Fluorescent probes for superresolution imaging of lipid domains on the plasma membrane†
Hideaki
Mizuno
*ab,
Mitsuhiro
Abe
c,
Peter
Dedecker‡
b,
Asami
Makino
c,
Susana
Rocha
b,
Yoshiko
Ohno-Iwashita
d,
Johan
Hofkens
b,
Toshihide
Kobayashi
c and
Atsushi
Miyawaki
a
aCell Function and Dynamics, Brain Science Institute, RIKEN, 2-1 Hirosawa, Wako-city, Saitama 351-0198, Japan
bDepartment of Chemistry, Katholieke Universiteit Leuven, Celestijnenlaan 200F, 3001, Heverlee, Belgium. E-mail: hideaki.mizuno@chem.kuleuven.be; Fax: +32 16 32 79 90; Tel: +32 16 32 73 99
cLipid Biology Laboratory, RIKEN Advanced Science Institute, 2-1 Hirosawa, Wako-city, Saitama 351-0198, Japan
dFaculty of Pharmacy, Iwaki Meisei University, 5-5-1 Chuodai Iino, Iwaki City, Fukushima 970-8551, Japan
First published on 27th May 2011
Abstract
Accumulating evidence indicates that membrane lipids are not randomly distributed but rather form specific domains. In particular, raft-like microdomains composed of cholesterol and sphingolipids are attracting a lot of attention. These microdomains are thought to serve as platforms for signal transduction and molecular trafficking, but it is difficult to elucidate their detailed structure since their reported size is smaller than the resolution of light microscopy. To circumvent this limitation, we designed probes for cholesterol- and sphingolipid-enriched microdomains dedicated for superresolution microscopy, PALM. The probes utilise the affinity of the toxins, θ-toxin and lysenin, for the cholesterol- and sphingomyelin-enriched membranes, respectively. The toxicity can be avoided by using non-toxic domains that retain the specific binding to the aforementioned membranes. The probes can easily be produced in E. coli as recombinant protein domains of toxins fused to a photoswitchable fluorescent protein, Dronpa. PALM imaging with these probes revealed two types of cholesterol-enriched microdomains, line-shaped ones with widths of around 150 nm and round ones with an average radius of 118 nm. All sphingomyelin-enriched microdomains were round with an average radius of 124 nm. Both the cholesterol- and sphingomyelin-enriched microdomains vanished by the depletion of cholesterol. The sphingomyelin-enriched microdomains also vanished by the depletion of sphingomyelin whereas the cholesterol-enriched microdomains were unaffected. We conclude that cholesterol- and sphingomyelin-enriched domains occupy different regions on the plasma membrane.
Introduction
The plasma membrane is composed of a heterogeneous distribution of a variety of lipids and membrane proteins. Cholesterol and sphingolipids form specific lipid complexes in the plasma membrane, resulting in the formation of lipid microdomains often called membrane rafts. These microdomains are thought to provide platforms for protein interaction allowing protein trafficking and cell signaling.1 The size estimates of the microdomains range from a few nanometres to a few hundreds of nanometres.2 Diffraction-limited fluorescence microscopy has been used to visualize and study the microdomains,3,4 but was unable to elucidate their detailed structure since their apparent size is below the diffraction limit of light.Photoactivation localization microscopy (PALM)5,6 and stochastic optical reconstruction microscopy (STORM)7 are novel fluorescence microscopy modalities to circumvent the diffraction limit (superresolution microscopy). We considered these techniques ideal methods to analyze the distribution and clustering of molecules on the plasma membrane. So far clustering of several proteins, hemagglutinin from the influenza virus, T cell receptor-pathway kinase Lck, Src kinase, T cell antigen receptor and linkers for activation of T cells have been observed on the plasma membrane by PALM/STORM,8 but the lipid components themselves have not been observed with these new forms of microscopy. Stimulated emission depletion microscopy (STED)9 in combination with fluorescence correlation microscopy (FCS-STED) has been used recently to determine the size of specific lipid nanodomains via the diffusion of labeled sphingolipid analogs,10 but did not image these structures directly.
To visualize clustering of the membrane components by PALM, we made probes for cholesterol- and sphingomyelin-enriched regions on the plasma membrane. These probes are composed of a photochromic fluorescent protein, Dronpa11,12 and two different protein toxins. θ-toxin binds to cholesterol on the plasma membrane and disrupts cells.13 The C-terminal domain (θ-D4) is sufficient for the binding to cholesterol but does not cause cellular damage.14Lysenin specifically binds to sphingomyelin and induces cytolysis.15 N-terminal truncated lysenin (NT-Lys) binds to sphingomyelin but does not cause cell death.4 With these probes, we performed superresolution imaging of cholesterol- and sphingomyelin-enriched microdomains on the plasma membrane of HeLa cells.
