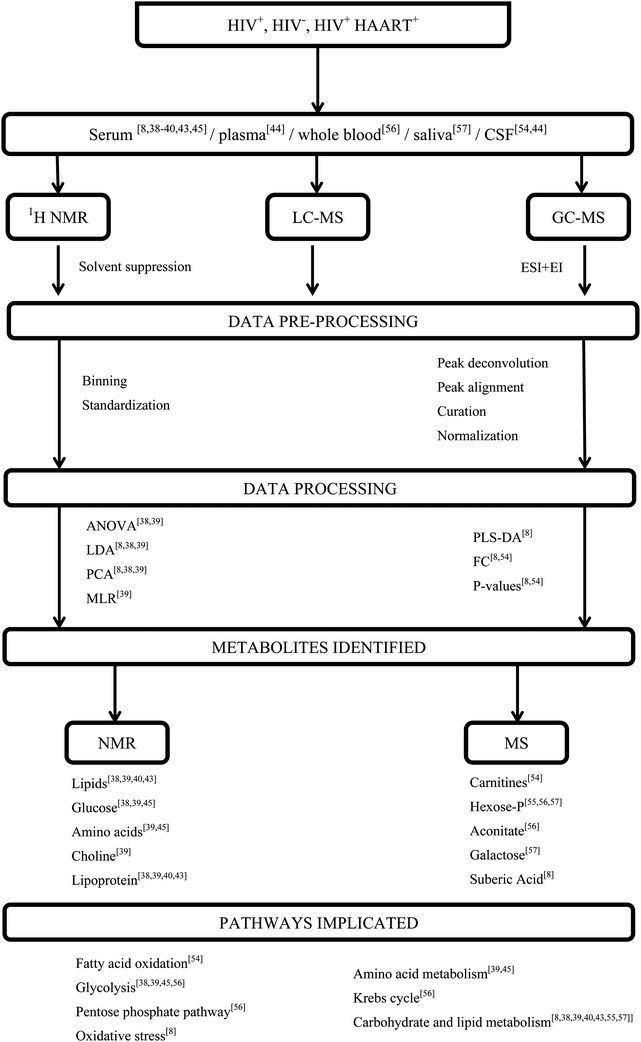 | ||
| Fig. 2 A schematic representation of metabonomics workflow applied to HIV infected biofluids. | ||
Both NMR and MS have distinct advantages and disadvantages. The major advantages of NMR include; its non-biased metabolite detection, quantitative nature, reproducibility and minimal sample preparation. The major disadvantage however, is its low sensitivity. In addition, NMR metabonomic analysis of biofluids requires sophisticated approaches for the suppression of the water resonance. MS on the other hand is highly sensitive, allows high separation efficiency, high spectral resolution and provides information that aids in the elucidation and identification of unknown compounds. In contrast, the major disadvantages of MS are sample destruction and multiple sample preparation steps. In relation to HIV/AIDS, the use of each technique separately or in combination allows an untargeted holistic study of the metabolites affected by the virus and/or its treatment, as opposed to classical biochemical methods that focus on predefined metabolites. As far as data detection is concerned, every peak on a mass spectrum represents a metabolite while a proton NMR spectrum presents many peaks per metabolite (dependent upon metabolite structure). There are several databases providing structural information of already detected and in some instances validated metabolites. These databases are more advanced for NMR analysis of biofluids as compared to MS. To date, only a few metabonomics investigations on biofluids infected with HIV/simian immunodeficiency virus (SIV) have been done. These studies are reviewed in the next section along with the most common metabolites and pathways affected. The basic trends of existing metabonomic analysis are summarised (in Tables 1 and 2) and presented with common chemometric choices and metabolites deemed to be affected. The tables are discussed (in detail) in the following sections.
