Freeze-dried cationic calcium phosphate nanorods as versatile carriers of nucleic acids (DNA, siRNA)
Jan
Klesing
,
Svitlana
Chernousova
and
Matthias
Epple
*
Inorganic Chemistry and Center for Nanointegration Duisburg-Essen (CeNIDE), University of Duisburg-Essen, Universitaetsstr. 5-7, D-45117, Essen, Germany. E-mail: matthias.epple@uni-due.de; Fax: +49 201 1832621; Tel: +49 201 2402
First published on 18th October 2011
Abstract
Functionalized calcium phosphate (hydroxyapatite) nanorods were freeze-dried in the presence of the cryoprotectant trehalose, giving a storable and easily redispersible system which can adsorb nucleic acids for transfection and gene silencing. The nanorods were first surface-functionalized with a layer of polyethyleneimine (PEI), purified by ultracentrifugation and redispersion, and freeze-dried in the presence of trehalose. The nanorods can be easily redispersed in water. The concentration-dependent adsorption of nucleic acids (DNA and siRNA) onto the surface of the redispersed cationic nanorods was measured by dynamic light scattering (particle diameter and zeta potential). The transfection efficiency on epithelial cells (HeLa) and osteoblasts (MG-63) was systematically determined for increasing amounts of added nucleic acid, up to a charge reversal by the anionic nucleic acids. Both transfection and gene silencing efficiency increased with increasing amount of nucleic acid, but went through minimum around the point of zero charge where the particles agglomerated. The application of an additional outer layer of polyethyleneimine around the hydroxyapatite/PEI/nucleic acid-nanoparticles reversed the charge back to positive, resulting in even higher transfection rates with almost complete cell viability.
Introduction
Calcium phosphate (CaP) nanoparticles are biocompatible and therefore suitable as drug-delivery systems, especially for the transfection of cells with nucleic acids.1–5 This method can be used to turn on (transfection, DNA) or to turn off (gene silencing, siRNA) the production of a specific protein.6,7 A major challenge is the reproducibility of the preparation of such nanoparticulate systems and their storability. Typically, colloidal dispersions of calcium phosphate tend to agglomerate and to precipitate after a few days or weeks.8 Cellular delivery is still the major challenge in non-viral gene therapy.9–11Here we report about the extension of our earlier work where we have coated hydroxyapatite nanorods with DNA and converted them into a paste that can be used for transfection.12 The transfection efficiency was low, probably due to DNA damage after ultrasonication for redispersion. We have now dispersed the nanorods by a cationic polymer poly(ethyleneimine) (PEI) to obtain positively charged hydroxyapatite nanorods. These nanorods adsorbed DNA or siRNA on their surface and can then be used as transfection systems in vitro. A similar approach was used for gold nanoparticles, which are biocompatible, but not biodegradable.13–16 The adsorption of nucleic acids onto the positively charged surface of calcium phosphate avoids the damage of DNA by ultrasonication during redispersion.
To enhance the storability of these dispersions, they were freeze-dried in the presence of trehalose. This is a versatile system for the storage of hydroxyapatite nanorods that can adsorb any kind of nucleic acid for cellular transport after redispersion in water.
Experimental
Synthesis of cationic CaP–PEI nanorods
1 g of a 6% dispersion of Ostim® (aap, Dieburg, Germany) (dispersion A) in pure water containing 60 mg of hydroxyapatite nanorods12 was diluted with pure water to 20 mL and stirred for 1 min, giving dispersion B. 2 mL containing 6 mg hydroxyapatite of this dispersion were then added to 18 mL of a polyethyleneimine solution (PEI, cationic; Aldrich, molecular weight 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) with a concentration of 2 g L−1. This dispersion C was ultrasonically dispersed for 3 min (Hielscher UP50H, sonotrode 7, 50% amplitude, pulse 0.8). The polymer excess of this dispersion was removed by ultracentrifugation for 30 min at 66
000 g mol−1) with a concentration of 2 g L−1. This dispersion C was ultrasonically dispersed for 3 min (Hielscher UP50H, sonotrode 7, 50% amplitude, pulse 0.8). The polymer excess of this dispersion was removed by ultracentrifugation for 30 min at 66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. The centrifuged nanoparticles were then redispersed in 20 mL of pure water for 2 min under ultrasonication, giving dispersion D. For freeze-drying, 10 mg of D-(+)-trehalose dihydrate (Aldrich) was added to 1 mL of the cationic dispersion D which was then frozen under liquid nitrogen. This system was then lyophilized for 20 h at 0.31 mbar and −7 °C.