Results and discussion
Design and evaluation of a probe for cholesterol-enriched membrane domains
We constructed a gene encoding θ-D4 fused to Dronpa on a bacterial expression vector, pET28 (Fig. 1). The plasmid contains T7 promoter accompanied with the lac operator at the upstream of the gene encoding Dronpa-θ-D4. With this plasmid, we transformed an E. coli strain with λ(DE3), BL21(DE3), and induced the production of recombinant Dronpa-θ-D4 by addition of isopropyl-β-D-thiogalactopyranoside (IPTG) at the logarithmic phase of bacterial growth.§ The recombinant Dronpa-θ-D4 was connected to a histidine-tag on its N-terminus, which makes it possible to isolate the recombinant protein from the bacterial lysate by metal chelate affinity chromatography. | ||
| Fig. 1 Constructs of plasmid encoding the dedicated PALM probes targeted for cholesterol- or sphingomyelin-enriched membrane microdomains. The gene encoding the C-terminal domain of θ toxin (θ-D4) or N-terminus truncated lysenin (NT-Lys) was fused to Dronpa and constructed to the multiple cloning site of pET28, in frame to the histidine-tag and thrombin cleavage site (His6-throm) of the vector. The plasmid has the T7 promoter (T7pro), lac operator (lacO), and ribosome binding site (rbs) at the upstream of the gene encoding the fusion protein, and the T7 terminator (T7ter) at its downstream. | ||
Dronpa-θ-D4 was shown to bind to multilamellar vesicles (MLVs) composed of cholesterol/phosphatidylcholine (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), but not to MLVs composed of only phosphatidylcholinevia centrifugal sedimentation followed by Western blotting (Fig. 2A). An enzyme-linked immunosorbent assay (ELISA) revealed that the binding of Dronpa-θ-D4 to lipids was specific to cholesterol (Fig. 2C). Incubation of HeLa cells with Dronpa-θ-D4 resulted in stained cells as visualized in Fig. 2E. Dronpa-θ-D4 lost its ability to stain the cells by 30 min pre-incubation with the MLVs composed of phosphatidylcholine/cholesterol (1
1), but not to MLVs composed of only phosphatidylcholinevia centrifugal sedimentation followed by Western blotting (Fig. 2A). An enzyme-linked immunosorbent assay (ELISA) revealed that the binding of Dronpa-θ-D4 to lipids was specific to cholesterol (Fig. 2C). Incubation of HeLa cells with Dronpa-θ-D4 resulted in stained cells as visualized in Fig. 2E. Dronpa-θ-D4 lost its ability to stain the cells by 30 min pre-incubation with the MLVs composed of phosphatidylcholine/cholesterol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (Fig. 2E). Dronpa-θ-D4 did not stain cholesterol-depleted HeLa cells which were prepared by 30 min incubation with methyl-β-cyclodextrin (MβCD) (Fig. 2F). It is known that θ-toxin binds only to cholesterol-enriched regions, with negligible binding to regions containing less than 20 mol% cholesterol.14 Hence Dronpa-θ-D4 can probe only cholesterol-enriched domains but not dispersed cholesterol.13 The probe does not display self-assembly, since it does not contain domains 1 and 3 of θ-toxin, which are essential for self-assembling and pore formation, respectively.16 Since Dronpa is a monomeric fluorescent protein, it is unlikely to induce clustering. Taking into account the above-mentioned properties, Dronpa-θ-D4 is clearly a suitable probe for the investigation of cholesterol-enriched membrane domains with PALM.
1) (Fig. 2E). Dronpa-θ-D4 did not stain cholesterol-depleted HeLa cells which were prepared by 30 min incubation with methyl-β-cyclodextrin (MβCD) (Fig. 2F). It is known that θ-toxin binds only to cholesterol-enriched regions, with negligible binding to regions containing less than 20 mol% cholesterol.14 Hence Dronpa-θ-D4 can probe only cholesterol-enriched domains but not dispersed cholesterol.13 The probe does not display self-assembly, since it does not contain domains 1 and 3 of θ-toxin, which are essential for self-assembling and pore formation, respectively.16 Since Dronpa is a monomeric fluorescent protein, it is unlikely to induce clustering. Taking into account the above-mentioned properties, Dronpa-θ-D4 is clearly a suitable probe for the investigation of cholesterol-enriched membrane domains with PALM.
