“Click-made” biaryl-linker improving efficiency in protein labelling for the membrane target protein of a bioactive compound†
Yoko
Nakamura
a,
Sho
Inomata
a,
Makoto
Ebine
a,
Yoshiyuki
Manabe
a,
Izumi
Iwakura
b and
Minoru
Ueda
*a
aDepartment of Chemistry, Tohoku University, 6-3 Aramaki-aza Aoba, Aoba-ku, Sendai 980-8578, Japan. E-mail: ueda@m.tohoku.ac.jp; Fax: +81 22 795 6553; Tel: +81 22 795 6553
bInnovative Use of Light and Materials/Life, PREST, JST, 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan
First published on 17th November 2010
Abstract
We report on the design, synthesis and assessment of a novel biaryl-linked (BArL) molecular probe for the exploration of low-abundant target proteins for bioactive compounds based on the activity based protein profiling (ABPP) approach. Surprisingly, the performance of the BArL probe was better than that of the stepwise tagging approach that is considered to be the most effective method used in ABPP study.
Activity-based protein profiling (ABPP)1 has become a powerful strategy for understanding biological processes in living cells and organisms. Because most target proteins for bioactive natural products are unknown, the prospect of using ABPP with a bioactive natural product to induce a physiologically intriguing phenomenon would launch a new field of chemical biology.
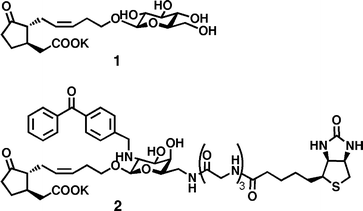
Chemical biology on jasmonate, an important plant hormone, has evolved into one of the cutting-edge topics in plant science since the identification of COI1-JAZs as a core signalling module.2 We identified β-D-glucopyranosyl 12-hydroxyjasmonic acid 1 as a bioactive metabolite which controls plant nyctinasty, a rhythmic movement of plant leaves controlled by a biological clock (Figure S1).3 Recently, our ABPP approach using probe 2, which is designed according to a previous FITC-probe used in a fluorescence imaging study,4 identified a membrane target protein for jasmonateglucoside 1 (MTJG).5 MTJG is expected to be involved in shrinking of the motor cell which leads to nyctinastic leaf-closure.6 The MTJG is completely different from COI1,7 a cytosolic receptor of jasmonate. This result implies the existence of an unknown mode of action on the jasmonates. However, the limited availability of trace MTJG makes its identification difficult. Certain classes of physiologically important proteins, including membrane-associated and low-abundance proteins, are difficult targets for chemical biology because of their low efficiency in specific chemical tagging. For example, it is well known that photolabelling efficiency for cross-link formation between a photoaffinity probe and the target protein is always as low as a few percent.8 Thus, it is essential to develop a high-performance molecular probe for efficient tagging of low-abundance membrane proteins. In this paper, we report the advantages of a biaryl-linked (BArL) molecular probe which is of synthetic utility using CuAAC9 “click” chemistry and provides remarkably high performance in labelling of membrane target proteins.
We employed the CuAAC reaction for a tractable probe synthesis. CuAAC streamlines probe synthesis with concomitant construction of a rigid biaryl linker moiety (Scheme 1). The use of a “rigid” linker usually causes a difficulty in probe synthesis, such as insolubility, difficult purification, and low yield.
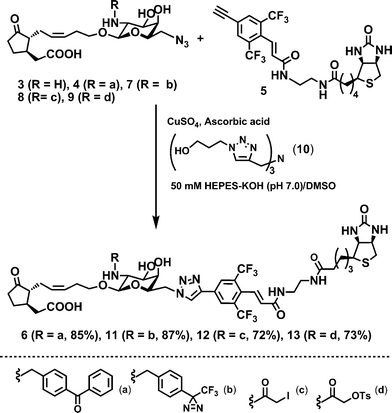 | ||
| Scheme 1 CuAAC synthesis of BArL probes: a = benzophenone; b = trifluoromethyldiazirine; c = iodoacetyl; d = tosyloxyacetyl. | ||
CuAAC strategy enables these difficulties to be circumvented. Alkylation of azide-containing 3 and subsequent CuAAC using a biotin-containing ethynylaryl unit105 with triazole ligand1110 gave BArL probe 6 without byproducts. In photoaffinity labelling experiments against living motor cells of Samanea saman, BArL probe 6 provided a distinct improvement of labelling efficiency over triglycyl 25 (Fig. 1a). BArL probe 6 biotinylated MTJG in a concentration-dependent manner between 1 and 100 μM, and the efficiency in biotinylation was estimated to be twenty times as much as that of 2. The specific binding was confirmed by competitive inhibition using an excess amount of naturally occurring 1 (Figure S2).