000g. The centrifuged nanoparticles were then redispersed in 20 mL of pure water for 2 min under ultrasonication, giving dispersion D. For freeze-drying, 10 mg of D-(+)-trehalose dihydrate (Aldrich) was added to 1 mL of the cationic dispersion D which was then frozen under liquid nitrogen. This system was then lyophilized for 20 h at 0.31 mbar and −7 °C.
Synthesis of CaP–PEI–DNA nanorods
For the loading of the nanoparticles with DNA, 1 mL of the cationic dispersion D was mixed with 6.25 μL, 12.5 μL, 18.8 μL, 25 μL, 31.3 μL, 37.5 μL, 62.5 μL, 75 μL, 125 μL, 150 μL, 200 μL, 250 μL, or 350 μL of a DNA solution, respectively. For the physicochemical characterization of the nanoparticles, model DNA (crude oligonucleotides from herring sperm, <50 bp, 1 mg mL−1, pH ≈ 7.7; Sigma) was used. For the transfection experiments, functional plasmid DNA was used (see below).The DNA concentration in each sample after addition of x μL of DNA solution was w(DNA) = x/(1 + x/1000) μg mL−1, e.g., w(DNA) = 350/(1 + 350/1000) = 259 μg mL−1 for the addition of 350 μL DNA solution. As each mL of dispersion D contained 300 μg hydroxyapatite, the effective concentration of hydroxyapatite after DNA addition was w(hydroxyapatite) = 300/(1 + x/1000) μg mL−1, i.e. 222 μg mL−1 for the addition of 350 μL DNA solution. The mass ratio between DNA and hydroxyapatite was w(DNA)/w(hydroxyapatite) = x/300, i.e. 350/300 = 1.17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for the addition of 350 μL DNA solution.
1 for the addition of 350 μL DNA solution.
For the transfection experiments, the cationic nanoparticles were mixed with the same amounts as above with pcDNA3-EGFP solution (1 mg mL−1). Plasmid DNA (pcDNA3-EGFP) coding for enhanced green fluorescent protein (EGFP) was prepared from Escherichia coli using Nucleobond endotoxin free plasmid DNA (Macherey-Nagel, Dueren, Germany).
Synthesis of CaP–PEI–siRNA nanorods
For the gene silencing experiments, 1 mL of the cationic dispersion D was mixed with 1.28 μL, 2.55 μL, 3.84 μL, 5.10 μL, 6.39 μL, 7.65 μL, 10.2 μL, 12.8 μL, 15.3 μL, 25.5 μL, 30.6 mL, 40.8 μL, 51.0 μL or 71.4 μL of siRNA (350 μM; 4.9 mg mL−1), respectively. Desalted, double-stranded anti-EGFP siRNA from Invitrogen (Paisley, UK), sense, 5′-GCAAGCUGACCCUGAAGUUCAU-3′ and antisense, 5′-AUGAACUUCAGGGUCAGCUUGC-3′, was used (M = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 021.4 g mol−1).
021.4 g mol−1).
The siRNA concentration in each sample after addition of x μL of siRNA solution was w(siRNA) = 4.9x/(1 + x/1000) μg mL−1, e.g. w(siRNA) = 4.9 × 71.4/(1 + 71.4/1000) = 350/1.0714 = 327 μg mL−1 for the addition of 71.4 μL siRNA solution. As each mL of dispersion D contained 300 μg hydroxyapatite, the effective concentration of hydroxyapatite after siRNA addition was w(hydroxyapatite) = 300/(1 + x/1000) μg mL−1, i.e. 280 μg mL−1 for the addition of 71.4 μL siRNA solution. The mass ratio between DNA and hydroxyapatite was w(DNA)/w(hydroxyapatite) = 4.9x/300, i.e. 350/300 = 1.17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for the addition of 71.4 μL siRNA solution.