 | ||
Fig. 2 Characterization of photoswitchable fluorescent probes for lipid-enriched membrane microdomains. A) and B) Binding of probes to multilamellar vesicles (MLVs) composed of phosphatidylcholine/cholesterol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (PC/chol), only phosphatidylcholine (PC), or sphingomyelin/cholesterol (1 1) (PC/chol), only phosphatidylcholine (PC), or sphingomyelin/cholesterol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (SM/chol). Proteins bound to MLVs were analyzed by Western blotting. Dronpa-θ-D4 and Dronpa-NT-Lys were subjected to the analysis, and full-length lysenin served as a control that binds to MLVs and assembles to form oligomers. C) and D) Specificity of Dronpa-θ-D4 (C) and Dronpa-NT-Lys (D) binding to phospholipids analyzed by ELISA. SM, sphingomyelin; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PI phosphatidylinositol; PG, phosphatidylglycerol; GalCer, galactosylceramide; LacCer, lactosylceramide; GM1, monosialotetrahexosylganglioside; Cer, ceramide; Chol, cholesterol. E) and G) Preabsorption experiment of the probes to MLVs. Dronpa-θ-D4 (E) or Dronpa-NT-Lys (G) were incubated with MLVs composed of PC/chol, PC, or SM/chol prior to applying them to HeLa cells for staining. F) and H) Staining of cholesterol- or sphingomyelin-depleted HeLa cells with Dronpa-θ-D4 (F) or Dronpa-NT-Lys (H). Cholesterol or sphingomyelin were depleted by 30 min incubation with MβCD or Sphingomyelinase from Staphylococcus aureus, respectively prior to the staining. Z-projection of confocal images (upper) and DIC images (lower) were shown in E, F, G, and H. Scale bars indicate 10 μm. 1) (SM/chol). Proteins bound to MLVs were analyzed by Western blotting. Dronpa-θ-D4 and Dronpa-NT-Lys were subjected to the analysis, and full-length lysenin served as a control that binds to MLVs and assembles to form oligomers. C) and D) Specificity of Dronpa-θ-D4 (C) and Dronpa-NT-Lys (D) binding to phospholipids analyzed by ELISA. SM, sphingomyelin; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PI phosphatidylinositol; PG, phosphatidylglycerol; GalCer, galactosylceramide; LacCer, lactosylceramide; GM1, monosialotetrahexosylganglioside; Cer, ceramide; Chol, cholesterol. E) and G) Preabsorption experiment of the probes to MLVs. Dronpa-θ-D4 (E) or Dronpa-NT-Lys (G) were incubated with MLVs composed of PC/chol, PC, or SM/chol prior to applying them to HeLa cells for staining. F) and H) Staining of cholesterol- or sphingomyelin-depleted HeLa cells with Dronpa-θ-D4 (F) or Dronpa-NT-Lys (H). Cholesterol or sphingomyelin were depleted by 30 min incubation with MβCD or Sphingomyelinase from Staphylococcus aureus, respectively prior to the staining. Z-projection of confocal images (upper) and DIC images (lower) were shown in E, F, G, and H. Scale bars indicate 10 μm. | ||
Visualization of cholesterol-enriched membrane domains
We stained HeLa cells by 10 min incubation with Dronpa-θ-D4 in order to observe the detailed structure of cholesterol-enriched microdomains on the plasma membrane. Confocal images showed that regions stained with Dronpa-θ-D4 are mainly distributed over the upper surface of the cell (the plasma membrane distant from the bottom) (Fig. 3A). Primary cilia were strongly stained, consistent with the high levels of cholesterol in the ciliary membrane.17 Overall the images appeared as a collection of small dots, the size of which is around the expected diffraction limit calculated according to the Rayleigh criterion (corresponding to 226 nm for emission at 518 nm and an NA value of the objective lens of 1.40).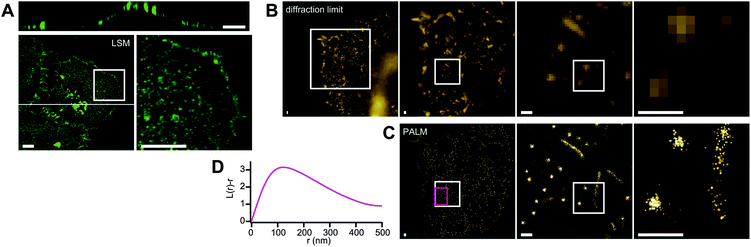 | ||
| Fig. 3 Cholesterol-enriched domains on the plasma membrane of HeLa cells. A) 3D confocal image. Bottom panels show z-projections. The right bottom panel is the expansion of the region indicated with the box on the left. The top panel is an xz-section of the cell at the position indicated with the line on the bottom left. B) Diffraction-limited image. Expansion of the region indicated with the box is shown on the right side of the respective panels. C) PALM image of the same regions as B. The spots representing a detected single molecule switching event, and the size of the respective spots corresponds to the precision of the fitting. D) Representative Ripley's K-function analysis. The analysis was applied at the region indicated with the magenta box on the PALM image. This region contains only round-shaped clusters. We analyzed 12 independent images and obtained essentially same results. Scale bars indicate 10 μm (A) or 500 nm (B and C). | ||
In the PALM image, two types of clustering/nanodomains could be observed (Fig. 3B and C). The first type of clusters appeared as lines with widths of around 150 nm and lengths of about 0.7 to 5.5 μm, which we attributed to microvilli. This is consistent with the facts that the upper surface of HeLa cells are enriched with microvilli,18 and microvilli involve a cholesterol-based lipid raft.19 The other type of clusters manifested themselves as round shapes, which we identified as cholesterol-enriched membrane domains. The average radius of the round structures was determined to be 118 nm by applying Ripley's K-function analysis20 (Fig. 3D).