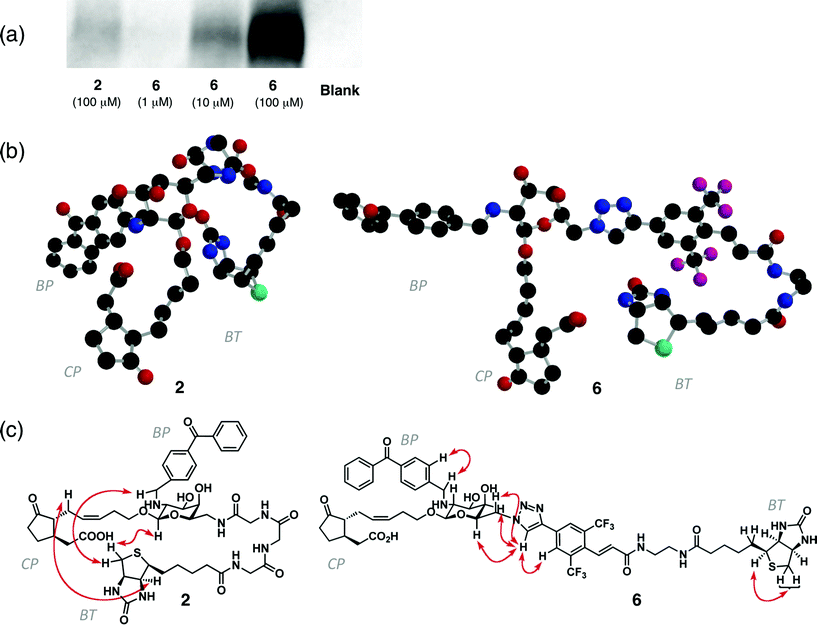 | ||
| Fig. 1 (a) Photoaffinity labelling and chemiluminescence detection of MTJG by 2 (100 μM) or 6 (1–100 μM).(b) Stereostructures of 2 and 6 (CP: cyclopentanone; BP: benzophenonyl; BT: biotinyl). The plausible conformer was obtained by MM and DFT(B3LYP/6-31G*) calculations. Each atom is represented by colors as follows: carbon (black), oxygen (red), nitrogen (blue), sulfur (green), fluorine (purple). (c) Important ROE correlations in 2 and 6(See also Figures S4 and S5). | ||
We found that this enhancement was due to the stronger affinity of 6 with MTJG than that of 2. Thus, when shrinking rates of motor cells, induced by structure recognition of 1 by MTJG,5 were compared between 2 and 6, greater shrinking was observed for 6 (Figure S3). The high performance of the BArL probe may come from the rigidity of its molecular shape due to the insertion of a biaryl linker system which projects the pharmacophore away from the molecular tag enough for the efficient binding with its target protein.12 This model was supported by a comparison between the most stable conformations of 2 and 6 obtained by MM and DFT calculations13 (Fig. 1b) coupled with ROESY experiments (Fig. 1c, S4 and S5). Probe 2 has a globular conformation in which the benzophenone group, molecular tag, and pharmacophore are folded in close proximity. On the other hand, probe 6 has a unique T-shaped conformation in which the benzophenone group, molecular tag, and pharmacophore are spatially separate from each other. This is because rigid biaryl linker moiety prevents the formation of folded conformation. In addition to the ease of synthesis, the BArL probe had a marked improvement in protein labelling efficiency because of its unique T-shaped conformation.