| Approach (Untargeted/targeted) | Biofluid | ART+/ART− | Primary statistics used | Key metabolites affected | Key pathway(s) affected | Reference |
|---|---|---|---|---|---|---|
All samples were at the chronic phase of HIV infection. Solvent used in all the references is Deuterium oxide. Targeted metabonomics measures specific/known metabolites, while untargeted measures all detectable metabolites in the metabolome. CD4 count (cells per mm3) median: Hewer et al. (not listed), Philippeos et al. (HIV+ HAART: >200, HIV+ non-HAART: <200), Riddler et al. (HIV+ HAART: 507, HIV+ non-HAART: 527), Swanson et al. (not listed), Maher et al. (HIV+ non-HAART: <350), Duprez et al. (HIV+ HAART: >350, HIV+ non-HAART: <400). Viral load (copies per mL) median: Hewer et al. (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 > 25), Philippeos et al. (not listed), Riddler et al. (HIV+ HAART: <50, HIV+ non-HAART: 12 000 > 25), Philippeos et al. (not listed), Riddler et al. (HIV+ HAART: <50, HIV+ non-HAART: 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340), Swanson et al. (not listed), Maher et al. (<50), Duprez et al. (not listed). 340), Swanson et al. (not listed), Maher et al. (<50), Duprez et al. (not listed). | ||||||
| Untargeted | Serum | ART−/ART+ | ANOVA | Lipids, glucose, amino acids | Lipid metabolism, glycolysis | Hewer et al. (2006) |
| LDA | ||||||
| Untargeted | Serum | ART−/ART+ | ANOVA | Lipids, glucose, amino acids, choline | Lipid metabolism, glycolysis, amino acid metabolism | Philippeos et al. (2009) |
| LDA | ||||||
| MLR | ||||||
| Targeted | Serum | ART−/ART+ | Quantile regression | Lipids, proteins | Lipid metabolism, protein metabolism | Riddler et al. (2008) |
| Targeted | Serum | ART+ | One-way ANOVA | Lipids | Lipid metabolism | Swanson et al. (2009) |
| Pearson's correlation Chi-square test | ||||||
| Targeted | Serum | ART+ | OPLS | Valine, glutamine, lipids | Glycolysis, amino acid metabolism/synthesis | Maher et al. (2010) |
| SHY | ||||||
| Targeted | Blood plasma | ART+ | Conditional logistic regression | Lipids | Lipid metabolism | Duprez et al. (2009) |
| Technique (s) used | Ionization method (s) | Biofluid | ART+/ART− | Extraction method/solvent | Primary statistics used | Key metabolites affecteda | Key pathway(s) affecteda | Ref. |
|---|---|---|---|---|---|---|---|---|
a An extensive list of the metabolites and the affected pathways is not provided; please refer to the respective articles for a comprehensive view of the metabolic changes. All samples were at the chronic phase of HIV infection, except for Wikoff et al. 2008. All references used the untargeted approach except for Williams et al. 2011.b Abbreviations defined: fold change (FC), electron impact (EI), effect sizes (ES), 5-hydroxyeicosatetraenoic acid (5-HETE), 5-hydroxyeicosapentaenoic acid (5-HEPE), fructose-1,6-biphosphate (FBP), glycerol-3-phosphate (G3P). CD4 count (cells per mm3) median: Wikoff et al. (not listed), Cassol et al. (HIV+ non-HAART: <181), Hollenbaugh et al. (not listed), Ghannoum et al. (HIV+ HAART: >600–1000, HIV+ non-HAART: <5–1000), Williams et al. (HIV+ non-HAART: 342.20–376.45). Viral load (copies per mL) median: Wikoff et al. (4.5–8.5), Cassol et al. (not listed), Hollenbaugh et al. (not listed), Ghannoum et al. (HIV+ HAART: 40–80, HIV+ non-HAART: 1100–18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), Williams et al. (HIV+ non-HAART: 3 500), Williams et al. (HIV+ non-HAART: 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 81 81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 542 (77 542 (77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 641–9 641–9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 51 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 995) −72 995) −72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 740 (2328–1 740 (2328–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 78 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 260)). 260)). | ||||||||
| Capillary reverse phase LC-MS/MS TOF | ESI+ | CSF | ART− | Methanol | FC T tests/P values | ↑ Carnitines ↑ Acyl-carnitines ↑ Fatty acids ↑ Phospholipids | Fatty acid oxidation | Wikoff et al. 2008a |
| High performance LC and GC-MS | ESI+ and ESI− EI | Plasma | ART+ | No indication | Random forest analysis Hierarchical clustering FC T tests/P values | ↑ Pyruvate ↑ α-Ketoglutarate ↑ Malate ↑ Fumarate ↓ Tryptophan ↓ Asparatate ↓ Cysteine ↓ 5-HEPE ↓ 5-HETE ↓ Leukotriene B4 | Krebs cycle Carbohydrate Amino acid and Lipid metabolism | Cassol et al. 2011 (Poster) |
| CD4 cells | ||||||||
| LC-MS/MS | No indication | Blood-derived CD4 T cells Macrophage cell line | ART− | Methanol | FC T-tests/P values | ↑ Glucose uptake ↑ Aconitate ↑ Isocitrate ↑ Hexose-P ↑ FBP ↑ Ribose P | Carbohydrate metabolism Krebs cycle Pentose phosphate pathway | Hollenbaugh et al. 2011 |
| Macrophage cell line | ||||||||
| ↓ Glucose uptake ↑ Pyruvate ↑ Malate ↓ Hexose-P ↓ FBP ↓ G3P | ||||||||
| LC-MS/MS and GC-MS | ESI+ and ESI− EI | Mouth wash (saliva) | ART− and ART+ | Organic and aqueous extractions Acetonitrile | Nonparametric Wilcoxon rank sum test (p-value) Spearman rank correlation | ↑ Phenylalanine ↑ Tyramine ↑ Tryptophan ↓ Fucose ↓ Galactose ↓ Pyruvate | Amino acid metabolism Carbohydrate metabolism | Ghannoum et al. 2011 |
| GC-MS | Serum | ART− | Acetyl acetate and diethyl ether | PCA PLS-DA ES FC T tests/P values | ↑ Adipic acid ↑ Suberic acid ↑Arachidonic acid ↑ Stearic acid ↑ Pyroglutamic acid | Mitochondrial metabolism Lipid metabolism Oxidative stress | Williams et al. 2011a | |
NMR-based metabonomics of HIV-infected biofluid