A crucial parameter in PALM imaging is the precision with which a single fluorescent molecule can be localized. This depends on the number of photons detected from each fluorophore as well as on the signal to noise ratio.5 In Fig. 3 and Fig. 4, the coordinates of individual localized molecules are rendered with a spot size that corresponds to the localization precision, and with higher intensities for more precise localizations. We applied a threshold on the image by including only those localizations with an estimated precision better than a given value. Different threshold levels produced similar images, and in the end we chose a threshold of 18 nm as a good compromise between signal intensity and resolution. This precision is adequate to visualize microdomains with an average radius of 118 nm.
In this experiment, we fixed the cells before the imaging. After fixing, the diffusion coefficient of the probe was determined to be 0.013 μm2s−1 by single particle tracking (Fig. S1, ESI†). This means that even after fixation, the probes are still slightly mobile. The reported diffusion coefficient corresponds to a mean displacement of 46 nm between consecutive frames, which is smaller than the size of microdomains visualized. Furthermore, the trajectories of all molecules were confined in an area of the studied microdomain (Fig. S1 C and D).
Multiple localizations of the same fluorophore are a major and general concern in PALM imaging. In this experiment, we employed Dronpa, which shows reversible photoswitching and thus might appear multiple times in a single experiment. While multiple observations of the same molecule could indeed give rise to apparent clusters with dimensions on the order of the localization precision, the fact that the stained regions are also clearly observable by diffraction-limited microscopy shows that the lipid microdomains have been stained with a large number of individual labels. Therefore the repeated observation of only one or a few labels is very unlikely.
Evaluation of a probe for sphingomyelin-enriched membrane domains
We similarly constructed a gene encoding NT-Lys fused to Dronpa on pET28 and made a recombinant Dronpa-NT-Lys as a probe for sphingomyelin-enriched membrane domains applicable for PALM (Fig. 1). Dronpa-NT-Lys was found to bind to MLVs composed of sphingomyelin/cholesterol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), but not to those composed of phosphatidylcholine/cholesterol (1
1), but not to those composed of phosphatidylcholine/cholesterol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (Fig. 2B). The apparent molecular weight of Dronpa-NT-Lys recovered from the MLVs of sphingomyelin/cholesterol was around 45 kDa, which corresponds to the size of monomeric Dronpa-NT-Lys (41.6 kDa), indicating that Dronpa-NT-Lys remains a monomer after binding to sphingomyelin on the MLVs. This is in sharp contrast with full-length lysenin, which showed a shift in apparent molecular weight to over 250 kDa due to oligomer formation on the MLVs. This is consistent with the report that the N-terminal truncated mutant loses the property of lysenin to oligomerize upon sphingomyelin binding.4 Specific binding of Dronpa-NT-Lys to sphingomyelin was confirmed by ELISA (Fig. 2D). HeLa cells were stained by incubation with Dronpa-NT-Lys (Fig. 2G). Dronpa-NT-Lys lost the ability to stain HeLa cells by preincubation with MLVs composed of sphingomyelin/cholesterol (Fig. 2G). Sphingomyelin-depleted HeLa cells by sphingomyelinase treatment were not stained with Dronpa-NT-Lys (Fig. 2H).