1 for the addition of 71.4 μL siRNA solution.
Synthesis of cationic CaP–PEI–(DNA or siRNA)–PEI nanorods
Nanoparticles with a core of hydroxyapatite, a first layer of PEI, a second layer of DNA, and a third layer of PEI were synthesized by adding 108 μL of pcEGFP-DNA (1 mg mL−1) to 600 μL of dispersion D. After 30 min, the charge was reversed by the addition of 40 μL of PEI. The resulting concentration of hydroxyapatite was 300 × 600/748 = 240 μg mL−1, and the resulting concentration of DNA was 108/0.748 = 144 μg mL−1. The resulting amount of DNA was 5.4 μg per well.Nanoparticles with a core of hydroxyapatite, a first layer of PEI, a second layer of siRNA, and a third layer of PEI were synthesized by adding 10 μL of siRNA (4.9 mg mL−1) to 300 μL of dispersion D. After 30 min, the charge was reversed by the addition of 20 μL of PEI. The resulting concentration of hydroxyapatite was 300 × 300/330 = 273 μg mL−1, and the resulting concentration of siRNA was 4.9 × 10/0.330 = 149 μg mL−1. The amount of siRNA was 5.55 μg per well.
Characterisation
The particles were characterised by thermogravimetric analysis (STA 409 EP Netzsch; heating from 30 to 1200 °C with 1 K min−1 in a dynamic air atmosphere). Scanning electron microscopy coupled with energy-dispersive X-ray spectroscopy (SEM) was carried out with an ESEM Quanta 400 FEG instrument (FEI, gold–palladium [80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20]-sputtered samples). Dynamic light scattering (DLS) and zeta potential determinations were carried out with a Zetasizer nanoseries instrument (Malvern Nano-ZS, laser: λ = 532 nm). Freeze-drying (lyophilization) was performed with a Christ, Alpha 2-4 LSC instrument.
20]-sputtered samples). Dynamic light scattering (DLS) and zeta potential determinations were carried out with a Zetasizer nanoseries instrument (Malvern Nano-ZS, laser: λ = 532 nm). Freeze-drying (lyophilization) was performed with a Christ, Alpha 2-4 LSC instrument.
Cell culture
HeLa (an epithelial cell line), MG-63 (a human osteoblastic cell line) and HeLa-EGFP (a genetically modified transformed cervix epithelial cell line expressing enhanced green fluorescent protein, EGFP) cells were transfected according to the protocols reported earlier.12,17,18 HeLa and MG-63 cells were cultivated in DMEM media, supplemented with 10% fetal bovine serum (FBS), 100 U mL−1penicillin, and 100 U mL−1streptomycin at 37 °C in humidified atmosphere with 5% CO2. The HeLa-EGFP cells were cultured in DMEM supplemented with 10% FBS and the antibiotic G418-sulfate (final concentration in medium 50 mg mL−1) at 37 °C under 10% CO2 atmosphere. Approximately 12 h before the transfection, the cells were trypsinized and seeded in 24-well plates with a density of 5 × 104cells per well.For the transfection with DNA-functionalized nanoparticles or for gene silencing with siRNA-functionalized nanoparticles, 500 μL of cell culture medium were mixed with 40 μL of the nanorod dispersion. Then, 500 μL of this mixture were added to the cells.
For transfection with PolyFect®, either 2 μg DNA or 4.9 μg siRNA were dissolved in 100 μL medium without FBS to which 22 μL PolyFect® solution were added. 5 min after mixing, 800 μL of the cell culture medium were added. The culture medium of the cells was removed, and 400 μL of fresh cell culture medium and 200 μL of the transfection mixture were used per well. The amount of DNA per well was 0.43 μg and that of siRNA was 1.07 μg per well.
Transfection with Lipofectamine™ 2000 was performed as recommended by the manufacturer in 24-well plates. The duration of the transfection was 7 h. After that, the transfection medium was replaced by fresh cell culture medium. The amount of DNA per well was 1.0 μg and that of siRNA was 0.279 μg per well.