The stepwise tagging approach using on-the-cell CuAAC14 is also known as a powerful method for tagging of a membrane protein (Figure S6). Protein labelling by the sterically minimal azide probe and subsequent introduction of tag using CuAAC was expected to improve its affinity to the target protein and enhance labelling efficiency.15 Surprisingly, the labelling performance of BArL probe was better than that of the stepwise tagging approach in protein labelling. BP-containing azide4 was used for affinity-based introduction of an azide handle to MTJG on a living cell. Successive CuAAC with alkyne5 was carried out at 4 °C to provide specific tagging of MTJG. The reaction conditions of CuAAC were determined on the basis of in vitro examination using 4 and 5 (Figure S7). Considering the toxicity of copper to the living cell,16 CuAAC conditions that require 500 μM copper(I) were employed.14,17 Copper ion provokes a decrease in the number of living cells during CuAAC reactions. The proteins released from the dead cells easily form insoluble copper aggregates during CuAAC. Thus, it is important to maintain the number of living cells during the reaction. The copper ion will increase the loss of the target protein because of formation of insoluble aggregates. A remarkable decrease in cell viability was observed for CuAAC at a higher temperature (30 °C) (data not shown). We compared the labelling performance with that of the BArL probe approach using 6. Unexpectedly, the BArL probe 6 turned out to give labelling efficiency twice as good as stepwise tagging using 4 with 5 (Fig. 2a). Thus, it was concluded that the BArL probe exhibited the best performance in the labelling of the membrane target among three different approaches using 2, 4 and 6.
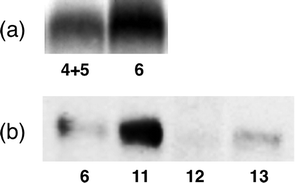 | ||
| Fig. 2 (a) Photoaffinity labelling and chemiluminescence detection of MTJG by a stepwise approach using 4 with 5 or the BArL approach using 6. (b) Chemiluminescence detection of labeled MTJG using 6, 11, 12 and 13. | ||
Next, we synthesized four biotin-bearing BArL probes with different reactive functionalities, such as benzophenone (BP: 6), trifluoromethyldiazirine (TFMD: 11), iodoacetyl (IA: 12), and tosyloxyacetyl (Ts: 13) and compared MTJG-labelling performances among them. Alkylation/acylation of common intermediate 3 and subsequent CuAAC with alkyne units gave a panel comprising four probes (Scheme 1). All the CuAAC couplings proceeded in good yields (ca. 70–90%).
We compared the performance of these four BArL probes in labelling efficiency for MTJG (Fig. 2b). Labelling reactions by four BArL probes (6, 11–13) using living cells were carried out in as mild conditions as possible. Chemical labelling using carbon electrophile3,1812 or 13 was carried out at 4 °C for 30 min with gentle incubation, and photoaffinity labelling using 6 or 11 was executed at 4 °C by UV irradiation (365 nm) for 30 min. As shown in the chemiluminescence detection of SDS-PAGE, photoaffinity groups gave better results than carbon electrophiles (Fig. 2b). The ratio in intensity of chemiluminescence was 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 = 1.0
13 = 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.4
3.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2
0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.7. Two photoaffinity probes, BP 6 and TFMD 11, labeled MTJG selectively, whereas IA 12 largely gave non-specific binding with 13 and 27 kDa proteins (Figure S8). Ts 13 gave a moderate result without non-specific binding. Generally, photoaffinity labelling is considered to be harmful for living cells, whereas gentle substitution labelling using carbon electrophiles is considered to be better for living cells. However, in this case, TFMD probe 11 gave the best result among these four probes (Fig. 2b). In this case, the labeling efficiency of TFMD functionality may be superior to those of BP and IA because there would be little difference among the affinity of three photoaffinity probes (11–13) with MTJG. The use of the BArL probe and screening for the best reactive functionality realized as much as ca. 70-fold enhancement in labelling efficiency compared to 2.
0.7. Two photoaffinity probes, BP 6 and TFMD 11, labeled MTJG selectively, whereas IA 12 largely gave non-specific binding with 13 and 27 kDa proteins (Figure S8). Ts 13 gave a moderate result without non-specific binding. Generally, photoaffinity labelling is considered to be harmful for living cells, whereas gentle substitution labelling using carbon electrophiles is considered to be better for living cells. However, in this case, TFMD probe 11 gave the best result among these four probes (Fig. 2b). In this case, the labeling efficiency of TFMD functionality may be superior to those of BP and IA because there would be little difference among the affinity of three photoaffinity probes (11–13) with MTJG. The use of the BArL probe and screening for the best reactive functionality realized as much as ca. 70-fold enhancement in labelling efficiency compared to 2.