NMR spectroscopy's routine application in the identification and quantification of chemical compounds is now being expanded upon by its use in metabonomics. Hewer et al. (2006) were the first to demonstrate that a distinction could be made between HIV-1 positive sera from patients on antiretroviral treatment (ART), HIV-1 positive sera naive of ART and HIV-1 negative sera based on the NMR metabolic profiles. These findings revealed significant dissimilarity (p < 0.01) in NMR spectral regions representing lipids, glucose and amino acids. Changes in lipids and glucose are known to be linked to the ART-associated disorders of lipodystrophy,6,28,33 hyperlipidaemia34,35 and hyperglycemia.36,37 Classification of the samples into three separate groups was therefore possible with a 95.2% correct classification using LDA.38 These results were later substantiated by Philippeos et al.,39 in a subsequent NMR metabonomics investigation using 300- and 600-(mega hertz) MHz NMR instruments and multinomial logistic regression (MLR) for data processing.39Chemical shifts corresponding to glucose, lipids and amino acids again showed significant differences (p < 0.05) when spectra of infected and uninfected individuals were compared. In addition, significant differences (p < 0.05) in glycerol and choline (which has a possible role in the AIDS dementia complex) were also detected. The authors concluded that the detection of choline required further investigation and verification by, among others, quantifying the choline concentrations and confirming related clinical evidence in the patients concerned.In one Multicenter AIDS Cohort study (N = 1072), Riddler and co-workers40 sought to measure the distribution of lipoprotein subclasses among three different groups of men along with examining the effects of antiretroviral therapy. HIV positive men on treatment showed increased very low density lipoprotein particles (VLDL-p) and (small) LDL-particles with lowered (large) LDL-p and high density lipoprotein particle (HDL-p) as compared to HIV negative men. HIV positive treatment-naive men were associated with lower concentrations of both the (small) and (large) LDL-p as well as lowered HDL-p concentrations.40 The authors' observations correspond to previously published work where the frequency of cardiovascular disease (CVD) in HIV infected patients on treatment appeared to be increased.41 Reasons for this increase in CVD could be the increased lipid serum levels brought on by HIV42 and/or its treatment.41 In a study to characterize the lipid profile of an HIV-infected group, Swanson et al.43 also found that protease inhibitor (PI)-containing treatment regimens had an association with greater (small) LDL-p concentration.43 This association contributes to the consideration of HIV-infected patients at risk of coronary heart disease (CHD). In contrast, Duprez et al.44 found that, neither VLDL-p nor LDL-p concentrations were associated with CVD.44 In general it's clear that HIV infection and subsequent treatment interfere with lipid metabolism to increase the possibility of cardiovascular complications for the patients. Although some drugs (PI) can be linked to CHD, more research is needed to determine the mechanism of action of other HIV treatments in relation to cardiovascular issues and other metabolic abnormalities.
The application of metabonomics to HIV-associated neurological complications has also gained interest.3 In their study, Maher et al. (2010) utilized NMR and MRS to identify metabolic correlations between HIV-infected blood plasma and CSF of the same individuals. Using both orthogonal projection to latent structures (OPLS) and statistical heterospectroscopy (SHY); the group observed high correlations for valine, glutamine, polyethylene glycol and lipids when comparing NMR data obtained from both blood plasma and CSF.45 These observations support the use of blood plasma instead of the more invasive CSF for predicting neurological changes related to HIV/AIDS, using metabonomic techniques. The integration of NMR blood plasma and MRS scans however, revealed no significant correlation.45
From the NMR-based investigations reported in Table 1, it can be deduced that the pathway most affected by HIV-infection and/or treatment involves lipid metabolism which explains the increased cases of lipodystrophy and dyslipidemia among HIV-infected individuals. The most used biofluid tends to be serum, probably given its easy means of collection and special relevance in all HIV research. From Table 1 it is also apparent that the most common statistical methods used for NMR metabonomic data interpretation are ANOVA, LDA and regression analysis. No references could be found for the quantitation of HIV influenced/affected metabolites. Recommendations on protocols and statistical considerations for quantitation of metabolites in biofluids or tissues (related to other diseases but applicable to infectious diseases) can be found in Serkova and Nieman61 where long relaxation times for example are considered paramount.
MS-based metabonomics of HIV-infected biofluid
Mass spectrometry is one of the most common techniques used for metabonomics investigations. In comparison to low molecular weight metabolites, proteins have been extensively investigated in the MS literature to investigate SIV and/or HIV's effect on the host proteome.46–50 Another area where the technology has been applied includes the investigation of antiretroviral drug concentrations in infected individuals after drug administration.51–53 Detecting and monitoring drug concentrations with MS allows for confirmation of compliance and yields information on the interactions of the various regimens within a system. MS has also been used to characterize virus and peptide structures as well as virus and protein interactions.49 In addition to the limited publications on HIV biofluid MS metabonomics, a few studies where this technique is applied to the study of HIV/AIDS but the work is not strictly metabolite analysis are referenced here to highlight the contribution of these studies in furthering our understanding of HIV infection and its treatment.Employing a global MS metabolomics approach; Wikoff et al.54 investigated the metabolic profile of CSF from SIV-infected monkeys before and after infection, with the aim of identifying biomarkers associated with neuroAIDS complications. The results indicated that the carnitines, acyl-carnitines, fatty acids and phospholipids were primarily elevated following SIV-induced encephalitis. The increase in fatty acids (e.g. palmitic acid) and lysophospholipids were associated with an increase in phospholipase activity and thus lipid breakdown processes. The authors also found that the detected metabolites had different structural and chemical characteristics. This led them to conclude that there was no single biochemical mechanism underlying the molecules' increase but that there was a biochemical relationship between these molecules through the fatty acid oxidation pathway.
