Intrinsically disordered regions as affinity tuners in protein–DNA interactions†
Dana
Vuzman
and
Yaakov
Levy
*
Department of Structural Biology, Weizmann Institute of Science, Rehovot, 76100, Israel. E-mail: koby.levy@weizmann.ac.il
First published on 15th September 2011
Abstract
Intrinsically disordered regions, terminal tails, and flexible linkers are abundant in DNA-binding proteins and play a crucial role by increasing the affinity and specificity of DNA binding. Disordered tails often undergo a disorder-to-order transition during interactions with DNA and improve both the kinetics and thermodynamics of specific DNA binding. The DNA search by proteins that interact nonspecifically with DNA can be supported by disordered tails as well. The disordered tail may increase the overall protein–DNA interface and thus increase the affinity of the protein to the DNA and its sliding propensity while slowing linear diffusion. The exact effect of the disordered tails on the sliding rate depends on the degree of positive charge clustering, as has been shown for homeodomains and p53 transcription factors. The disordered tails, which may be viewed as DNA recognizing subdomains, can facilitate intersegment transfer events that occur via a “monkey bar” mechanism in which the domains bridge two different DNA fragments simultaneously. The “monkey bar” mechanism can be facilitated by internal disordered linkers in multidomain proteins that mediate the cross-talks between the constituent domains and especially their brachiation dynamics and thus their overall capability to search DNA efficiently. The residue sequence of the disordered tails has unique characteristics that were evolutionarily selected to achieve the optimized function that is unique to each protein. Perturbation of the electrostatic characteristics of the disordered tails by post-translational modifications, such as acetylation and phosphorylation, may affect protein affinity to DNA and therefore can serve to regulate DNA recognition. Modifying the disordered protein tails or the flexibility of the inter-domain linkers of multidomain proteins may affect the cross-talk between the constituent domains so as to facilitate the search kinetics of non-specific DNA sequences and increase affinity to the specific sequences.
 Dana Vuzman | Dana Vuzman received her BSc in the field of Chemistry-Biology from the Tel-Aviv University and her MSc in the field of Organometallic Chemistry from the Weizmann Institute of Science where she worked on Platinum pincer complexes under the supervision of Prof. David Milstein. She started her PhD in 2008 in the Weizmann Institute of Science under the supervision of Dr Yaakov Levy. Her current research concentrates on the characteristics and effects of disordered regions of proteins on DNA search. |
 Yaakov Levy | Yaakov Levy received his PhD in chemical physics from Tel-Aviv University, Israel, in 2002, where he worked with Profs. Joshua Jortner and Oren Becker on energy landscape characterization of polypeptides. He then joined the labs of Profs. José Onuchic and Peter Wolynes at the University of California at San Diego and the Center for Theoretical Biological Physics. In his postdoctoral studies, he focused on understanding the first principles of biological self-organization processes using minimalist models. In 2006, he joined the Dept. of Structural Biology at the Weizmann Institute of Science where he studies various problems in molecular biophysics using theoretical and computational tools. In particular, his group is focused on the effect on post-translational modifications on the biophysics of protein folding and the molecular determinants that control the speed and efficiency of DNA search. |
Introduction
The conventional structure–function paradigm assumes that function significantly depends on a well characterized 3D structure. However, in recent years it has become clear that intrinsically disordered protein segments or even whole proteins can perform key roles in the cell1–3 generally associated with regulatory functions.1–12 Disordered proteins are abundant in a range of cells and their frequency rises as the complexity of the organism increases.13,14 Unlike globular proteins, disordered segments are rich in charged amino acids, deficient in hydrophobic residues, and have a low degree of complexity.15,16 Disordered regions can adopt collapsed structures17–19 whose degree of compaction can be modulated by charge interactions.18,20–24 Not much is known about the sequence–structure–function relationship in disordered regions, although it is recognized as essential with respect to structured proteins. To overcome this knowledge gap, studies must be undertaken of the sequence properties and evolutionary conservation of the structure and function in intrinsically disordered proteins.25,26The activities of disordered regions are frequently related to molecular recognition,27 which is often coupled with disorder-to-order transitions.28 However, disordered proteins may also retain their flexibility, and even remain partly disordered, in the bound state.29 In general, the binding process is rapid due to the large capture radius of disordered regions, which supports the fly-casting mechanism.30–32 The faster binding may come at the expense of slower folding kinetics33 (Fig. 1A). The interactions of disordered regions may be realized via multiple segments34,35 that have a relatively low overall DNA affinity. Protein–protein interactions involving disordered regions are described by several related recognition models.36–39 Intrinsically unstructured regions assist protein-–DNA interactions in various mechanisms,40 yet the detailed biophysical aspects of disordered regions in protein–DNA recognition are less well understood than in protein–protein recognition.4,39 Disorder-to-order transitions improve the specificity of DNA binding41 by a mechanism that remains to be elucidated. A number of studies have demonstrated that disordered tails can contribute to selective sequence binding specificity,42–44 but there are only a few hypotheses for the target selection mechanism.45–48 Although it is common to focus on the globular part of DNA-binding proteins to evaluate affinity and selectivity to DNA, it is becoming apparent that the disordered regions (as tails at the termini or as internal regions) modulate several essential features in protein–DNA affinity.40,45
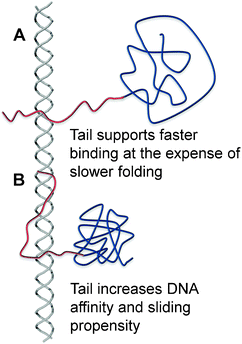 | ||
| Fig. 1 A schematic illustration of the effect of the disordered tail on the interplay between folding and binding. (A) The disordered tail promotes fast binding to DNAvia the fly-casting mechanism but slows protein folding. Protein folding will take place after the protein is at least partially bound to the DNA.33 (B) The folded protein has a greater affinity to the DNA because its interface is larger as a result of the extended conformation of the positively charged disordered tail when it interacts with DNA. The tighter binding to the DNA may result in a larger sliding propensity.64 | ||
Transcription factors recognize specific DNA sequences (typically 6–12 base pairs) and their binding to the target sites is necessary for gene activation or repression at the gene-regulation level.49 Although transcription factors must find their target DNA sites efficiently and rapidly to ensure a quick response to cellular stimuli, the target search can be a time-consuming process.50–54 Following the studies of Berg and von Hippel,54,55 it is widely accepted that the target search process on DNA is facilitated by the combination of a 3D diffusion and other search modes performed in a lower dimensional space:51–67 sliding, hopping, and intersegment transfer (jumping). The disordered tail of a DNA binding protein can be viewed as another subdomain that interacts with the DNA and increases the affinity to DNA (Fig. 1B). Although many eukaryotic gene-regulatory proteins contain multiple DNA-binding domains tethered by a disordered linker for higher specificity and affinity,68 the presence of multiple DNA-binding domains can also slow down translocation of the proteins on DNA because the protein molecules have to transiently break all interactions with the DNA molecule whenever the molecules move from one DNA site to another.69
In this review, we focus on the effect of disordered regions on nonspecific protein–DNA interactions, and particularly their role in DNA recognition, with the aim of understanding how disordered regions contribute to the specificity and affinity of DNA binding in general and to sliding and intersegment transfer in particular. We describe the molecular details of specific binding, 1D diffusion by sliding and hopping, the microscopic mechanism of and criteria for intersegment transfer, and test the effect that proteins with disordered tails and multiple domains tethered via a disordered linker may have on search efficiency. In addition, we describe the role of disordered tails in gene regulation viapost-translational modifications.