1) (Fig. 2B). The apparent molecular weight of Dronpa-NT-Lys recovered from the MLVs of sphingomyelin/cholesterol was around 45 kDa, which corresponds to the size of monomeric Dronpa-NT-Lys (41.6 kDa), indicating that Dronpa-NT-Lys remains a monomer after binding to sphingomyelin on the MLVs. This is in sharp contrast with full-length lysenin, which showed a shift in apparent molecular weight to over 250 kDa due to oligomer formation on the MLVs. This is consistent with the report that the N-terminal truncated mutant loses the property of lysenin to oligomerize upon sphingomyelin binding.4 Specific binding of Dronpa-NT-Lys to sphingomyelin was confirmed by ELISA (Fig. 2D). HeLa cells were stained by incubation with Dronpa-NT-Lys (Fig. 2G). Dronpa-NT-Lys lost the ability to stain HeLa cells by preincubation with MLVs composed of sphingomyelin/cholesterol (Fig. 2G). Sphingomyelin-depleted HeLa cells by sphingomyelinase treatment were not stained with Dronpa-NT-Lys (Fig. 2H).
Visualization of sphingomyelin-enriched membrane domains
We subjected HeLa cells to staining with Dronpa-NT-Lys by 10 or 60 min of incubation, followed by fixation to observe sphingomyelin-enriched membrane microdomains. As was the case with Dronpa-θ-D4, confocal imaging after 10 min incubation revealed that only the upper cell surface was covered with a collection of fluorescent dots (Fig. 4A and C). During the 60 min incubation, some of the sphingomyelin-enriched domains were endocytosed (Fig. 4B and C), in contrast with the cholesterol-enriched domains which remained on the plasma membrane (Fig. 4D). | ||
| Fig. 4 Sphingomyelin-enriched domains in a HeLa cell. A) and B) Z-projection of confocal images. C) xz-sections of the cell at the position indicated with the white line on A and B. HeLa cells were stained by 10 min (A) or 60 min (B) of incubation with Dronpa-NT-Lys. D) Cholesterol-enriched regions stained with Dronpa-θ-D4 for 60 min. An xz-section of the cell is shown. E) Diffraction limited image of the sphingomyelin-enriched domains of the plasma membrane visualized by 10 min incubation with Dronpa-NT-Lys. The white box shows the region used in the PALM image. F) PALM image of the cell. Expansion of the region indicated with the white box is shown on the right side of respective panels. G) Representative Ripley's K-function analysis at the region of the magenta box on the PALM image. We analyzed 8 independent images and obtained essentially same results. Scale bars indicate 10 μm (A–D) or 500 nm (E and F). | ||
The sphingomyelin-enriched regions stained by 10 min incubation were readily observed on the upper surface by PALM, with an average radius of the domains calculated to be 124 nm (Fig. 4E–G). No microvilli structures were observed. Lysenin selectively binds to sphingomyelin-enriched domains but not to dispersed sphingomyelin,4,15 and thus might not visualize membrane domains with low local sphingomyelin concentration. The difference in shape and number of clusters/domains between cells stained with Dronpa-NT-Lys and Dronpa-θ-D4 together with the difference in endocytotic property and in the effect of the sphingomyelinase treatment strongly suggest that the cholesterol- and sphingomyelin-enriched regions detectable with our probes occupy different places on the plasma membrane, though there might be some overlap. Interestingly no sphingomyelin-enriched regions could be observed after cholesterol depletion although these regions were spatially distinct from the cholesterol-enriched regions (Fig. 2H), pointing out that a small amount of cholesterol might be required for the formation of these domains.
During preparation of this manuscript, a paper reporting the mobility of membrane molecules after chemical fixation was published by Tanaka et al.21 They investigated the mobility of molecules after the fixation by single particle tracking and claimed that some types of membrane molecules were inefficiently immobilized by chemical fixations, and this imperfect immobilization can induce antibody-induced artificial clustering. Notably, neither cholesterol nor sphingolipid were immobilized under any of the conditions they applied. While we have aimed to visualize the cholesterol- and sphingomyelin-enriched microdomains in this study, we believe that the regions we observed were not artificial clusters for the following reasons. First, we stained cholesterol and sphingomyelin with proteinous probes before the fixation. The probes can hence crosslink together as well as with other endogenous proteins in the plasma membrane during the formaldehyde fixation. Secondly, we did not use antibodies to stain the microdomains. We also confirmed that our proteinous probes themselves do not assemble to form artificial oligomers (Fig. 2A and B). Lastly, we confirmed by single particle tracking that the diffusion rate of the probes was slow enough for PALM imaging after the fixation, and that the mobility of the molecules was confined in the studied microdomains (Fig. S1, ESI†).