The metabolites detected by Wikoff et al.54 include; stearic acid, vaccenic acid and arachidonic acid. These metabolites are constituents of phospholipids. Vaccenic acid is a structural component of the cardiolipins (bisphosphatidyl glycerol), which are important components of the inner mitochondrial membrane. Vaccenic acid increases in individuals with mental disorders and suggests neurological complications to be associated with HIV infection. Arachidonic acid is a membrane glycerophospholipid and serves as a substrate for phospholipase A2 (PLA2,62). Adequate levels of this metabolite are usually required for proper neurological function. A disruption in arachidonic acid metabolism is thus associated with neurological dysfunction.
A poster presentation by Cassol et al.55 reported on the metabolomic investigation of plasma from HIV-infected individuals receiving ART. In their study, the authors wanted to illustrate that “omics”-based technologies i.e. liquid and gas chromatography MS can inform on mechanisms regulating immune reconstitution and inflammation following ART administration. Changes in carbohydrate, lipid and amino acid metabolism were primarily detected. Krebs cycle intermediates were especially elevated. Several molecules having a role in neutrophil recruitment, natural killer cell activity and complement-mediated killing were decreased, suggesting a dysregulation in immune function. Using metabolomics the authors were able to demonstrate that multiple metabolic pathways were affected in HIV-infected individuals undergoing therapy. Insight into ART-induced metabolic abnormalities was therefore gained and associations between metabolites and immune molecules which regulate the immune response and inflammatory cytokine production revealed.
Given that some cells infected by HIV are long-lived (e.g. macrophage cell line) compared to others (primary CD4 cells); Hollenbaugh et al.56 employed a metabolomics approach using LC-MS/MS to show virus-induced metabolic changes in these cell types and that the detected metabolic profiles for the cell types differed. The primary CD4 T cells that had been infected with HIV in vitro, reflected an increase in glucose uptake and produced an increase in glycolytic metabolites whilst the chronically infected macrophage cell line demonstrated the opposite. The authors thus concluded a difference in energy requirements for these cell types. Generally, cell lines have a greater energy requirement than primary cells; however, the Hollenbaugh study56 demonstrated the opposite. A possible explanation is that the chronically infected macrophage cell line having been continuously passaged perhaps experienced a loss in cell viability or cell growth patterns over time. The cells harbouring HIV were depleted in number causing the virus' survival rate to decline. It is known that the virus makes use of the host machinery to produce new viral particles and sustains its survival. This host cell and virus decrease would imply that less energy was required for the survival of the pathogen. The cell line therefore took up less of the energy precursor namely; glucose and halted glycolytic processes. Metabolites of the pentose phosphate pathway, the Krebs cycle and several UDP-sugars were also affected as well as nucleoside triphosphate pools and oxidation phosphorylation ratios.
In addition to the metabolomic investigations performed on CSF and blood-derived products as described above, Ghannoum et al.57 showed metabolomics to be capable of characterizing the oral metabolome of treatment naive and treatment experienced HIV-infected patients. Mouth wash samples were collected, solvent extraction performed and the extracts analysed using both GC and LC-MS. The authors found a greater amount of metabolic alterations in treated compared to untreated patients. Metabolites having a role in amino acid metabolism were mostly elevated whilst metabolites linked to carbohydrate metabolism were mainly lowered. The elevated phenylalanine:tyrosine ratio in treatment naive patients was found by the authors to be potentially of use for monitoring immune status and therapy compliance.
Whilst the above-mentioned investigations were mainly untargeted (entire metabolome analysed), Williams et al.8 applied a targeted GC-MS metabonomics approach to characterize for the first time, organic acid changes in the serum of treatment naive patients. The organic acid metabolome was investigated because these metabolites are associated with mitochondrial dysfunction, a pathological consequence of HIV infection which was also confirmed through the detection of apoptosis8 and cytokine production.60 Organic acids having a role in mitochondrial metabolism, lipid metabolism and oxidative stress pathways were mostly elevated in infected patient sera.
From the MS-based investigations summarized in Table 2 it is evident that LC-MS is the technique of choice and that solvent extraction was primarily used for isolating the metabolites. The data in Table 2 indicate that chronic HIV infections were investigated more frequently than SIV animal models. There was also a tendency to work with biofluids which are less of an infection risk and can be obtained in a less invasive manner than serum/plasma for example; urine and mouth wash samples (see Table 2). To identify differences in metabolite levels between uninfected individuals, treatment naive and treatment experienced patients, an array of statistical methods was used of which the t-test and multivariate classification tools were most common (Table 2).