Results and discussion
The role of the disordered tail in DNA search
We have previously shown that disordered tails comprise ∼70% of human DNA-binding proteins and only ∼50% of non-DNA-binding proteins (Fig. 2A and B).70,71 The occurrence of a single tail (at either the N- or C-terminus) or two tails (at both termini) is higher in DNA-binding proteins than in other proteins in the human genome (Fig. 2A). Furthermore, the tails of DNA-binding proteins are longer than those of other proteins (∼60% of the tails of DNA-binding proteins are longer than 5 residues while only 30% of non-DNA-binding proteins have tails longer than 5 residues, Fig. 2B). The disordered tails of DNA-binding proteins have a larger net positive charge and these charges tend to be much more clustered in the tails of DNA-binding proteins than in the tails of non-DNA binding proteins. We note that the composition of positively charged residues and their degree of clustering in the structured parts of DNA-binding proteins and non-DNA binding proteins are very similar (Fig. 2C and D).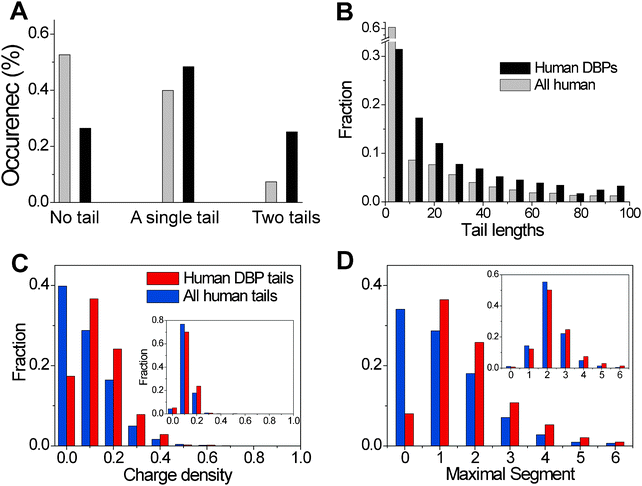 | ||
Fig. 2 Bioinformatic analysis of the properties of the disordered tails of DNA-binding proteins in the human genome. (A) The occurrence in the human genome (which totals 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 334 proteins) of DNA-binding proteins (DBP, a total of 864 proteins) with no tail, a single tail (either at the C- or N-termini), and two tails. (A tail is defined as a disordered region predicted by IUPred.) For comparison, a similar analysis was performed for all proteins in the human genome. (B) Histograms of the tail lengths in human DBPs and all human proteins. (C) Distributions of the positive charge density in the tails of DBPs and (D) distributions of the maximal absolute charged segment of positively charged residues in DBPs. The insets in (C) and (D) refer to the corresponding distributions in the globular regions of the proteins. 334 proteins) of DNA-binding proteins (DBP, a total of 864 proteins) with no tail, a single tail (either at the C- or N-termini), and two tails. (A tail is defined as a disordered region predicted by IUPred.) For comparison, a similar analysis was performed for all proteins in the human genome. (B) Histograms of the tail lengths in human DBPs and all human proteins. (C) Distributions of the positive charge density in the tails of DBPs and (D) distributions of the maximal absolute charged segment of positively charged residues in DBPs. The insets in (C) and (D) refer to the corresponding distributions in the globular regions of the proteins. | ||
The importance of the N-terminal disordered tail for protein folding and for specific or non-specific interactions with DNA was studied recently for several homeodomain transcription factors, both experimentally and computationally. These disordered tails, which are positively charged and highly disordered in solution, can cause thermodynamic destabilization or stabilization of the protein in the absence or presence of DNA, respectively.33,45,72 In the absence of DNA, the tail containing the larger fraction of charged residues may interact with other charged residues and perturb folding by preventing native contacts in the core of the protein. In the presence of DNA, the tail serves to anchor the protein to DNA with electrostatic forces that modulate the folding of the homeodomain and, in particular, stabilize the folded state. The tails of the homeodomain may fold upon binding to the DNA in the minor groove or remain partly flexible or disordered.42
A recent study based on coarse-grained molecular dynamic simulations of three homeodomain proteins with different tail lengths and net charges (Table 1) in which the interactions between proteins and DNA are governed solely by electrostatic forces demonstrated the role of the disordered tail in facilitating DNA search,64 in agreement with kinetic NMR studies.62,73,74 The presence of an N-tail increases the affinity of the protein to the DNA, and therefore enhances its sliding propensity at the expense of hopping and 3D diffusion (Fig. 3A). However, better sliding has its price: the linear diffusion coefficient of the protein moving along the DNA is lower, which results in a slower search (Fig. 3B).64 The increased use of the sliding search mode accompanied by the decreased use of linear diffusion in the presence of an N-tail is more pronounced for the Antp than for the HoxD9 homeodomain, which is correlated with the existence of a longer tail and a higher net positive charge for Antp than HoxD9 (Table 1 and Fig. 3A and B).
| Tail length | Net charge | Sequence of the disordered tail | |
|---|---|---|---|
 —acetylation. —acetylation.  —phosphorylation. —phosphorylation. |
|||
| HoxD9 | 9 | +2 |
|
| Antp | 10 | +4 |
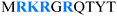
|
| NK-2 | 17 | +5 |

|
| p53 | 31 | +5 |

|
| PTMs | −2 | ||
| H3 | 40 | +13 |

|
| PTMs | +5 | ||
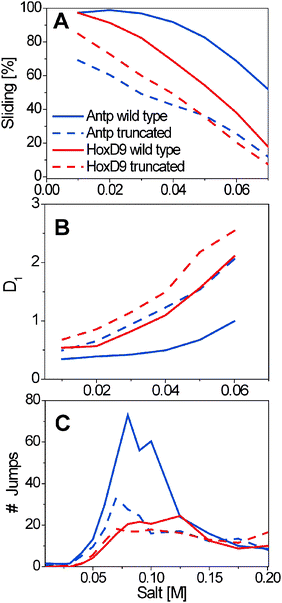 | ||
| Fig. 3 The effect of the disordered tail on the features of DNA search. The search performed by the Antp (red) and HoxD9 (blue) homeodomains and their tail truncated variants (dashed lines) is characterized by the sliding propensity (A), the speed of linear diffusion along the DNA, D1, calculated as the mean square displacement MSD per time step (Å2 per time step) according to ref. 63 (B), and the number of intersegment transfer jumps during 108 time steps between two parallel DNA molecules positioned 60 Å apart (C). The N-tails of Antp and HoxD9 comprise 10 and 9 residues (their net charge is 4 and 2), respectively.64 | ||
In homeodomains, the N-tail can also assist intersegment transfer, which is known to enhance DNA search when the protein interacts nonspecifically with DNA.75–78 Direct relocation from one nonspecific DNA sequence to another occurs without accumulation of free protein during the intersegment transfer of transcription factors.62,64,70,71 The tailed homeodomain translocates between DNAs through an intermediate in which the recognition helix of the protein is adsorbed onto one DNA fragment and the N-tail is adsorbed onto the second, suggesting a “monkey bar” mechanism where the transfer occurs using a motion that resembles that of a child brachiating along “monkey bars”64 (Fig. 4A). Alternatively, the transfer may be described as following the fly-casting mechanism.31,32,79,80 In this mechanism, the disordered tail reaches relatively distant DNA regions that lie beyond those near the structured region of the protein, and therefore fly-casting suggests an increase in the kinetic specificity of the search.32 The tradeoff between the entropy lost during binding-induced folding81,82 and the entropy gained from the hydrophobic effect may increase the specificity of the homeodomains for binding sites on DNA and modulate their sequence selectivity. Thus, in both types of intersegment transfer, the act of transfer is facilitated by the disordered tail. Tail length can significantly affect the propensity to engage in intersegmental transfer, such that more jumping events are seen for the Antp homeodomain, whose tail is longer and more positively charged, than for the HoxD9 homeodomain, whose tail is shorter and with lower net charge (Fig. 3C).