Conclusions
In summary, we have found important advantages of a “click”-made BArL probe, which has remarkably high performance in protein labelling, attributed to its unique T-shaped conformation. The efficiency surpassed that of the stepwise tagging approach for the MTJG protein. “Click”-made BArL probes promise to make ABPP-based exploration of trace membrane targets for bioactive compounds a powerful method.Notes and references
- N. Jessani and B. F. Cravatt, Curr. Opin. Chem. Biol., 2004, 8, 54–59 CrossRef CAS; M. J. Evans and B. F. Cravatt, Chem. Rev., 2006, 106, 3279 CrossRef CAS; U. Hillaert, M. Verdoes, B. I. Florea, A. Saragliadis, K. L. L. Habets, J. Kuiper, S. V. Calenbergh, F. Ossendrop, G. A. van derMarel, C. Driessen and H. S. Overkleeft, Angew. Chem., Int. Ed., 2009, 48, 1629 CrossRef CAS; S. Tsukiji, M. Miyagawa, Y. Takaoka, T. Tamura and I. Hamachi, Nat. Chem. Biol., 2009, 3, 341 CrossRef.
- C. Wasternack and E. Kombrink, ACS Chem. Biol., 2010, 5, 63 CrossRef CAS; S. Fonseca, A. Chini, M. Hamberg, B. Adie, A. Porzel, R. Kramell, O. Miersch, C. Wasternack and R. Solano, Nat. Chem. Biol., 2009, 5, 344 CrossRef CAS; B. Thines, L. Katsir, M. Melotto, Y. Niu, A. Mandakar, G. Liu, K. Nomura, S. Y. He, G. A. Howe and J. Browse, Nature, 2007, 661, 448; A. Chini, S. Fonseca, G. Fernandez, B. Adie, J. M. Chico, O. Lorenzo, G. G.-Casado, I. L.-Vidriero, F. M. Lozano, M. R. Ponce, J. M. Micol and R. Solano, Nature, 2007, 448, 666 CrossRef CAS.
- M. Ueda, M. Okazaki, K. Ueda and S. Yamamura, Tetrahedron, 2000, 56, 8101 CrossRef CAS; M. Ueda and S. Yamamura, Angew. Chem., Int. Ed., 2000, 39, 1400 CrossRef CAS.
- Y. Nakamura, R. Miyatake, A. Matsubara, H. Kiyota and M. Ueda, Tetrahedron, 2006, 62, 8805 CrossRef CAS and references cited therein.
- Y. Nakamura, R. Miyatake and M. Ueda, Angew. Chem., Int. Ed., 2008, 47, 7289 CrossRef CAS.
- N. Moran, FEBS Lett., 2007, 581, 2337 CrossRef CAS.
- B. J. F. Feys, C. E. Benedetti, C. N. Penfold and J. G. Turner, Plant Cell, 1994, 6, 751 CAS; J. Yan, C. Zhang, M. Gu, Z. Bai, W. Zhang, T. Qi, Z. Cheng, W. Peng, H. Luo, F. Nan, Z. Wang and D. Xie, Plant Cell, 2009, 21, 2220 CrossRef CAS.
- M. Hashimoto and Y. Hatanaka, Eur. J. Org. Chem., 2008, 2513–2523 CrossRef CAS.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004 CrossRef CAS; C. W. Tornøe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057 CrossRef CAS.
- Y. Manabe, M. Mukai, S. Ito, N. Kato and M. Ueda, Chem. Commun., 2010, 46, 469 RSC.
- T. R. Chan, R. Hilgraf, K. B. Sharpless and V. V. Fokin, Org. Lett., 2004, 6, 2853 CrossRef CAS.
- Pharmacophore of 1 was identified from previous SAR studies: Y. Nakamura, R. Miyatake, S. Inomata and M. Ueda, Biosci., Biotechnol., Biochem., 2008, 72, 2867 Search PubMed.
- Gaussian 03 (Revision D.02), M. J. Frisch, et al.Gaussian, Inc., Wallingford CT, 2004. See SI ref. 11 Search PubMed.
- A. J. Link and D. A. Tirrell, J. Am. Chem. Soc., 2003, 125, 11164 CrossRef CAS; A. E. Spencer, G. C. Adam and B. F. Cravatt, J. Am. Chem. Soc., 2003, 125, 4686 CrossRef CAS.
- L. Ballell, K. J. Alink, M. Slijper, C. Versluis, R. M. J. Liskamp and R. J. Pieters, ChemBioChem, 2005, 6, 291 CrossRef CAS.
- L. M. Gaetke and C. K. Chow, Toxicology, 2003, 189, 147 CrossRef CAS.
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974 CrossRef CAS.
- E. Weerapana, G. M. Simon and B. Cravatt, Nat. Chem. Biol., 2008, 4, 405 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S8, materials and methods, experimental procedures and NMR spectra. See DOI: 10.1039/c0ob00843e |
| This journal is © The Royal Society of Chemistry 2011 |