Similar to the NMR investigations summarized in Table 1, the MS investigations highlighted in Table 2 were relative rather than quantitative. Cells mainly experienced alterations in carbohydrate, lipid and amino acid metabolism. The findings that metabolites linked to carbohydrate and lipid metabolism were mainly affected (Tables 1 and 2) support the increased risk of diabetes mellitus, hyperlipidaemia and cardiovascular risk in HIV positive individuals whether treatment naive or treatment experienced. From Tables 1 and 2 it is clear that the virus affects metabolites which have roles in multiple pathways. This in turn explains the lack of identification of a single mechanism which induces metabolic change, an observation first made by Wikoff et al.54 Based on the pathways highlighted in the two Tables, it appears that the virus not only causes metabolic abnormalities as a consequence of infection but that it also induces metabolic change as a means to sustain its continued multiplication. This is supported by the fact that HIV is parasitic and dependent on the host infrastructure for energy and macromolecular precursors.32 Hollenbaugh et al.56 observed that as the metabolic requirements of the virus increased, so did the number of affected metabolic pathways, further supporting the lack of a single mechanism responsible for inducing metabolic change.
Impact of metabolites on disease progression
To comment on whether the metabonomics detected metabolites identified thus far, can be implicated in disease progression one must correlate the metabolites with existing markers of AIDS; CD4 and viral load. Not all studies reviewed here can be compared to the same extent in this regard, because some analyses were on biofluids while others used supernatant of cells infected in vitro or chronically. Furthermore some studies were on SIV biofluids rather than HIV. Most importantly, not all studies provided information on viral load and CD4 count and when provided these values varied extensively. The most complicated aspect was the fact that most studies reported multiple metabolites associated with either viral load and others with CD4 count or at times both. Because the study design and focus differed, the metabolites detected mostly differed. When looking at the studies on an individual basis, it appears that some metabolites can be associated with viral load and others with CD4 count. In their study, Riddler et al.40 observed that HIV positive men on HAART (with CD4 cell count below 200 cells per mm3) had the lowest levels of all lipoprotein particles, this possibly means that if viral load is controlled by treatment, amongst men on HAART with poor clinical status, the metabolic markers that are influenced are lipoproteins. The study by Williams et al.8 demonstrated through CD4 cell count and viral load that most patients in this study were not defined as having AIDS and were therefore relatively healthy, with CD4 cell counts of above 600 cells per mm.3 These authors therefore concluded that the majority of metabolites (organic acids) identified as being associated with HIV-induced mitochondrial damage was not associated with disease progression.The bigger problem for commenting on disease progression and associated metabolites is that the identified metabolites have not yet been quantified. Knowing metabolite levels (increase/decrease or disappearance) in the presence or absence of infection and correlating these to viral load and/or CD4 count will more specifically highlight the metabolite's importance. Pearson correlations (for example) between the metabolites, the viral load and CD4 counts will only be possible if quantitative information is available for all three variables.
Mass spectrometry data were able to identify metabolites more specifically than NMR, probably because of the nature of the analyses. NMR spectral regions can be linked to metabolites and pathways while MS can more easily identify metabolites specifically. The primary metabonomics methodologies (NMR and MS) did not detect exactly the same metabolites (so direct comparison across studies was not possible), possibly because of differences in experimental design and research approach. Also, the nature of NMR analysis means that there's overlap in some metabolite peaks, making it difficult to clearly distinguish certain metabolites (for example glutamine has resonance peaks near 2.14 ppm, 2.45 ppm and 3.78 ppm; the 3.78 ppm peak overlaps with peaks from both glutamate and glucose because of overlapping spectral regions). In NMR experiments, this is one of the areas where improvements in data collection (e.g. longer relaxation times) are under development and is also why metabolite validation is recommended. Quantitation of metabolites may also contribute in this instance, for clearly distinguishing different metabolites.
Concluding remarks
Global and targeted HIV/AIDS biofluid metabonomic studies using MS and/or NMR are possible and easily distinguish experimental groups (with the help of routine chemometric analysis). This distinction is based on multiple metabolites and metabolite groups rather than a single molecule. From the presented data it is clear that HIV mainly disrupts lipid and carbohydrate metabolism. This is not surprising since immune system cells depend on glucose as one of its primary fuel sources and HIV disrupts immune system function. There is also sufficient evidence suggesting HIV to be a stimulant of glycolysis. Alternatively, the activation of the glycolytic cycle could be a response mechanism by the cells to survive HIV infection. These opposing possibilities can be investigated through metabonomics approaches as has been done for cancer studies. Based on the review data in Tables 1 and 2, it is not likely that a single metabolite (occurring in single or multiple pathways) will be detected as being responsible for HIV/AIDS induced metabolic change. Instead, the finding of synergistic molecules is more probable. This is in agreement with data from several studies including but not limited to Wikoff,54 Cassol,55 Williams8 and Hollenbaugh56 who showed multiple metabolites to be affected by the virus and/or treatment and that of Lin et al.59 who suggested the use of multiple biomarkers to be more advantageous over a single marker because multiple markers could provide biological information on disease mechanisms. In many studies the identities of the molecules were not always validated (e.g. comparing spectral patterns in biofluids with those of standard solutions of commercially available samples of identified metabolites), neither were the statistical models used to distinguish between groups. Assigning particular metabolic changes to specific drug regimens using metabonomics also remains to be done. Homogeneity of samples for metabonomic experiments has to be maintained in order to rule out CD4 count, viral load, age, gender and drug regimens as contributing to the detection of metabolic anomalies. On the other hand, CD4 counts and viral load can be linked to specific metabolite groups/pathways which allows for conclusions on specific pathways that can be linked to disease progression. To make these conclusions of greater value, metabolite quantitation is a necessity. With enough preliminary and proof of concept studies published, the next step must be to quantify metabolites instead of referring to relative changes in concentration occurring during HIV infection, disease and treatment. Continuing research is also needed in the validation of the methodology used for specific metabolite detection and linking specific drug classes with particular metabolic complications.Acknowledgements
This work was supported by grants from the Technology Innovation Agency (TIA) of South Africa.References
- M. J. T. Hommes, J. A. Romijn, M. H. Godfried, J. K. Schattenkerk, W. A. Buurman, E. Endert and H. P. Sauerwein, Metab., Clin. Exp., 1990, 39, 1186–1190 Search PubMed.