Experimental
Preparation of lipid domain probes
pET28/Dronpa-θ-D4 expressing Dronpa-θ-D4 was constructed by replacement of EGFP on a plasmid expressing His-EGFP-D414 with Dronpa.11pET28/Dronpa-NT-Lys was generated by replacing θ-D4 on pET28/Dronpa-θ-D4 with NT-Lys.4 Recombinant proteins were expressed in E. coli strain BL21(DE3) and purified using HisTrap FF crude columns (GE Healthcare).ELISA
Binding of Dronpa-θ-D4 and Dronpa-NT-Lys to various lipids was analyzed by ELISA according to a protocol reported elsewhere.15,22 Mouse anti penta-His antibody (Qiagen) and horse radish peroxidase (HRP)-conjugated sheep anti mouse IgG (Amersham) were used as primary and secondary antibodies, respectively.Binding of the probes to MLVs
Phospholipids were completely dried and then resuspended in Dulbecco's phosphate buffered saline (PBS) by vortex to prepare MLVs. The MLVs were incubated with Dronpa-θ-D4 or Dronpa-NT-Lys at 37 °C for 30 min, and then sedimented by centrifugation and subjected for SDS-PAGE followed by Western blotting. The proteins were detected with mouse anti penta-His antibody (Qiagen) as a primary antibody and HRP-conjugated goat anti mouse IgG (Thermo Scientific) as a secondary antibody. Amersham ECL plus (GE Healthcare) was used as a substrate of HRP reaction to emit chemiluminescence. The luminescent image was acquired with a lumino image analyzer LAS-4000 (Fujifilm, Tokyo, Japan).Cell staining
HeLa cells were cultured on a glass base dish (Asahi Techno Glass, Tokyo, Japan) in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. Cells were incubated with PBS containing 3.5% of bovine serum albumin (Gibco) and 0.1 μM of either Dronpa-θ-D4 or Dronpa-NT-Lys for 10 or 60 min at room temperature, washed quickly in ice-cold Hanks’ balanced salt solution (HBSS) (Gibco) for 3 times, and fixed with 4% formaldehyde (Thermo Scientific) in PBS on ice for 1 h.For the preabsorption experiment, 0.1 μM of either Dronpa-θ-D4 or Dronpa-NT-Lys was incubated with 2 mM of MLVs at 37 °C for 30 min prior to apply them to HeLa cells.
Confocal imaging
We used a confocal microscope LSM510 with a Plan-Apochromat 63x/1.40 objective lens (Carl Zeiss).PALM/dSTORM imaging
PALM imaging was done in PBS. Gold colloids (100 nm, BBI international, Cardiff, UK) were loaded to the sample as fiduciary markers. We used a prototype Carl Zeiss PALM microscope with dedicated software. All the PALM data were processed with this software, which has been developed based on the theory reported in the PALM paper by Betzig et al.5 The objective lens used was an αPlan-APOCHROMAT 100x/1.46 (Carl Zeiss). To observe upper surface of the cells, we used highly inclined laminated optical sheet (HILO) illumination23 instead of total internal reflection fluorescence (TIRF). For PALM with Dronpa, we used a 489-nm laser (61 W cm−2) for excitation and turning-off the fluorescence, and relied on the spontaneous recovery for the stochastic turning-on of the fluorescence.24 Only localizations with a precision of 18 nm or better were used for image reconstruction. The pixel size of the reconstructed images corresponds to 5 nm.Cholesterol or sphingomyelin depletion from HeLa cells
HeLa cells were incubated at 37 °C for 30 min in HBSS containing 10 mM of MβCD or 2.5 units of sphingomyelinase from Staphylococcus aureus (Sigma Aldrich) to deplete cholesterol or sphingomyelin, respectively.Conclusions
We have developed and demonstrated probes for cholesterol- and sphingomyelin-enriched membrane regions dedicated to superresolution microscopy. Clustering of lipid molecules in nanodomains was visualized by PALM with a spatial resolution one order of magnitude better than the diffraction limit and a sufficiently dense labeling to resolve the detailed structure of the nanodomains. The concept can be easily extended to other photoswitchable fluorescent proteins such as KikGR, Dendra2, mEOS2, PA-GFP, and PAmCherry1. We envision that the developed probes in combination with superresolution microscopy will be powerful tools in exploring the role and functionality of lipid micro/nano-domains on the plasma membrane.Acknowledgements
We thank Carl Zeiss MicroImaging GmbH for technical support, Drs. R. Ishitsuka, T. Fukano, A. Sakaue-Sawano, Mss. T. Kogure, R. Nakazawa, Y. Ichikawa for fruitful discussion and technical assistance, and Dr R. Neely for careful reading of the manuscript. The authors acknowledge financial support from the Molecular Ensemble Program and Lipid Dynamics Program at RIKEN, the Japan MEXT Grant-in Aid for Scientific Research on priority areas, the Research Foundation Flanders (FWO; Grants G.0402.09 G.0413.10 G.0181.10), the K.U.Leuven Research Fund (GOA2006/2, Center of Excellence INPAC, ‘Inter-Disciplinair Onderzoek’ (IDO/07/010)), the Flemish Government (long term structural funding- Methusalem funding CASAS), the Federal Science Policy of Belgium (IAP-VI/27), and the Hercules foundation (HER/08/021). PD is a postdoctoral fellow of the FWO.Notes and references
- K. Simon and E. Ikonen, Nature, 2000, 387, 569–572 CrossRef CAS; D. A. Brown and E. London, Annu. Rev. Cell Dev. Biol., 1998, 14, 111–136 CrossRef; R. G. Anderson and K. Jacobson, Science, 2002, 296, 1821–1825 CrossRef; S. Mayor and M. Rao, Traffic, 2004, 5, 231–240 CrossRef; L. J. Pike, J. Lipid Res., 2006, 47, 1597–1598 CrossRef; M. F. Hanzal-Bayer and J. F. Hancock, FEBS Lett., 2007, 581, 2098–2104 CrossRef.