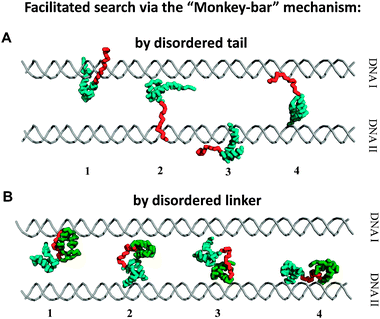 | ||
| Fig. 4 Intersegment transfer via the “monkey bar” mechanism. Snapshots of intersegment transfer between two parallel DNA molecules by DNA-binding proteins with a disordered tail64 (A) and when two domains are tethered by a disordered linker71 (B). The brachiation dynamics between two DNA molecules is promoted by splitting the protein into two regions (a structured domain and tail or two domains tethered by linker) that allow facilitated jumps. | ||
The three homeodomain proteins, HoxD9, Antp, and NK-2 (see ref. 64) search DNA quite differently although they have very similar globular regions. The presence of an N-tail in these homeodomains modulates the characteristics of the DNA search. Although the efficiency of the DNA search is correlated with tail length and net charge, the linkage is more complex than that. A comparison of the tail sequences of these three homeodomains reveals that, while the lengths and net charges are different for each, the positive residues are clustered into positive segments in all of them. Our recent study of more than 600 homeodomain variants that have different net charges and positive charge distributions on the tails addresses the question of the tail properties required to achieve efficient DNA search.70 The intersegment transfer search mode is most populated for moderately charged segments (Fig. 5A) and for positive charges that are clustered together in the middle of the tail or towards its N terminus (Fig. 5B). A bioinformatic analysis of 1384 wild-type homeodomains illustrates that the N-tails are highly positively charged, their positive charges are clustered, and the spacing between the charge clusters is smaller than between clusters of the globular region of homeodomains, thus consistent with an optimal charge pattern to promote intersegment transfer and therefore an overall rapid search. All together, the results propose that the intrinsically disordered tails of DNA-binding proteins were evolutionary selected for optimal DNA search.
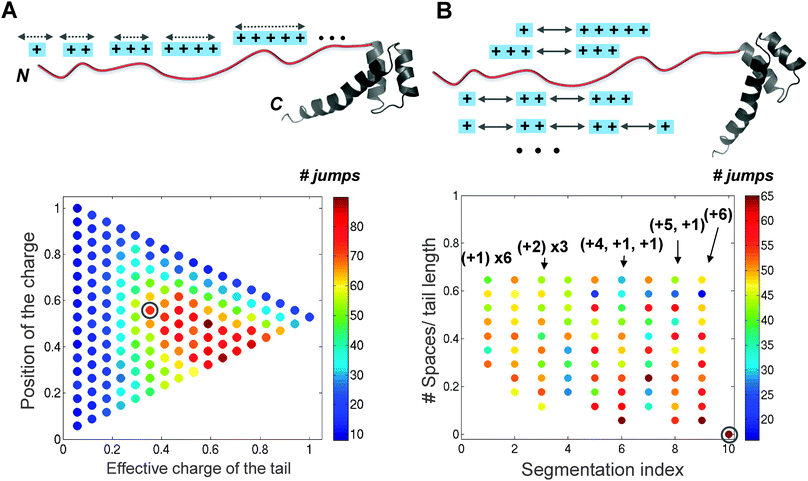 | ||
| Fig. 5 The effect of the charge composition and distribution in the intrinsically disordered tail on the properties of DNA search by the homeodomain. (A) The influence of the effective charge and the average position of the charged segment within the tail on the number of intersegment transfer events (i.e., jumps) during 108 time steps performed by the 153 variants of the NK-2 homeodomain between two DNA fragments separated by 60 Å. (B) Quantitative characteristics of intersegment transfer by 253 NK-2 variants that include six positive charges that are divided into different charge segments and positioned at different locations along the tail. The jumps were followed between two DNA molecules separated by 60 Å at a salt concentration of 0.07 M. The gray circle indicates the corresponding values of the N-tail of wild-type NK-2, suggesting that its sequence evolved to promote the “monkey bar” mechanism (for more details see70). | ||
An elegant example for the importance of the disordered tails in the interaction of proteins with DNA is transcription factor p53, which plays a major role in preventing tumor development and has been intensively studied over the last 30 years.83,84 The p53 protein has N-terminal and C-terminal tails with different functions.85–89 The N-tails, which are negatively charged and essential for the functioning of p53, are exposed to the solvent and do not interact with either the protein or the DNA and therefore have little effect on the sliding search mode.88 The C-tails are positively charged (Table 1) and thus strongly interact with DNA and can affect sliding features very significantly (Fig. 6). The C-tails interact differently with the DNA depending on whether p53 slides as two dimers or as a tetramer (Fig. 6A–C). Recent single-molecule experiments90 and a coarse-grained simulation study88 showed that the C-tails of p53 increase its sliding speed along DNA. In the truncated version, the sliding speed of p53 slows significantly. To better understand the capability of the C-tails to speed up linear motion along DNA, we studied the sliding of several variants of the C-tails of p53. Clustering of the 8 positive residues into one segment in the middle of the C-tail and neutralizing the 3 negative charges at the C-edge result in an additive increase in sliding propensity but slower linear diffusion (Fig. 6D and E). The large capture radius of the C-tails allows interactions with remote DNA segments, which may enhance the localization and affinity of p53 to DNA (Fig. 6).88 The positively charged residues of the C-tails are therefore essential for modulating interactions with DNA. These findings support previous studies on the significance of net charge to the biophysics and function of intrinsically disordered proteins, in general, and to the modulation of protein–DNA interactions, in particular.18,20–23,70
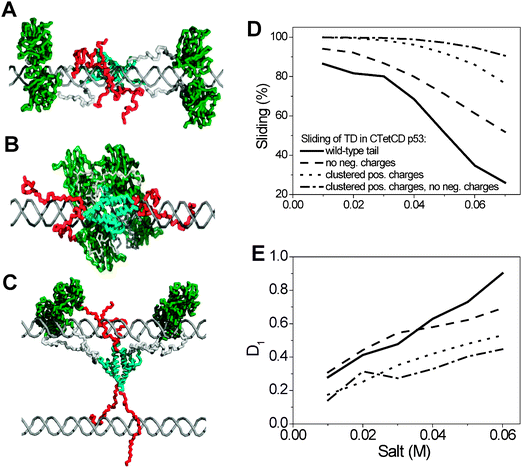 | ||
| Fig. 6 The importance of the C-tails of p53 in its sliding along DNA. Snapshots of the sliding of the CTetCD model of p53 (a truncated tetrameric p53 protein comprised of the core domain, Tet domain (TD), and a C-tail (residues 94–393)) along DNA. The p53 protein can slide in various oligomeric states. (A) p53 slides while the DNA binding domains are organized as two dimers. (B) p53 slides while the core domains form a tetramer. The C-tails are stretched along the DNA when the core domain diffuses as a tetramer but they are placed in between the two primary dimers when the secondary interface is not formed. (C) The sliding of p53 at a high concentration of DNA in which the C-tails can interact with different DNA molecules. The complex dynamics of the C-tails can take place in various oligomeric states of the core domains. (D and E) The interplay between the features of the C-tails on the sliding propensity and the speed of linear diffusion, D1. Clustering the charges into a single large cluster or neutralizing the negatively charged residues of the C-tails results in a larger sliding propensity and slower sliding. | ||
Moreover, the disordered C-tails mediate the interactions between the DNA-binding-domain (the core domain) and DNA, and dictate the orientation of the tetramerization domain relative to the DNA, although this orientation is also dependent on the local cellular environment.88 These results support the hypothesis that the tails can further promote a conformational change in the p53–DNA complex. Therefore, the disordered tails have a role not only in searching for the location of binding sites but also in promoting the specific interactions with the DNA (e.g., DNA bending, as observed in the crystal structure of the specific complex of p53 with some of its response elements).91–93
Post-translational modifications to disordered tails serve as DNA affinity tuners with major biological impact