- S. Gurunathan, R. El Habib, L. Baglyos, C. Meric, S. Plotkin, B. Dodet, L. Corey and J. Tarraglia, Vac. J., 2009, 27, 1997–2015 Search PubMed.
- G. Pendyala, E. J. Want, W. Webb, G. Siuzdak and H. S. Fox, J. Neuroimmune Pharmacol., 2007, 2, 72–80 Search PubMed.
- R. G. Jain, E. S. Furfine, L. Pedneault, A. L. White and J. M. Lenhard, Antiviral Res., 2001, 51, 151–177 CrossRef CAS.
- I. Schuster, G. J. Thoni, S. Edehys, G. Walther, S. Nottin, A. Vinet, F. Boccara, M. Khireddine, P. M. Girard and J. M. Mouboussin, Am. J. Cardiol, 2008, 101, 1213–1217 Search PubMed.
- F. Villarroya, P. Domingo and M. Viralt, Trends Pharmacol. Sci., 2004, 2, 88–93 Search PubMed.
- N. Friis-Moller, R. Weber, P. Reiss, R. Thiebaut, O. Kirk, A. D'Arminio Monforte, C. Pradier, L. Morfeldt, S. Mateu, M. Law, W. El-Sadr, S. De Wit, C. A. Sabin, A. N. Phillips and J. D. Lundgren, AIDS, 2003, 17, 1179–1193 Search PubMed.
- A. Williams, G. Koekemoer, Z. Lindeque, C. Reinecke and D. Meyer, Metabolomics, 2012, 8, 804–818 Search PubMed.
- G. Pendyala and H. S. Fox, Genome Med., 2010, 2, 22 Search PubMed.
- D. S. Wishart, D. Tzur, C. Knox, R. Eisner, A. C. Guo, N. Young, D. Cheng, K. Jewell, D. Arndt, S. Sawhney, C. Fung, L. Nikolai, M. Lewis, M. A. Coutouly, I. Forsythe, P. Tang, S. Shrivastava, K. Jeroncic, P. Stothard, G. Amegbey, D. Block, D. D. Hau, J. Wagner, J. Miniaci, M. Clements, M. Gebremedhin, N. Guo, Y. Zhang, G. E. Duggan, G. D. Macinnis, A. M. Weljie, R. Dowlatabadi, F. Bamforth, D. Clive, R. Greiner, L. Li, T. Marrie, B. D. Sykes, H. J. Vogel and L. Querengesser, Nucleic Acids Res., 2007, 35, D521–6 CrossRef CAS.
- L. Slama, C. Le Camus, L. Serfaty, G. Pialoux, J. Capeau and S. Gharakhanian, Diabetes Metab., 2009, 35, 1–11 Search PubMed.
- K. G. Alberti, P. Zimmet and J. Shaw, Diabetic Med., 2006, 23, 469–480 Search PubMed.
- W. Powderly, AIDS Patient Care STDS, 2004, 18, 431–435 Search PubMed.
- A. Martin and S. Emery, Expert Rev. Clin. Pharmacol., 2009, 2, 381–390 Search PubMed.
- B. J. Lane and M. Provost-Craig, J. Women's Health Gender Based Med., 2000, 9, 321–327 Search PubMed.
- J. Salas-Salvadó and P. García-Lorda, Clin. Nutr., 2001, 20, 379–391 Search PubMed.
- S. Pascal, L. Resnick, W. W. Barker, D. Loewenstein, F. Yoshii, J. Chang, T. Boothe, J. Sheldon and R. Duara, J. Nucl. Med., 1991, 32, 1725–1729 Search PubMed.
- Z. Hattingh, C. Walsh, F. J. Veldman and C. J. Bester, S. Afr. J. Clin. Nutr., 2009, 22, 23–28 Search PubMed.
- R. Polo, S. Martinez, P. Madrigal and M. Gonzalez-Munoz, J. Acquired Immune Defic. Syndr., 2003, 34, 32–36 Search PubMed.