The transfection efficiency was determined approximately 48–72 h after transfection by transmission light microscopy and fluorescence microscopy (Carl Zeiss MicroImaging, Göttingen, Germany; magnification 200×). The transfection efficiency of the DNA-functionalized nanorods was calculated by the ratio of the fluorescing cells (in which EGFP was expressed) to the total examined number of cells.
The efficiency of gene silencing with the siRNA-functionalized nanorods was computed as:19
| [(percentage of not fluorescing cells after transfection) − (percentage of not fluorescing cells in the control)]/(percentage of fluorescing cells in the control) × 100%. |
The cell viability was analyzed by an MTT assay 48–72 h after the transfection. MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma, Taufkirchen, Germany) was dissolved in PBS (5 mg mL−1) and then diluted to 1 mg mL−1 in the cell culture medium. The cell culture medium of the transfected cells was replaced by 300 μL of the MTT solution and incubated for 1–2 h at 37 °C under 5% CO2 in humidified atmosphere. 300 μL DMSO were added to the cells. After 30 min, a 100 μL aliquot was taken for spectrophotometric analysis with a Multiscan FC (Thermo Fisher Scientific, Vantaa, Finland) at λ = 570 nm. The absorption of transfected cells was normalized to that of control (untransfected) cells, thereby indicating the relative level of cell death.
Results and discussion
Cationic hydroxyapatite–PEI nanorods
Hydroxyapatite nanorods were used as starting material. The unfunctionalized hydroxyapatite nanorod dispersion (6%) contained high amounts of agglomerates as shown by dynamic light scattering. These agglomerates were dispersed by adding PEI, followed by 3 min ultrasonication, and excess polymer was removed by ultracentrifugation and redispersion. Dynamic light scattering showed a dispersion with an average particle size of 190 nm without any agglomerates (see below, Fig. 7) and electrosteric stabilization (zeta potential +45 mV). Scanning electron micrographs showed nanorods with an average length of 100–200 nm (Fig. 1). The particles contained about 6 wt% of PEI as determined by thermogravimetry.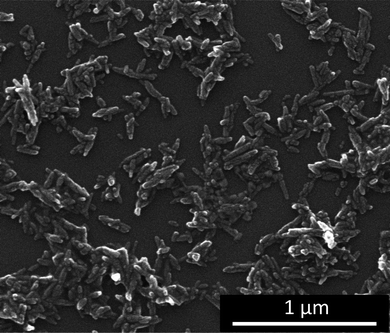 | ||
| Fig. 1 Scanning electron micrograph of hydroxyapatite nanorods after functionalization with polyethyleneimine (PEI). | ||
Adsorption of DNA and siRNA onto hydroxyapatite/PEI nanorods
To examine the adsorption capacity of the cationic nanorods, we added different amounts of DNA to the dispersion and studied the dispersions by dynamic light scattering. The negatively charged DNA changed the cationic character of the nanoparticles as shown by the decreasing zeta potential. Fig. 2 shows the colloidal stability of the nanorod dispersion during the addition of DNA. The cationic and colloidally stable character of the PEI-functionalized nanorods was preserved up to 50 μg mL−1nucleic acid. Upon adding higher amounts of nucleic acids (60–125 μg mL−1) the colloidal system collapsed due to bridging agglomeration. The resulting zeta potential in this range was around zero, i.e. the electrostatic stabilization was absent. If even higher amounts (150–350 μg mL−1) of nucleic acids were rapidly added, they adsorbed onto the cationic surface, reversed the particle charge and formed an outer shell as obvious from the negative zeta potential. This shell led to a colloidally stable system which was negatively charged and electrostatically stabilized in aqueous media, according to the concept of layer-by-layer stabilization.20–22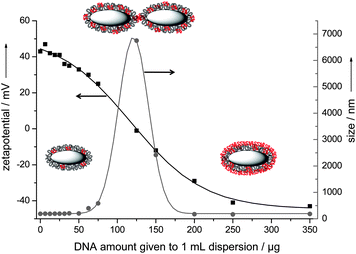 | ||