The capability of disordered tails to modulate specific and nonspecific interactions with DNA is tightly linked to the fact that the tails are rich in positively charged residues. The high content of charged residues and their non-random distribution along the tail call for increased understanding of the sequence–structure–function relationship in disordered regions. Perturbing the composition and distribution of charged residues in the disordered regions, for example by post-translational modifications such as phosphorylation and acetylation,94 may impede the capability of the tail to interact nonspecifically with DNA by reducing its DNA affinity. Phosphorylation, a typical post-translational modification that often occurs on disordered regions,9,95 is common to many DNA-binding proteins and may take place at multiple sites until protein activity or inhibition is reached.96,97 A recent NMR experiment demonstrated that multiple signal-dependent phosphorylations within the disordered tail of the transcription factor Ets-1 accumulate additively to regulate DNA-binding affinity in a graded manner.98 Additive phosphorylation progressively shifts Ets-1 from an active state in which the protein is dynamic and poised to bind DNA, to a folded inhibited conformation. Analogous to Ets-1, phosphorylation of FACT (facilitates chromatin transcription)99 and of PC4 (positive cofactor 4)100 gradually decreases their DNA binding affinity. The unstructured tails of these proteins show the significance of disordered segments in the variable regulation of DNA-binding and protein activity.The p53 protein is regulated mostly through covalent modifications101,102 that occur on both the C- and N-tails (the modification sites of the C-tail of p53 are shown in Table 1).103,104 Dynamic p53 acetylation and deacetylation events were observed in response to DNA damage.105 The acetylation events within the C-tail significantly enhance site-specific DNA-binding activity.105,106 Moreover, C-tail acetylation levels are well correlated with p53-mediated activationin vivo.105Phosphorylation of the C-tail positively regulates DNA binding and tetramerization of p53.107,108Activation of p53 DNA binding may be critical to the regulation of its cell growth arrest function. Indeed, combinations of various post-translational modifications are involved in the stabilization and activation of p53 and were shown to have an important role in tumorigenesis.101
A well known example of the importance of post-translational modifications of the disordered tail in modulating interactions with DNA is effects of post-translational modifications on histone tails. The core histone proteins (i.e. H3, H4, H2A, H2B, and H1) package the DNA into repeating nucleosomal units that are folded into higher order chromatin fibers.109–113 The disordered tails, whose sequences are among the most highly conserved in eukaryotes, are located at the N-terminus of all four core histone proteins and the C-terminus of histone H2A.114 The positively charged tails (see the sequence of H3's N-tail in Table 1) bind DNA with high affinity at the minor groove115 and are relatively exposed to the solvent and thus readily accessible to chemical modification.116,117 The tails were shown to have an important role in the stable folding of nucleosomes into condensed chromatin fibers and higher order structures, and modulation of DNA accessibility within the nucleosome.114
Chromatin is subject to a variety of post-translational modifications on core histone disordered tails (the modification sites of a H3 tail are shown in Table 1). Acetylation, in synergy with the effects mediated by other chromatin remodeling factors, increases the accessibility of nucleosomal DNA by weakening histone–DNA interactions.118–120 The exchange to repressive chromatin is initiated by deacetylation, which promotes the condensation of the nucleosome and provokes repressive factors.118 A combination of modifications on the histone flexible tails (Table 1) influences transcriptional regulation, replication, site specific recombination, and DNA repair,120,121 and is therefore considered the “histone-code”, which expands the information potential of the genetic code.122,123
Facilitated DNA search by multidomain transcription factors tethered via a disordered linker
More than two-thirds of eukaryotic proteins are composed of multiple domains.124,125 In many cases, these domains lack any interface and can cooperate only via a disordered, flexible linker. Understanding the degree of cooperation between domains tethered by a linker that allows proteins to search DNA efficiently is essential for understanding cellular network at the molecular level. Flexible linkers are crucial to several types of functions in protein–DNA recognition.126 The linker can attach unfolded domains and keep the protein as a folded dimer that can bind to DNA, such as in the designed single-chain Arc repressor.126,127 In other cases, the disordered linker can tether the DNA-binding domain with a dimerization domain as in the λ repressor,126,128 or with a tetramerization domain, as in p53,129 and enhance the binding affinity.128 The disordered linker was shown to be crucial for DNA binding and recognition for Oct-1130 and for the human replication protein A (RPA).131 We focus on linked domains, such as are found in the Pax6,71Oct-1,69,71 and p5388transcription factors, in which the disordered linkers that connect between the different DNA-binding domains increase their affinity to DNA over that of the isolated domains.Our recent study of the molecular mechanisms of DNA search by two-DNA-binding-domain transcription factors demonstrated that when Pax6 and Oct-1 engage in nonspecific binding, their individual domains exhibit significantly different affinities to DNA:71 this finding is supported by experiments.69 Tethering the two domains via a flexible linker increases their affinity to the DNA,126 resulting in a higher propensity for sliding along the DNA, which is more significant for the domain with the weaker DNA-binding affinity.71 The domain that binds DNA more tightly anchors the multidomain protein to the DNA and, via the linker, increases the local concentration of the weak DNA-binding domain, at the expense of slower linear diffusion than is exhibited by the isolated domains.
Computational71 and NMR78,132,133 studies showed that multi-domain transcription factors exchange occurs between two DNA molecules via a bridged intermediate where each domain is attached to a different DNA segment (Fig. 4B), suggesting the “monkey bar” mechanism.64 The intersegmental transfer is facilitated by tethering individual domains by a flexible linker, where the electrostatic affinity of each domain to DNA significantly affects the number of jumping events. Greater facilitation of the jumping search mode is achieved when one domain has a higher affinity to DNA and acts as a strong anchor while the complementary domain has a lower DNA binding affinity and serves as an explorer for binding sites on adjacent DNA molecules, as was shown for the Pax-671 and Oct-171,78,133transcriptions factors.
In the p53 protein, cross-talk was demonstrated between the DNA-binding domain and the tetramerization domain to which it was tethered by a flexible linker.88 The tetramerization domain has a low affinity to DNA, whereas the isolated DNA-binding domain performs DNA search by a combination of sliding, hopping, and the 3D diffusion search modes. Tethering the DNA-binding domain to the tetramerization domain results in a tetrameric core domain that uses sliding as an exclusive search mode at most salt concentrations and whose tetramerization domain interacts less tightly with DNA and slides more slowly than its isolated counterpart.88
The disordered linker mediates cross talk between two domains when there is no physical interface between them. Tethering by the linker can lead to a significant improvement in the search efficiency that strongly depends on the relative affinities of the domains to DNA. Comparison of the electrostatic contributions to the protein–DNA binding energy made by the individual domains that comprise several multidomain proteins revealed that tethered domains tend to have different DNA-binding affinities, which may imply that both specific and nonspecific DNA binding has biological significance.71 The efficient DNA search observed for multidomain proteins with different affinities to DNA and the polarized DNA-binding affinities found for multidomain proteins suggest that there is an evolutionary drift toward tethering of domains with different affinities to DNA by a flexible linker.