- G. Garrabou, S. Lopez, C. Moren, E. Martinez, J. Fontdevila, F. Cardellach, J. M. Gatell and O. Miro, AIDS, 2011, 5, 165–170 Search PubMed.
- S. Safrin and C. Grunfeld, AIDS, 1999, 13, 2493–2505 Search PubMed.
- E. R. Feeney and P. W. G. Mallon, Best Pract. Res., Clin. Endocrinol. J., 2011, 25, 443–458 Search PubMed.
- J. Falutz, Curr. Opin. Clin. Nutr. Metab. Care, 2011, 14, 255–260 Search PubMed.
- G. McComsey, AIDS Res. Rev., 2002, 4, 140–147 Search PubMed.
- R. H. Haubrich, S. A. Riddler, A. G. DiRienzo, L. Komarow, W. G. Powderly, K. Klingman, K. W. Garren, D. L. Butcher, J. F. Rooney, D. W. Haas, J. W. Mellors and D. V. Havlir, AIDS, 2009, 23, 1109–1118 Search PubMed.
- M. Caron-Debarle, C. Lagathu, C. Boccara, C. Vigoroux and J. Capeau, Trends Mol. Med., 2010, 16, 218–229 Search PubMed.
- H. J. M. Hofstede, D. M. Burger and P. P. Koopmans, Neth. J. Med., 2003, 12, 393–403 Search PubMed.
- A. Carr, K. Samaras, S. Burton, M. Law, J. Freund, D. J. Chrisholm and D. A. Cooper, AIDS, 1998, 12, F51–F58 Search PubMed.
- M. Silva, P. R. Skolnik, S. L. Gorbach, D. Spiegelman, I. B. Wilson, M. G. Fernandez-DiFranco and T. A. Knox, AIDS, 1998, 12, 1645–1651 Search PubMed.
- P. Y. Wu, C. C. Hung, W. C. Liu, C. Y. Hsieh, H. Y. Sun, C. L. Lu, H. Wu and K. L. Chien, J. Antimicrob. Chemother., 2012, 64, 1001–1009 Search PubMed.
- G. A. Lee, J. M. Schwarz, S. Patzek, S. Kim, A. Dyachenko, M. Wen, K. Mulligan, M. Schambelan and C. Grunfeld, Antiviral Ther., 2007, 12, L46 Search PubMed.
- J. Munger, S. U. Bajad, H. A. Coller, T. Shenk and J. D. Rabinowitz, PLoS Pathog., 2006, 2, e132 Search PubMed.
- S. Grinspoon and K. Mulligan, Clin. Infect. Dis., 2003, 36, S69–S75 Search PubMed.
- K. L. Dong, L. L. Bausserman, M. M. Flynn, B. P. Dickinson, T. P. Flanigan, M. D. Mileno, K. T. Tashima and C. C. Carpenter, J. Acquired Immune Defic. Syndr., 1999, 21, 107–113 Search PubMed.
- K. Mulligan, C. Grunfeld, V. W. Tai, H. Algran, M. Pang, D. N. Chernoff, J. C. Lo and M. Schambelan, J. Acquired Immune Defic. Syndr., 2000, 23, 35–43 Search PubMed.
- S. H. Mehta, R. D. Moore, D. L. Thomas, R. E. Chaisson and M. S. Sulkowski, J. Acquired Immune Defic. Syndr., 2003, 33, 577–584 Search PubMed.
- M. P. Dube, D. L. Johnson, J. S. Currier and J. M. Leedom, Lancet, 1997, 350, 713–714 Search PubMed.
- R. Hewer, J. Vorster, F. E. Steffens and D. Meyer, J. Pharm. Biomed. Anal., 2006, 41, 1442–1446 CrossRef CAS.
- C. Philippeos, F. E. Steffens and D. Meyer, J. Biomol. NMR, 2009, 44, 127–137 Search PubMed.
- S. A. Riddler, Li. Xiuhong, J. Otvos, W. Post, F. Palella, L. Kingsley, B. Visscher, L. P. Jacobson and A. R. Sharrett, J. Acquired Immune Defic. Syndr., 2008, 48, 281–288 Search PubMed.
- J. S. Currier, J. D. Lundgren, A. Carr, D. Klein, C. A. Sabin, P. E. Sax, J. T. Schouten and M. Smieja, Circulation, 2008, 118, e29–e35 Search PubMed.
- M. Floris-Moore, A. A. Howard, Y. Lo, J. H. Arnsten, N. Santoro and E. E. Schoenbaum, HIV Med., 2006, 7, 421–430 Search PubMed.
- B. Swanson, B. E. Sha, J. K. Keithley, L. Fogg, J. Nerad, R. Novak and O. Adeyemi, J. Clin. Lipidol., 2009, 6, 379–384 Search PubMed.
- D. A. Duprez, L. H. Kuller, R. Tracy, J. Otvos, D. Cooper, J. Hoy, J. Neuhaus, N. Paton, N. Friis-Moller, F. Lampe, A. P. Liappis and J. D. Neaton, Atherosclerosis, 2009, 207(2), 524 Search PubMed.
- D. Maher, L. A. Cysique, B. J. Brew and C. D. Rae, J. Proteome Res., 2011, 10, 1737–1745 Search PubMed.