| Fig. 2 Change of colloid–chemical characteristics of hydroxyapatite–PEI nanoparticles after the rapid addition of model DNA. Grey: hydroxyapatite core; black: PEI molecules; red: DNA molecules. The arrows indicate the axes to which the data points refer. See the Experimental section for the effective concentrations of hydroxyapatite and DNA for each data point. | ||
Transfection with hydroxyapatite/PEI/DNA nanorods
For the transfection of HeLa cells, different amounts of EGFP-encoding DNA were added to the hydroxyapatite–PEI nanorods (Fig. 3).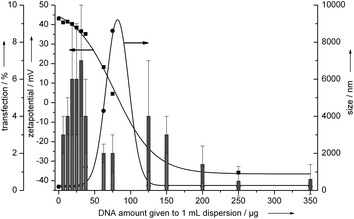 | ||
| Fig. 3 Transfection efficiency of hydroxyapatite–PEI–DNA nanoparticles on HeLa cells (epithelial cells). The vertical bars represent the transfection efficiency. The arrows indicate the axes to which the data points refer. See the Experimental section for the effective concentrations of hydroxyapatite and DNA for each data point. | ||
The transfection efficiency strongly depended on the charge of the particles. The cationic nanorods which contained only a small amount of DNA (left part of Fig. 3) showed higher transfection efficiency (7%) than the nanorods with large amounts of DNA and reversed (negative) charge (right part of Fig. 3). This can be explained with the higher affinity of a cationic delivery system to the negatively charged cell-membrane which leads to a higher endocytotic acceptance.
These transfection experiments were also performed with MG-63 cells (osteoblasts). In this case, a successful transfection of EGFP was only achieved with cationic nanorods, but with a transfection efficiency between 30 and 40% (Fig. 4). The transfection efficiencies were different for the two cell lines, as it is typically observed because cell type has its own susceptibility to transfection and gene silencing.10,23,24 Therefore, we have used a liposomal transfection agent (Lipofectamine™ 2000) and a polycationic transfection agent (PolyFect®) as control agents. With Lipofectamine, the transfection efficiency was 60 ± 11% for HeLa and 31 ± 8% for MG-63, and with Polyfect, it was 22 ± 6% for HeLa and 14 ± 3% for MG-63.
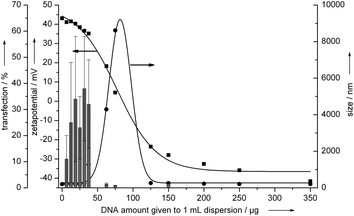 | ||
| Fig. 4 Transfection efficiency of hydroxyapatite–PEI–DNA nanoparticles on MG-63 cells (osteoblasts). The vertical bars represent the transfection efficiency. The arrows indicate the axes to which the data points refer. See the Experimental section for the effective concentrations of hydroxyapatite and DNA for each data point. | ||
The optimum transfection efficiency was observed for a partial coverage of the surface of the hydroxyapatite/PEI nanorods, i.e. for a situation where the zeta potential was still positive.
Gene silencing with hydroxyapatite/PEI/siRNA nanorods
For gene silencing experiments it was found that the knockdown efficiency of the cationic nanorods was in the range of 30–40% (Fig. 5) and comparable to the anionic triple shell CaP/siRNA/CaP/siRNA nanoparticles described earlier.19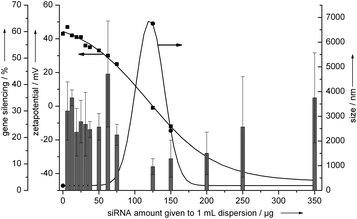 | ||