Conclusions
Intrinsically disordered tails are common in DNA-binding proteins and can modulate binding specificity and affinity through the formation of transient and dynamic interactions that can supplement the interactions between the folded protein and DNA. The disorder regions can be viewed as subdomains that should be taken into consideration when attempting to formulate a molecular code for protein–DNA recognition. Clearly, the dynamic nature of these intrinsically disordered subdomains may further complicate attempts to decipher this code.In addition to fine-tuning the specificity of protein–DNA interactions, the intrinsically disordered regions increase the affinity of the protein to DNA during nonspecific interactions and thus aid in DNA search. The flexible subdomain may improve search efficiency on nonspecific DNA by increasing the affinity of the globular domain to the DNA and thus promoting sliding dynamics. Flexible subdomains often facilitate the search by supporting a brachiation motion during intersegment transfer. Similarly, constructing a protein by tethering two structured domains via a flexible linker is another approach, evolution might exploit to promote brachiation dynamics between different DNA segments.
The ability of the disordered tail to promote jumping depends on its length and net charge. Yet, it is not only the composition of the charged residues of the tail but also their arrangement that control jumping using “monkey bar” dynamics. We found that higher numbers of intersegment transfers are achieved when the tail is significantly but moderately charged and when the charges are concentrated in the middle of the tail or closer to its N terminus. Increased clustering of the charges, i.e., the absence of large or numerous spaces between charged residues, enhances the “monkey bar” mechanism. A bioinformatic investigation has shown that disordered tails are highly positively charged, their positive charges are highly clustered, and the spacing between the charge clusters is much smaller than between clusters of positive charges in the globular regions. Overall, our results strengthen recent reports that the sequences of intrinsically disordered regions have unique characteristics that govern their structure and function and suggest that the tails of DNA-binding proteins have been evolutionarily selected to have the charge pattern for optimal DNA search.
Nature manipulates the composition and distribution of charges within the intrinsically disordered regions to regulate the strength of protein–DNA interactions as well. Post-translational modifications (e.g., acetylation or phosphorylation), which often take place on intrinsically disordered regions, can change the electrostatic properties of the flexible subdomain and thus the nonspecific or specific affinity of the protein to the DNA. Accordingly, the flexible subdomains have not only the ability to fine tune the affinity of protein–DNA interactions, but may also act as real-time tuners with the ability to change affinity to DNA depending on the need to regulate cellular processes.
Acknowledgements
We are grateful to Netaly Khazanov for fruitful discussions on p53 dynamics on DNA. This work was supported by the Kimmelman Center for Macromolecular Assemblies and the Minerva Foundation with funding from the Federal German Ministry for Education and Research. Y.L. is the incumbent of the Lillian and George Lyttle Career Development Chair.References
- P. E. Wright and H. J. Dyson, Intrinsically unstructured proteins: re-assessing the protein structure–function paradigm, J. Mol. Biol., 1999, 293, 321–331 CrossRef CAS.
- A. K. Dunker, J. D. Lawson, C. J. Brown, R. M. Williams and P. Romero, Intrinsically disordered protein, J. Mol. Graphics Modell., 2001, 19, 26–59 CrossRef CAS.
- P. Tompa, Intrinsically unstructured proteins, Trends Biochem. Sci., 2002, 27, 527–533 CrossRef CAS.
- H. J. Dyson and P. E. Wright, Intrinsically unstructured proteins and their functions, Nat. Rev. Mol. Cell Biol., 2005, 6, 197–208 CrossRef CAS.
- V. N. Uversky, What does it mean to be natively unfolded?, Eur. J. Biochem., 2002, 269, 2–12 CrossRef CAS.
- J. G. Liu, N. B. Perumal, C. J. Oldfield, E. W. Su, V. N. Uversky and A. K. Dunker, Intrinsic disorder in transcription factors, Biochemistry, 2006, 45, 6873–6888 CrossRef CAS.
- T. Mittag, L. E. Kay and J. D. Forman-Kay, Protein dynamics and conformational disorder in molecular recognition, J. Mol. Recognit., 2010, 23, 105–116 CAS.
- A. K. Dunker, I. Silman, V. N. Uversky and J. L. Sussman, Function and structure of inherently disordered proteins, Curr. Opin. Struct. Biol., 2008, 18, 756–764 CrossRef CAS.
- J. Gsponer, M. E. Futschik, S. A. Teichmann and M. M. Babu, Tight regulation of unstructured proteins: from transcript synthesis to protein degradation, Science, 2008, 322, 1365–1368 CrossRef CAS.
- M. M. Babu, R. van der Lee, N. S. de Groot and J. Gsponer, Intrinsically disordered proteins: regulation and disease, Curr. Opin. Struct. Biol., 2011, 21, 432–440 CrossRef CAS.
- Y. Wang, J. C. Fisher, R. Mathew, L. Ou, S. Otieno, J. Sublet, L. Xiao, J. Chen, M. F. Roussel and R. W. Kriwacki, Intrinsic disorder mediates the diverse regulatory functions of the Cdk inhibitor p21, Nat. Chem. Biol., 2011, 7, 214–221 CrossRef CAS.
- T. Hagai, A. Tóth-Petróczy, A. Azia and Y. Levy, Intrinsic Disorder in Ubiquitination Substrates, J. Mol. Biol., 2011, 412, 319–324 CrossRef CAS.
- A. K. Dunker, C. J. Brown, J. D. Lawson, L. M. Iakoucheva and Z. Obradovic, Intrinsic disorder and protein function, Biochemistry, 2002, 41, 6573–6582 CrossRef CAS.
- J. J. Ward, J. S. Sodhi, L. J. McGuffin, B. F. Buxton and D. T. Jones, Prediction and functional analysis of native disorder in proteins from the three kingdoms of life, J. Mol. Biol., 2004, 337, 635–645 CrossRef CAS.
- V. N. Uversky, J. R. Gillespie and A. L. Fink, Why are “natively unfolded” proteins unstructured under physiologic conditions?, Proteins: Struct., Funct., Genet., 2000, 41, 415–427 CrossRef CAS.
- P. Romero, Z. Obradovic, X. H. Li, E. C. Garner, C. J. Brown and A. K. Dunker, Sequence complexity of disordered protein, Proteins: Struct., Funct., Genet., 2001, 42, 38–48 CrossRef CAS.
- H. T. Tran, A. Mao and R. V. Pappu, Role of backbone—solvent interactions in determining conformational equilibria of intrinsically disordered proteins, J. Am. Chem. Soc., 2008, 130, 7380–7392 CrossRef CAS.
- M. Borg, T. Mittag, T. Pawson, M. Tyers, J. D. Forman-Kay and H. S. Chan, Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 9650–9655 CrossRef CAS.
- D. Eliezer, Biophysical characterization of intrinsically disordered proteins, Curr. Opin. Struct. Biol., 2009, 19, 23–30 CrossRef CAS.
- J. A. Marsh and J. D. Forman-Kay, Sequence Determinants of Compaction in Intrinsically Disordered Proteins, Biophys. J., 2010, 98, 2383–2390 CrossRef CAS.
- A. H. Mao, S. L. Crick, A. Vitalis, C. L. Chicoine and R. V. Pappu, Net charge per residue modulates conformational ensembles of intrinsically disordered proteins, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 8183–8188 CrossRef CAS.
- S. Muller-Spath, A. Soranno, V. Hirschfeld, H. Hofmann, S. Ruegger, L. Reymond, D. Nettels and B. Schuler, Charge interactions can dominate the dimensions of intrinsically disordered proteins, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14609–14614 CrossRef CAS.