- J. R. Berger, M. Avison, Y. Mootoor and C. Beach, J. Neurovirol., 2005, 11, 557–562 Search PubMed.
- J. P. Laspiur, E. R. Anderson, P. Ciborowski, V. Wojna, W. Rozek, F. Duan, R. Mayo, E. Rodríguez, M. Plaud-Valentín, J. Rodríguez-Orengo, H. E. Gendelman and L. M. Meléndez, J. Neuroimmunol., 2007, 192, 157–170 Search PubMed.
- G. Pendyala, S. A. Trauger, E. Kalisiak, R. J. Ellis, G. Siuzdak and H. S. Fox, J. Proteome Res., 2009, 8, 2253–2260 Search PubMed.
- L. Zhang, X. Jia, X. Zhang, J. Sun, X. Peng, T. Qi, F. Ma, L. Yin, Y. Yao, C. Qiu and H. Lu, Proteome Sci., 2010, 8, 12 Search PubMed.
- G. Pendyala, S. A. Trauger, G. Siuzdak and H. S. Fox, AIDS Res. Hum. Retroviruses, 2011, 27, 179–182 Search PubMed.
- J. D. Moore, G. Valette, A. Darque, X. J. Zhou and J. P. Sommadossi, J. Am. Soc. Mass Spectrom., 2000, 11, 1134–1143 CrossRef CAS.
- A. Volosov, C. Alexander, L. Ting and S. J. Soldin, Clin. Biochem., 2002, 35, 99–103 CrossRef CAS.
- T. Koal, H. Burhenne, R. Romling, M. Svoboda, K. Resch and V. Kaever, Rapid Commun. Mass Spectrom., 2005, 19, 2995–3001 CrossRef CAS.
- W. R. Wikoff, G. Pendyala, G. Siuzdak and H. S. Fox, J. Clin. Invest., 2008, 118, 2661–2669 CAS.
- E. Cassol, V. Misra, A. Holman, A. Kamat and D. Gabuzda, 18th Conference on Retroviruses and Opportunistic Infections, 2011, Paper # 287 Search PubMed.
- J. A. Hollenbaugh, J. Munger and B. Kim, Virology, 2011, 415, 153–159 Search PubMed.
- M. A. Ghannoum, P. K. Mukherjee, R. J. Jurevic, M. Retuerto, R. E. Brown, M. Sikaroodi, J. Webster-Cyriaque and P. M. Gillevet, OMICS, 2012 DOI:10.1089/omi.2011.0035 [Epub ahead of print].
- A. Kanekar, J. Clin. Med. Res., 2010, 2, 55–61 Search PubMed.
- L. Lin, Z. Huang, Y. Gao, X. Yan, J. Xing and W. Hang, J. Proteome Res., 2011, 10(3), 1396–1405 CrossRef CAS.
- A. A. Williams, PhD thesis, University of Pretoria, Pretoria, 2012, http://upetd.up.ac.za/thesis/available/etd-05302012-202713/ Search PubMed.
- N. J. Serkova and C. U. Niemann, Expert Rev. Mol. Diagn., 2006, 5, 717–731 Search PubMed.
- P. A. Sandstrom, P. W. Tebbey, S. Van Cleave and T. M. Buttke, J. Biol. Chem., 1994, 269, 798–801 Search PubMed.
- J. S. Ladha, M. K. Tripathy and D. Mitra, Cell Death Differ., 2005, 12, 1417–1428 Search PubMed.
- J. E. Ricci, C. Munoz-Pinedo and P. Fitzgerald, Cell, 2004, 117, 773–786 CrossRef CAS.
- G. Hofhaus, D. R. Johns, O. Hurko, G. Attardi and A. Chomyn, J. Biol. Chem., 1996, 271, 13155–13161 Search PubMed.
- T. Niwa, K. Meada, T. Ohki, A. Saito and I. Tsuchida, J. Chromatogr., 1981, 225, 1–8 Search PubMed.
- A. Schulze-Bergkamen, J. G. Okun, U. Spiekerkotter, M. Lindner, D. Haas, D. Kohlmuller, E. Mayatepek, H. Schulze-Bergkamen, C. R. Greenberg, J. Zschocke, G. F. Hoffmann and S. Kolker, Pediatr. Res., 2005, 58, 873–880 Search PubMed.
- C. Reinecke, G. Koekemoer, d. W. van, R. Louw, J. Lindeque, L. Mienie and I. Smuts, Metabolomics, 2011, 1–20.
- B. Korzeniewski, Biochim. Biophys. Acta, 2001, 1504, 31–45 CAS.
- G. Guillemin, L. Wang and B. Brew, J. Neuroinflammation, 2005, 2, 16 Search PubMed.
- C. A. Wiley, J. NeuroVirol., 1995, 1, 328–29 Search PubMed.
- E. Peterhans, J. Nutr., 1997, 127, 962S–965S Search PubMed.
- I. Wolowczuk, C. Verwaerde, O. Viltart, A. Delanoye, M. Delacre, B. Pot and C. Grangette, J. Clin. Dev. Immunol., 2008, 2008, 63908 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c2mb25318f |
| This journal is © The Royal Society of Chemistry 2013 |