- K. Jacobson, O. G. Mouritsen and R. G. Anderson, Nat. Cell Biol., 2007, 9, 7–14 CrossRef CAS.
- K. Gaus, E. Gratton, E. P. Kable, A. S. Jones, I. Gelissen, L. Kritharides and W. Jessup, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 15554–15559 CrossRef CAS; S. Sugii, P. C. Reid, N. Ohgami, Y. Shimada, R. A. Maue, H. Ninomiya, Y. Ohno-Iwashita and T. Y. Chang, J. Lipid Res., 2003, 44, 1033–1041 CrossRef; R. Ishitsuka, A. Yamaji-Hasegawa, A. Makino, Y. Hirabayashi and T. Kobayashi, Biophys. J., 2004, 86, 296–307 CrossRef; P. C. Reid, N. Sakashita, S. Sugii, Y. Ohno-Iwashita, Y. Shimada, W. F. Hickey and T. Y. Chang, J. Lipid Res., 2004, 45, 582–591 CrossRef; A. Margineanu, J. Hotta, R. A. Vallée, M. Van der Auweraer, M. Ameloot, A. Stefan, D. Beljonne, Y. Engelborghs, A. Herrmann, K. Müllen, F. C. De Schryver and J. Hofkens, Biophys. J., 2007, 93, 2877–2891 CrossRef.
- E. Kiyokawa, T. Baba, N. Otsuka, A. Makino, S. Ohno and T. Kobayash, J. Biol. Chem., 2005, 280, 24072–24084 CrossRef CAS.
- E. Betzig, G. H. Patterson, R. Sougrat, O. W. Lindwasser, S. Olenych, J. S. Bonifacino, M. W. Davidson, J. Lippincott-Schwartz and H. F. Hess, Science, 2006, 313, 1642–1645 CrossRef CAS.
- S. T. Hess, T. P. Girirajan and M. D. Mason, Biophys. J., 2006, 91, 4258–4272 CrossRef CAS.
- M. J. Rust, M. Bates and X. Zhuang, Nat. Methods, 2006, 3, 793–795 CrossRef CAS; M. Heilemann, S. van de Linde, M. Schüttpelz, R. Kasper, B. Seefeldt, A. Mukherjee, P. Tinnefeld and M. Sauer, Angew. Chem., Int. Ed., 2008, 47, 6172–6176 CrossRef.
- S. T. Hess, T. J. Gould, M. V. Gudheti, S. A. Maas, K. D. Mills and J. Zimmerberg, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 17370–17375 CrossRef CAS; D. M. Owen, C. Rentero, J. Rossy, A. Magenau, D. Williamson, M. Rodriguez and K. Gaus, J. Biophotonics, 2010, 3, 446–454 CrossRef; B. F. Lillemeier, M. A. Mörtelmaier, M. B. Forstner, J. B. Huppa, J. T. Groves and M. M. Davis, Nat. Immunol., 2010, 11, 90–96 CrossRef.