- J. L. England and G. Haran, To fold or expand-a charged question, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14519–14520 CrossRef CAS.
- D. Potoyan and G. Papoian, Energy landscape analyses of disordered histones tails reveal special organization of their conformational dynamics, J. Am. Chem. Soc., 2011, 133, 7405–7415 CrossRef CAS.
- C. J. Brown, A. K. Johnson and G. W. Daughdrill, Comparing models of evolution for ordered and disordered proteins, Mol. Biol. Evol., 2010, 27, 609–621 CrossRef CAS.
- C. J. Brown, A. K. Johnson, A. K. Dunker and G. W. Daughdrill, Evolution and disorder, Curr. Opin. Struct. Biol., 2011, 21, 441–446 CrossRef CAS.
- P. Tompa, The interplay between structure and function in intrinsically unstructured proteins, FEBS Lett., 2005, 579, 3346–3354 CrossRef CAS.
- H. J. Dyson and P. E. Wright, Coupling of folding and binding for unstructured proteins, Curr. Opin. Struct. Biol., 2002, 12, 54–60 CrossRef CAS.
- P. Tompa and M. Fuxreiter, Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions, Trends Biochem. Sci., 2008, 33, 2–8 CrossRef CAS.
- B. W. Pontius, Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association, Trends Biochem. Sci., 1993, 18, 181–186 CrossRef CAS.
- B. A. Shoemaker, J. J. Portman and P. G. Wolynes, Speeding molecular recognition by using the folding funnel: The fly-casting mechanism, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 8868–8873 CrossRef CAS.
- E. Trizac, Y. Levy and P. G. Wolynes, Capillarity theory for the fly-casting mechanism, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 2746–2750 CrossRef CAS.
- A. Tóth-Petróczy, M. Fuxreiter and Y. Levy, Disordered tails of homeodomains facilitate DNA recognition by providing a trade-off between folding and specific binding, J. Am. Chem. Soc., 2009, 131, 15084–15085 CrossRef.
- K. Gunasekaran, C. J. Tsai and R. Nussinov, Analysis of ordered and disordered protein complexes reveals structural features discriminating between stable and unstable monomers, J. Mol. Biol., 2004, 341, 1327–1341 CrossRef CAS.
- B. Meszaros, P. Tompa, I. Simon and Z. Dosztanyi, Molecular principles of the interactions of disordered proteins, J. Mol. Biol., 2007, 372, 549–561 CrossRef CAS.
- M. Fuxreiter, I. Simon, P. Friedrich and P. Tompa, Preformed structural elements feature in partner recognition by intrinsically unstructured proteins, J. Mol. Biol., 2004, 338, 1015–1026 CrossRef CAS.
- V. Csizmok, M. Bokor, P. Banki, E. Klement, K. F. Medzihradszky, P. Friedrich, K. Tompa and P. Tompa, Primary contact sites in intrinsically unstructured proteins: the case of calpastatin and microtubule-associated protein 2, Biochemistry, 2005, 44, 3955–3964 CrossRef CAS.
- V. Neduva and R. B. Russell, Linear motifs: evolutionary interaction switches, FEBS Lett., 2005, 579, 3342–3345 CrossRef CAS.
- M. Fuxreiter, P. Tompa and I. Simon, Local structural disorder imparts plasticity on linear motifs, Bioinformatics, 2007, 23, 950–956 CrossRef CAS.
- M. Fuxreiter, I. Simon and S. Bondos, Dynamic protein–DNA recognition: beyond what can be seen, Trends Biochem. Sci., 2011, 36, 415–423 CrossRef CAS.
- R. S. Spolar and M. T. Record, Jr, Coupling of local folding to site-specific binding of proteins to DNA, Science, 1994, 263, 777–784 CAS.
- J. M. Gruschus, D. H. Tsao, L. H. Wang, M. Nirenberg and J. A. Ferretti, Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity, Biochemistry, 1997, 36, 5372–5380 CrossRef CAS.
- M. Billeter, Y. Q. Qian, G. Otting, M. Muller, W. Gehring and K. Wuthrich, Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex, J. Mol. Biol., 1993, 234, 1084–1093 CrossRef CAS.
- R. Joshi, J. M. Passner, R. Rohs, R. Jain, A. Sosinsky, M. A. Crickmore, V. Jacob, A. K. Aggarwal, B. Honig and R. S. Mann, Functional specificity of a Hox protein mediated by the recognition of minor groove structure, Cell, 2007, 131, 530–543 CrossRef CAS.
- C. Crane-Robinson, A. I. Dragan and P. L. Privalov, The extended arms of DNA-binding domains: a tale of tails, Trends Biochem. Sci., 2006, 31, 547–552 CrossRef CAS.
- W. L. Zeng, D. J. Andrew, L. D. Mathies, M. A. Horner and M. P. Scott, Ectopic Expression and Function of the Antp and Scr Homeotic Genes—the N-Terminus of the Homeodomain Is Critical to Functional Specificity, Development, 1993, 118, 339–352 CAS.
- P. L. Privalov, A. I. Dragan, C. Crane-Robinson, K. J. Breslauer, D. P. Remeta and C. A. Minetti, What drives proteins into the major or minor grooves of DNA?, J. Mol. Biol., 2007, 365, 1–9 CrossRef CAS.
- A. I. Dragan, Z. Li, E. N. Makeyeva, E. I. Milgotina, Y. Liu, C. Crane-Robinson and P. L. Privalov, Forces driving the binding of homeodomains to DNA, Biochemistry, 2006, 45, 141–151 CrossRef CAS.
- B. Lewin, Genes VII, Oxford Univ. Press, Oxford, 2000 Search PubMed.
- R. F. Bruinsma, Physics of protein-DNA interaction, Phys. A (Amsterdam, Neth.), 2002, 313, 211–237 CAS.
- S. E. Halford, An end to 40 years of mistakes in DNA-protein association kinetics?, Biochem. Soc. Trans., 2009, 37, 343–348 CrossRef CAS.
- S. E. Halford and J. F. Marko, How do site-specific DNA-binding proteins find their targets?, Nucleic Acids Res., 2004, 32, 3040–3052 CrossRef CAS.
- L. Mirny, M. Slutsky, Z. Wunderlich, A. Tafvizi, J. Leith and A. Kosmrlj, How a protein searches for its site on DNA: the mechanism of facilitated diffusion, J. Phys. A: Math. Theor., 2009, 42, 434013–434035 CrossRef.
- P. H. Von Hippel and O. G. Berg, Facilitated Target Location in Biological-Systems, J. Biol. Chem., 1989, 264, 675–678 CAS.
- O. G. Berg, R. B. Winter and P. H. Vonhippel, Diffusion-Driven Mechanisms of Protein Translocation on Nucleic-Acids. 1. Models and Theory, Biochemistry, 1981, 20, 6929–6948 CrossRef CAS.
- H. X. Zhou and A. Szabo, Enhancement of association rates by nonspecific binding to DNA and cell membranes, Phys. Rev. Lett., 2004, 93, 178101-1–18101-4 Search PubMed.
- S. E. Halford and M. D. Szczelkun, How to get from A to B: strategies for analysing protein motion on DNA, Eur. Biophys. J., 2002, 31, 257–267 CrossRef CAS.
- O. G. Berg and P. H. Von Hippel, Diffusion-Controlled Macromolecular Interactions, Annu. Rev. Biophys. Biophys. Chem., 1985, 14, 131–160 CrossRef CAS.
- K. V. Klenin, H. Merlitz, J. Langowski and C. X. Wu, Facilitated diffusion of DNA-binding proteins, Phys. Rev. Lett., 2006, 96, 018104 CrossRef.