| Fig. 5 Gene silencing efficiency of hydroxyapatite–PEI–siRNA nanoparticles on HeLa-EGFP cells. The vertical bars represent the gene silencing efficiency. The arrows indicate the axis to which the data points refer. See the Experimental section for the effective concentrations of hydroxyapatite and DNA for each data point. | ||
In all cases the viability of the transfected cells with the nanorods was between 80 and 100% (Fig. 6). In the case of the liposomal (Lipofectamine™ 2000) and polycationic (PolyFect®) transfection agents, the viability of the cells was 30% and 80%, respectively, in accordance with earlier reports on the cell toxicity of such compounds.25 This underscores the high biocompatibility of the nanoparticles, despite the presence of polyethyleneimine.18
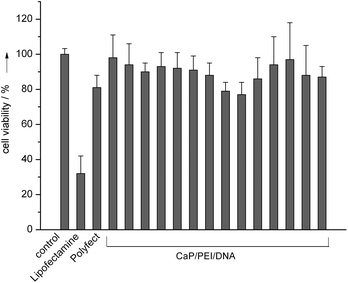 | ||
| Fig. 6 HeLa cell vitality after transfection with CaP/PEI/EGFP-DNA nanoparticles. The individual data points refer to the experiments shown in Fig. 3 from left to right, i.e. from 6.25 to 350 μg added DNA (see the Experimental section for details). | ||
Lyophilization of hydroxyapatite/PEI nanorods
To enhance the storability of nanoparticles in aqueous media, such dispersions can be freeze-dried (lyophilized) in the presence of cryoprotectant trehalose.26–30 This cryoprotectant is biocompatible and able to protect nanoparticles from agglomeration after redispersion. It was also observed that trehalose can increase the transfection efficiency of liposomal systems.31–33 By adding 10 mg of trehalose dihydrate to 1 mL of the cationic nanorod dispersion (hydroxyapatite-PEI), the resulting sugar cake after lyophilization was easily redispersible in water without any loss of the colloidal character (Fig. 7).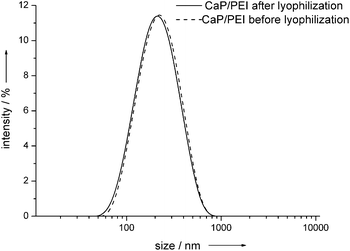 | ||
| Fig. 7 Dynamic light scattering of hydroxyapatite–PEI nanoparticles before and after lyophilization in the presence of trehalose. | ||
The transfection efficiency of the freeze-dried redispersions was the same as that of the native dispersions within the error of the experiment (data not shown). It is notable that earlier observations on lyophilized liposomes ascribed a beneficial effect to trehalose.31–33 The cell-viability after transfection with freeze-dried dispersions was also between 80 and 90% (data not shown).
Charge reversal: addition of an outer layer of PEI
To enhance the cellular uptake, the charge of the cationic nanoparticles was reversed by DNA or siRNA, followed by an additional layer of polyethyleneimine (Fig. 8). The size distribution of the nanoparticles was determined by dynamic light scattering, showing a monodisperse character between 190 and 210 nm. The charge reversal after the application of each layer was proven by zeta potential measurements.34 The value for the PEI-stabilized nanorods was +38 mV, after adding DNA it was −38 mV, and after adding PEI it was +28 mV. This positively charged carrier system led to excellent transfection efficiencies of about 50% for EGFP transfection (HeLA cells) and about 60% for gene silencing (anti-EGFP on HeLa-EGFP-cells; Fig. 9 and 10). This clearly demonstrates the effect of the particle charge for the cellular uptake of nanoparticles: With almost constant amount of applied DNA, these cationic nanoparticles showed much higher transfection ability than comparable negatively charged nanoparticles (particles on the right parts of Fig. 3, 4 and 5). The cell viability was between 80 and 90% for these nanoparticles (data not shown), i.e. the additional amount of PEI was not harmful for the cells.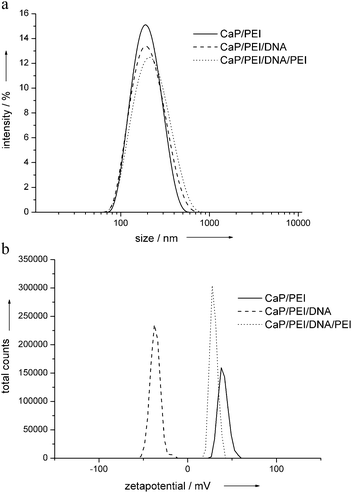 | ||