- K. I. Willig, S. O. Rizzoli, V. Westphal, R. Jahn and S. W. Hell, Nature, 2006, 440, 935–939 CrossRef CAS; K. I. Willig, R. R. Kellner, R. Medda, B. Hein, S. Jakobs and S. W. Hell, Nat. Methods, 2006, 3, 721–723 CrossRef; K. I. Willig, B. Harke, R. Medda and S. W. Hell, Nat. Methods, 2007, 4, 915–918 CrossRef; B. Hein, K. I. Willig and S. W. Hell, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14271–14276 CrossRef; J. Hotta, E. Fron, P. Dedecker, K. P. Janssen, C. Li, K. Müllen, B. Harke, J. Bückers, S. W. Hell and J. Hofkens, J. Am. Chem. Soc., 2010, 132, 5021–5023 CrossRef.
- C. Eggeling, C. Ringemann, R. Medda, G. Schwarzmann, K. Sandhoff, S. Polyakova, V. N. Belov, B. Hein, C. von Middendorff, A. Schönle and S. W. Hell, Nature, 2009, 457, 1159–1162 CrossRef CAS.
- R. Ando, H. Mizuno and A. Miyawaki, Science, 2004, 306, 1370–1373 CrossRef CAS.
- S. Habuchi, R. Ando, P. Dedecker, W. Verheijen, H. Mizuno, A. Miyawaki and J. Hofkens, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 9511–9516 CrossRef CAS; C. Flors, J. Hotta, H. Uji-i, P. Dedecker, R. Ando, H. Mizuno, A. Miyawaki and J. Hofkens, J. Am. Chem. Soc., 2007, 129, 13970–13977 CrossRef; H. Mizuno, T. K. Mal, M. Wälchli, A. Kikuchi, T. Fukano, R. Ando, J. Jeyakanthan, J. Taka, Y. Shiro, M. Ikura and A. Miyawaki, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 9227–9232 CrossRef.
- A. A. Waheed, Y. Shimada, H. F. Heijnen, M. Nakamura, M. Inomata, M. Hayashi, S. Iwashita, J. W. Slot and Y. Ohno-Iwashita, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 4926–4931 CrossRef CAS.
- Y. Shimada, M. Maruya, S. Iwashita and Y. Ohno-Iwashita, Eur. J. Biochem., 2002, 269, 6195–203 CrossRef CAS.
- A. Yamaji, Y. Sekizawa, K. Emoto, H. Sakuraba, K. Inoue, K. Kobayashi and M. Umeda, J. Biol. Chem., 1998, 273, 5300–5306 CrossRef CAS.
- J. Rossjohn, S. C. Feil, W. J. McKinstry, R. K. Tweten and M. W. Parker, Cell, 1997, 89, 685–92 CrossRef CAS.
- S. T. Christensen and C. M. Ott, Science, 2007, 317, 330–331 CrossRef CAS.
- H. W. Fisher and T. W. Cooper, J. Cell Biol., 1967, 34, 569–576 CrossRef CAS.
- K. Röper, D. Corbeil and W. B. Huttner, Nat. Cell Biol., 2000, 2, 582–592 CrossRef.
- P. G. Parton and J. F. Hancock, Trends Cell Biol., 2004, 14, 141–147 CrossRef.
- K. A. Tanaka, K. G. Suzuki, Y. M. Shirai, S. T. Shibutani, M. S. Miyahara, H. Tsuboi, M. Yahara, A. Yoshimura, S. Mayor, T. K. Fujiwara and A. Kusumi, Nat. Methods, 2010, 7, 865–866 CrossRef CAS.
- M. Umeda, K. Igarashi, K. S. Nam and K. Inoue, J. Immunol., 1989, 143, 2273–2279 CAS.
- C. A. Konopka and S. Y. Bednarek, Plant J., 2008, 53, 186–196 CrossRef CAS; M. Tokunaga, N. Imamoto and K. Sakata-Sogawa, Nat. Methods, 2008, 5, 159–161 CrossRef.
- H. Mizuno, P. Dedecker, R. Ando, T. Fukano, J. Hofkens and A. Miyawaki, Photochem. Photobiol. Sci., 2010, 9, 239–248 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c1sc00169h |
| ‡ Present address: Department of Pharmacology and Molecular Sciences, The Johns Hopkins University School of Medicine, Baltimore, Maryland, USA. |
| § An E. coli strain with λ(DE3) is a λ prophage carrying the T7 polymerase gene under the regulation of lac UV5 promoter. At the downstream of UV5 promoter, there is the lac operator to which the lac respressor binds and represses the transcription from the promoter. IPTG binding to the lac repressor induces dissociation of it from the lac operator and activates the transcription of T7 polymerase from the UV5 promoter. The lac repressor also binds to the lac operator on the pET28 vector next to the T7 promoter and represses the transcription from the T7 promoter. This repressor also dissociates upon IPTG binding. |
| This journal is © The Royal Society of Chemistry 2011 |