- J. Elf, G. W. Li and X. S. Xie, Probing transcription factor dynamics at the single-molecule level in a living cell, Science, 2007, 316, 1191–1194 CrossRef CAS.
- P. C. Blainey, A. M. van Oijent, A. Banerjee, G. L. Verdine and X. S. Xie, A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 5752–5757 CrossRef CAS.
- J. Iwahara, M. Zweckstetter and G. M. Clore, NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15062–15067 CrossRef CAS.
- O. Givaty and Y. Levy, Protein Sliding along DNA: Dynamics and Structural Characterization, J. Mol. Biol., 2009, 385, 1087–1097 CrossRef CAS.
- D. Vuzman, A. Azia and Y. Levy, Searching DNA via a “monkey bar” mechanism: the significance of disordered tails, J. Mol. Biol., 2010, 396, 674–684 CrossRef CAS.
- D. M. Gowers, G. G. Wilson and S. E. Halford, Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 15883–15888 CrossRef CAS.
- M. Slutsky and L. A. Mirny, Kinetics of protein–DNA interaction: facilitated target location in sequence-dependent potential, Biophys. J., 2004, 87, 4021–4035 CrossRef CAS.
- Z. Wunderlich and L. A. Mirny, Spatial effects on the speed and reliability of protein-DNA search, Nucleic Acids Res., 2008, 36, 3570–3578 CrossRef CAS.
- J. C. Venter, M. D. Adams, E. W. Myers, P. W. Li and R. J. Mural, et al. The sequence of the human genome, Science, 2001, 291, 1304–1351 CrossRef CAS.
- J. D. Klemm and C. O. Pabo, Oct-1 POU domain DNA interactions: cooperative binding of isolated subdomains and effects of covalent linkage, Genes Dev., 1996, 10, 27–36 CrossRef CAS.
- D. Vuzman and Y. Levy, DNA search efficiency is modulated by charge composition and distribution in the intrinsically disordered tail, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 21004–21009 CrossRef CAS.
- D. Vuzman, M. Polonsky and Y. Levy, Facilitated DNA search by multidomain transcription factors: cross talk via a flexible linker, Biophys. J., 2010, 99, 1202–1211 CrossRef CAS.
- P. L. Privalov, A. I. Dragan, C. Crane-Robinson, K. J. Breslauer, D. P. Remeta and C. A. S. A. Minetti, What drives proteins into the major or minor grooves of DNA?, J. Mol. Biol., 2007, 365, 1–9 CrossRef CAS.
- J. Iwahara and G. M. Clore, Detecting transient intermediates in macromolecular binding by paramagnetic NMR, Nature, 2006, 440, 1227–1230 CrossRef CAS.
- J. Iwahara and G. M. Clore, Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy, J. Am. Chem. Soc., 2006, 128, 404–405 CrossRef CAS.
- T. Hu and B. I. Shklovskii, How a protein searches for its specific site on DNA: the role of intersegment transfer, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2007, 76, 051909 CrossRef.
- M. Sheinman and Y. Kafri, The effects of intersegmental transfers on target location by proteins, Phys. Biol., 2009, 6, 016003-1–016003-17 CrossRef.
- C. Loverdo, O. Benichou, R. Voituriez, A. Biebricher, I. Bonnet and P. Desbiolles, Quantifying Hopping and Jumping in Facilitated Diffusion of DNA-Binding Proteins, Phys. Rev. Lett., 2009, 102, 188101-1–188101-4 CrossRef.
- M. Doucleff and G. M. Clore, Global jumping and domain-specific intersegment transfer between DNA cognate sites of the multidomain transcription factor Oct-1, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 13871–13876 CrossRef CAS.
- A. G. Turjanski, J. S. Gutkind, R. B. Best and G. Hummer, Binding-induced folding of a natively unstructured transcription factor, PLoS Comput. Biol., 2008, 4, e1000060 Search PubMed.
- J. H. Chen, Intrinsically Disordered p53 Extreme C-Terminus Binds to S100B(beta beta) through “Fly-Casting”, J. Am. Chem. Soc., 2009, 131, 2088–2089 CrossRef CAS.
- D. Ganguly and J. Chen, Topology-based modeling of intrinsically disordered proteins: balancing intrinsic structural propensities and intermolecular interactions, Proteins: Struct., Funct., Bioinf., 2011, 79, 1251–1266 CrossRef CAS.
- T. Click, D. Ganguly and J. Chen, Intrinsically disordered proteins in a physics-based world, Int. J. Mol. Sci., 2010, 11, 5292–5309 CrossRef CAS.
- A. J. Levine and M. Oren, The first 30 years of p53: growing ever more complex, Nat. Rev. Cancer, 2009, 9, 749–758 CrossRef CAS.
- K. H. Vousden and C. Prives, Blinded by the Light: The Growing Complexity of p53, Cell, 2009, 137, 413–431 CrossRef CAS.
- J. Ahn and C. Prives, The C-terminus of p53: the more you learn the less you know, Nat. Struct. Biol., 2001, 8, 730–732 CrossRef CAS.
- R. L. Weinberg, S. M. V. Freund, D. B. Veprintsev, M. Bycroft and A. R. Fersht, Regulation of DNA binding of p53 by its C-terminal domain, J. Mol. Biol., 2004, 342, 801–811 CrossRef CAS.
- K. McKinney, M. Mattia, V. Gottifredi and C. Prives, p53 linear diffusion along DNA requires its C terminus, Mol. Cell, 2004, 16, 413–424 CrossRef CAS.
- N. Khazanov and Y. Levy, Sliding of p53 along DNA Can Be Modulated by Its Oligomeric State and by Cross-Talks between Its Constituent Domains, J. Mol. Biol., 2011, 408, 335–355 CrossRef CAS.
- M. Sauer, A. C. Bretz, R. Beinoraviciute-Kellner, M. Beitzinger, C. Burek, A. Rosenwald, G. S. Harms and T. Stiewe, C-terminal diversity within the p53 family accounts for differences in DNA binding and transcriptional activity, Nucleic Acids Res., 2008, 36, 1900–1912 CrossRef CAS.
- A. R. Fersht, A. Tafvizi, F. Huang, L. A. Mirny and A. M. van Oijen, A single-molecule characterization of p53 search on DNA, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 563–568 CrossRef.
- Y. P. Pan and R. Nussinov, P53-induced DNA bending: the interplay between p53-ONA and p53-p53 interactions, J. Phys. Chem. B, 2008, 112, 6716–6724 CrossRef CAS.
- Y. P. Pan and R. Nussinov, Structural basis for specific p53 binding-induced DNA bending, Biophys. J., 2007, 233a Search PubMed.
- A. K. Nagaich, V. B. Zhurkin, S. R. Durell, R. L. Jernigan, E. Appella and R. E. Harrington, p53-induced DNA bending and twisting: p53 tetramer binds on the outer side of a DNA loop and increases DNA twisting, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 1875–1880 CrossRef CAS.
- D. Vuzman, Y. Hoffman, Y. Levy, Modulating Protein–DNA Interactions by Post-Translational Modifications at Disordered Regions, submitted.
- Madhusudan, E. Trafny, N. Xuong, J. Adams, L. Ten Eyck, S. Taylor and J. Sowadski, Camp-Dependent Protein-Kinase—Crystallographic Insights into Substrate Recognition and Phosphotransfer, Protein Sci., 1994, 3, 176–187 CrossRef CAS.
- C. Salazar and T. Hofer, Allosteric regulation of the transcription factor NFAT1 by multiple phosphorylation sites: a mathematical analysis, J. Mol. Biol., 2003, 327, 31–45 CrossRef CAS.