| Fig. 8 The addition of another layer of polyethyleneimine onto hydroxyapatite–PEI–DNA nanorods leads to a positive particle charge. Top: particle size; bottom: zeta potential. Model DNA was used (see the Experimental section). | ||
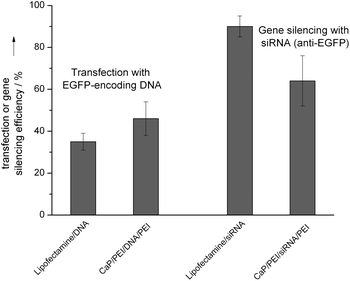 | ||
| Fig. 9 Transfection efficiencies (HeLa cells) and gene silencing efficiencies (HeLa-EGFP cells) with cationic hydroxyapatite/PEI/DNA/PEI nanorods in comparison with the liposomal transfection agent Lipofectamine. | ||
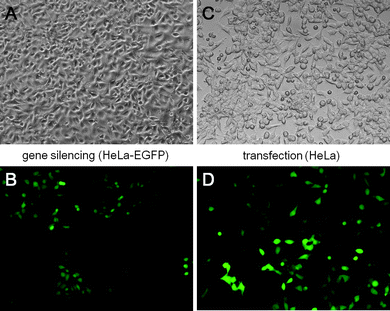 | ||
| Fig. 10 Transmission light microscopy (top) and fluorescence microscopy (bottom) of HeLa-EGFP cells after gene silencing with cationic CaP/PEI/siRNA/PEI nanoparticles (A and B) and of HeLa cells after transfection with CaP/PEI/DNA/PEI nanoparticles (C and D) (magnification in all cases: 200×). | ||
Conclusions
We have developed a biocompatible and cationic delivery system based on hydroxyapatite nanorods for the transfection of cells with different nucleic acids. The hydroxyapatite/PEI nanorods can be freeze-dried in the presence of trehalose and stored. Before use, they can be easily redispersed in water and loaded either with DNA for transfection or with siRNA for gene silencing. The transfection efficiencies are comparable to liposomal or cationic transfection agents, but the cell viability after transfection is almost 80 to 90%, in contrast to the liposomal transfection agent. Thus, the system is applicable for the delivery of nucleic acids into cells in gene therapy.Acknowledgements
We thank the Bundesministerium for Bildung und Forschung (BMBF) for financial support of the project no. 13N10035/BIONANOPLANT. We thank aap Biomaterials GmbH, Dieburg, for the provision of the hydroxyapatite nanorod dispersion (OSTIM®).Notes and references
- V. Uskokovic and D. P. Uskokovic, J. Biomed. Mater. Res., Part B, 2011, 96, 152–191 CrossRef.
- D. Olton, J. Li, M. E. Wilson, T. Rogers, J. Close, L. Huang, N. P. Kumta and C. Sfeir, Biomaterials, 2007, 28, 1267–1279 CrossRef CAS.
- R. Gonzalez-McQuire, D. W. Green, K. A. Partridge, R. O. C. Oreffo, S. Mann and S. A. Davis, Adv. Mater., 2007, 19, 2236–2240 CrossRef CAS.
- A. Maitra, Expert Rev. Mol. Diagn., 2005, 5, 893–905 CrossRef CAS.
- I. Roy, S. Mitra, A. Maitra and S. Mozumdar, Int. J. Pharm., 2003, 250, 25–33 CrossRef CAS.
- M. Epple, K. Ganesan, R. Heumann, J. Klesing, A. Kovtun, S. Neumann and V. Sokolova, J. Mater. Chem., 2010, 20, 18–23 RSC.
- V. Sokolova and M. Epple, Angew. Chem., Int. Ed., 2008, 47, 1382–1395 CrossRef CAS.
- V. Sokolova, O. Prymak, W. Meyer-Zaika, H. Cölfen, H. Rehage, A. Shukla and M. Epple, Materialwiss. Werkstofftech., 2006, 37, 441–445 CrossRef CAS.
- D. Reischl and A. Zimmer, Nanomedicine, 2009, 5, 8–20 CAS.
- M. Morille, C. Passirani, A. Vonarbourg, A. Clavreul and J. P. Benoit, Biomaterials, 2008, 29, 3477–3496 CrossRef CAS.
- Z. Racz and P. Hamar, Curr. Med. Chem., 2006, 13, 2299–2307 CrossRef CAS.