- S. Orlicky, X. J. Tang, A. Willems, M. Tyers and F. Sicheri, Structural basis for phospho-dependent substrate selection and orientation by the SCF(Cdc4) ubiquitin ligase, Cell, 2003, 112, 243–256 CrossRef CAS.
- M. A. Pufall, G. M. Lee, M. L. Nelson, H. S. Kang, A. Velyvis, L. E. Kay, L. P. McIntosh and B. J. Graves, Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region, Science, 2005, 309, 142–145 CrossRef CAS.
- K. Morikawa, Y. Tsunaka, J. Toga, H. Yamaguchi, S. Tate and S. Hirose, Phosphorylated Intrinsically Disordered Region of FACT Masks Its Nucleosomal DNA Binding Elements, J. Biol. Chem., 2009, 284, 24610–24621 CrossRef.
- H. R. A. Jonker, R. W. Wechselberger, M. Pinkse, R. Kaptein and G. E. Folkers, Gradual phosphorylation regulates PC4 coactivator function, FEBS J., 2006, 273, 1430–1444 CrossRef CAS.
- A. M. Bode and Z. G. Dong, Post-translational modification of p53 in tumorigenesis, Nat. Rev. Cancer, 2004, 4, 793–805 CrossRef CAS.
- L. Jayaraman and C. Prives, Covalent and noncovalent modifiers of the p53 protein, Cell. Mol. Life Sci., 1999, 55, 76–87 CrossRef CAS.
- E. Appella and C. W. Anderson, Post-translational modifications and activation of p53 by genotoxic stresses, Eur. J. Biochem., 2001, 268, 2764–2772 CrossRef CAS.
- X. Yang, Regulation of p53 responses by post-translational modifications, Cell Death Differ., 2003, 10, 400–403 CrossRef.
- J. Y. Luo, M. Y. Li, Y. Tang, M. Laszkowska, R. G. Roeder and W. Gu, Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 2259–2264 CrossRef CAS.
- W. Gu and R. G. Roeder, Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain, Cell, 1997, 90, 595–606 CrossRef CAS.
- T. R. Hupp, D. W. Meek, C. A. Midgley and D. P. Lane, Regulation of the Specific DNA-Binding Function of P53, Cell, 1992, 71, 875–886 CrossRef CAS.
- K. Sakaguchi, H. Sakamoto, D. Xie, J. W. Erickson, M. S. Lewis, C. W. Anderson and E. Appella, Effect of phosphorylation on tetramerization of the tumor suppressor protein p53, J. Protein Chem., 1997, 16, 553–556 CrossRef CAS.
- K. Luger and T. J. Richmond, The histone tails of the nucleosome, Curr. Opin. Genet. Dev., 1998, 8, 140–146 CrossRef CAS.
- J. Hayes and J. Hansen, Nucleosomes and the chromatin fiber, Curr. Opin. Genet. Dev., 2001, 11, 124–129 CrossRef CAS.
- C. Woodcock and S. Dimitrov, Higher-order structure of chromatin fiber, Curr. Opin. Genet. Dev., 2001, 11, 130–135 CrossRef CAS.
- R. D. Kornberg and Y. L. Lorch, Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome, Cell, 1999, 98, 285–294 CrossRef CAS.
- K. E. van Holde, Chromatin, ed. A. Rich, 1988, pp. 111–148 Search PubMed.
- C. Y. Zheng and J. J. Hayes, Structures and interactions of the core histone tail domains, Biopolymers, 2003, 68, 539–546 CrossRef CAS.
- P. D. Cary, C. Cranerobinson, E. M. Bradbury and G. H. Dixon, Effect of Acetylation on the Binding of N-Terminal Peptides of Histone-H4 to DNA, Eur. J. Biochem., 1982, 127, 137–143 CrossRef CAS.
- S. F. Lambert and J. O. Thomas, Lysine-Containing DNA-Binding Regions on the Surface of the Histone Octamer in the Nucleosome Core Particle, Eur. J. Biochem., 1986, 160, 191–201 CrossRef CAS.
- C. Hill and J. Thomas, Core histone–DNA interactions in Sea-Urchin sperm chromatin- the N-terminal tail of H2b interacts with linker DNA, Eur. J. Biochem., 1990, 187, 145–153 CrossRef CAS.
- A. Eberharter and P. B. Becker, Histone acetylation: a switch between repressive and permissive chromatin—second in review series on chromatin dynamics, EMBO Rep., 2002, 3, 224–229 CrossRef CAS.
- C. Fry and C. Peterson, Chromatin remodeling enzymes: who's on first?, Curr. Biol., 2001, 11, R185–R197 CrossRef CAS.
- S. Y. Roth, J. M. Denu and C. D. Allis, Histone acetyltransferases, Annu. Rev. Biochem., 2001, 70, 81–120 CrossRef CAS.
- A. P. Wolffe and J. J. Hayes, Chromatin disruption and modification, Nucleic Acids Res., 1999, 27, 711–720 CrossRef CAS.
- B. D. Strahl and C. D. Allis, The language of covalent histone modifications, Nature, 2000, 403, 41–45 CrossRef CAS.
- T. Jenuwein and C. D. Allis, Translating the histone code, Science, 2001, 293, 1074–1080 CrossRef CAS.
- S. A. Teichmann, J. Park and C. Chothia, Structural assignments to the Mycoplasma genitalium proteins show extensive gene duplications and domain rearrangements, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 14658–14663 CrossRef CAS.
- G. Apic, J. Gough and S. A. Teichmann, Domain combinations in archaeal, eubacterial and eukaryotic proteomes, J. Mol. Biol., 2001, 310, 311–325 CrossRef CAS.
- H. X. Zhou, The affinity-enhancing roles of flexible linkers in two-domain DNA-Binding proteins, Biochemistry, 2001, 40, 15069–15073 CrossRef CAS.
- M. Sieber and R. K. Allemann, Thermodynamics of DNA binding of MM17, a ‘single chain dimer’ of transcription factor MASH-1, Nucleic Acids Res., 2000, 28, 2122–2127 CrossRef CAS.
- H. X. Zhou, Single-chain versus dimeric protein folding: thermodynamic and kinetic consequences of covalent linkage, J. Am. Chem. Soc., 2001, 123, 6730–6731 CrossRef CAS.
- A. C. Joerger and A. R. Fersht, Structure-function-rescue: the diverse nature of common p53 cancer mutants, Oncogene, 2007, 26, 2226–2242 CrossRef CAS.
- H. C. vanLeeuwen, M. J. Strating, M. Rensen, W. deLaat and P. C. vanderVliet, Linker length and composition influence the flexibility of Oct-1 DNA binding, EMBO J., 1997, 16, 2043–2053 CrossRef CAS.
- P. D. Vise, B. Baral, A. J. Latos and G. W. Daughdrill, NMR chemical shift and relaxation measurements provide evidence for the coupled folding and binding of the p53 transactivation domain, Nucleic Acids Res., 2005, 33, 2061–2077 CrossRef CAS.
- Y. Takayama, D. Sahu and J. Iwahara, NMR Studies of Translocation of the Zif268 Protein between Its Target DNA Sites, Biochemistry, 2010, 49, 7998–8005 CrossRef CAS.
- Y. Takayama and G. M. Clore, Intra- and intermolecular translocation of the bi-domain transcription factor Oct1 characterized by liquid crystal and paramagnetic NMR, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, E169–E176 CrossRef.
Footnote |
| † Published as part of a Molecular BioSystems themed issue on Intrinsically Disordered Proteins: Guest Editor M. Madan Babu |
| This journal is © The Royal Society of Chemistry 2012 |