- J. Klesing, S. Chernousova, A. Kovtun, S. Neumann, L. Ruiz, J. M. Gonzalez-Calbet, M. Vallet-Regi, R. Heumann and M. Epple, J. Mater. Chem., 2010, 20, 6144–6148 RSC.
- D. A. Giljohann, D. S. Seferos, W. L. Daniel, M. D. Massich, P. C. Patel and C. A. Mirkin, Angew. Chem., Int. Ed., 2010, 49, 3280–3294 CAS.
- N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. R. Lytton-Jean, M. S. Han and C. A. Mirkin, Science, 2006, 312, 1027–1030 CrossRef CAS.
- A. C. Bonoiu, S. D. Mahajan, H. Ding, I. Roy, K. T. Yong, R. Kumar, R. Hu, E. J. Bergey, S. A. Schwartz and P. N. Prasad, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 5546–5550 CrossRef CAS.
- H. Ding, K. T. Yong, I. Roy, H. E. Pudavar, W. C. Law, E. J. Bergey and P. N. Prasad, J. Phys. Chem. C, 2007, 111, 12552–12557 CAS.
- V. Sokolova, S. Neumann, A. Kovtun, S. Chernousova, R. Heumann and M. Epple, J. Mater. Sci., 2010, 45, 4952–4957 CrossRef CAS.
- S. Neumann, A. Kovtun, I. D. Dietzel, M. Epple and R. Heumann, Biomaterials, 2009, 30, 6794–6802 CrossRef CAS.
- V. Sokolova, A. Kovtun, O. Prymak, W. Meyer-Zaika, E. A. Kubareva, E. A. Romanova, T. S. Oretskaya, R. Heumann and M. Epple, J. Mater. Chem., 2007, 17, 721–727 RSC.
- B. G. de Geest, G. B. Sukhorukov and H. Mohwald, Expert Opin. Drug Delivery, 2009, 6, 613–624 CrossRef CAS.
- T. Boudou, T. Crouzier, K. Ren, G. Blin and C. Picart, Adv. Mater., 2010, 22, 441–467 CrossRef CAS.
- W. Tong and C. Gao, J. Mater. Chem., 2008, 18, 3799 RSC.
- K. Kodama, Y. Katayama, Y. Shoji and H. Nakashima, Curr. Med. Chem., 2006, 13, 2155–2161 CrossRef CAS.
- I. R. Gilmore, S. P. Fox, A. J. Hollins and S. Akhtar, Curr. Drug Delivery, 2006, 3, 147–155 CrossRef CAS.
- H. T. Lv, S. B. Zhang, B. Wang, S. H. Cui and J. Yan, J. Controlled Release, 2006, 114, 100–109 CrossRef CAS.
- E. C. A. van Winden and D. J. A. Crommelin, J. Controlled Release, 1999, 58, 69–86 CrossRef CAS.
- P. Sundaramurthi and R. Suryanarayanan, J. Phys. Chem. Lett., 2010, 1, 510–514 CrossRef CAS.
- Y. Aso and S. Yoshioka, Chem. Pharm. Bull., 2005, 53, 301–304 CrossRef CAS.
- T. J. Anchordoquy, J. F. Carpenter and D. J. Kroll, Arch. Biochem. Biophys., 1997, 348, 199–206 CrossRef CAS.
- S. D. Allison and T. J. Anchordoquy, J. Pharm. Sci., 2000, 89, 682–691 CrossRef CAS.
- W. C. Tseng, C. H. Tang, T. Y. Fang and L. Y. Su, Biotechnol. Prog., 2007, 23, 1297–1304 CrossRef CAS.
- W. C. Tseng, C. H. Tang, T. Y. Fang and L. Y. Su, J. Gene Med., 2007, 9, 659–667 CrossRef CAS.
- T. Ahn and C. H. Yun, Anal. Biochem., 2004, 333, 199–200 CrossRef CAS.
- J. Schwiertz, W. Meyer-Zaika, L. Ruiz-Gonzalez, J. M. Gonzales-Calbet, M. Vallet-Regi and M. Epple, J. Mater. Chem., 2008, 18, 3831–3834 RSC.
| This journal is © The Royal Society of Chemistry 2012 |
