Aptamer-conjugated nanomaterials for bioanalysis and biotechnology applications
Tao
Chen
a,
Mohammed Ibrahim
Shukoor
a,
Yan
Chen
ab,
Quan
Yuan
a,
Zhi
Zhu
a,
Zilong
Zhao
a,
Basri
Gulbakan
a and
Weihong
Tan
*ab
aDepartment of Chemistry and Department of Physiology and Functional Genomics, Shands Cancer Center, Center for Research at Bio/Nano Interface, University of Florida Genetics Institute, and McKnight Brain Institute, University of Florida, Gainesville, FL 32611-7200
bState Key Laboratory of Chemo/Biosensing and Chemometrics, College of Biology, College of Chemistry and Chemical Engineering, Hunan University, Changsha 410082, P.R. China
First published on 25th November 2010
Abstract
In recent years, nanomaterials have captured the attention of scientists from a wide spectrum of domains. With their unique properties, nanomaterials offer great promise for numerous applications, ranging from catalysis to energy harvesting and information technology. Functionalized with the desired biomolecules, nanomaterials can also be utilized for many biomedical applications. This paper summarizes recent achievements in the use of aptamer-conjugated nanomaterials for bioanalysis and biotechnology applications. First, we discuss the features and properties of aptamers and then illustrate the use of aptamer-conjugated nanomaterials as sensing platforms and delivery vehicles, emphasizing how such integration can result in enhanced sensitivity and selectivity.
 Tao Chen | Ms. Tao Chen obtained her B. S. (2008) in Chemistry from the Wuhan University, China. Currently, she is a Ph.D. candidate majoring in Analytical Chemistry in the Department of Chemistry at the University of Florida. Her research interest includes constructing novel nucleic acid aptamer-conjugated nanostructures for rare protein capturing, biosensor development and targeted cancer therapy. |
 Mohammed Ibrahim Shukoor | Dr Mohammed Ibrahim Shukoor received his Ph.D. (2008) in Chemistry from the Johannes Guetenberg University, Germany, where he graduated suma cum laude. As a postdoctor in the Department of Chemistry at the University of Florida, he is currently involved with the development of smart novel hybrid nanomaterials for biosensors, cancer applications and diagnostics. |
 Yan Chen | Dr Yan Chen obtained her B.S. (2005) in Chemical Physics from the University of Science and Technology, China and Ph.D. (2010) in Analytical Chemistry with the academic women Madelyn Lockhart dissertation fellowship and the Chinese government award for outstanding self-financed student abroad from the University of Florida. She is currently a postdoctor in the Tan group and working on instrumental development, nanomaterial engineering for live cell mapping, biosensor development and energy harvesting. |
 Quan Yuan | Dr Quan Yuan received her B.S. (2004) from the Wuhan University, China and Ph.D. (2009) in Inorganic Chemistry from the Peking University, China. Currently, she is a postdoctor in the Department of Chemistry at the University of Florida. Her research interest is concentrated on metal nanoparticles, mesoporous materials, and their applications in biomedical fields. |
 Zhi Zhu | Ms. Zhi Zhu earned her B.S. (2006) majoring in Chemistry from the Peking University, China. She is currently studying for her Ph.D. degree in the Department of chemistry at the University of Florida. Her research interest is focused on the investigation of aptamer molecular engineering, and their applications in biosensing, diagnosis and cancer therapy. |
 Weihong Tan | Dr Weihong Tan, V. T. and Louis Jackson Professor of Chemistry and Professor of Physiology and Functional Genomics at the University of Florida, earned his Ph.D. (1993) in Chemistry at the University of Michigan. His research interest lies in molecular engineering, bionanotechnology and chemical biology. His group has engineered nucleic acid probes for biosensing platforms and DNA nanomotors. Tan group has also developed numerous bioconjugated nanostructures for molecular imaging, efficient cell separation and sensitive cell detection. With the development of cell-based SELEX, the Tan group has generated various aptamers for cancer cells as molecular tools for cancer treatment and biomarker discovery. |
Introduction
Nanomaterials are structures having a size of 100 nanometers or smaller in at least one dimension. Because of quantum effects stemming from the large surface area to volume ratio, nanomaterials possess unique optical,1 electronic,2 magnetic,3 mechanical,4 physical and chemical properties.5 Thus far, nanomaterials have generated widespread interest and applications in energy harvesting6 and information technology.7 In energy harvesting, nanomaterials are commonly applied to reduce energy consumption, improve energy production efficiency, and exploit energy systems compatible with the environment. Meanwhile, information technologists are mainly concerned with the use of nanomaterials for memory storage expansion, novel semiconductor development and optoelectronic device construction. Recently, however, investigators have prepared nanomaterials for use in biomedicine, where increasing applications have been found in biosensor development,8 molecular imaging,9drug delivery10 and cancer therapy.11Both enhanced biocompatibility and biofunctionality can be achieved by conjugating nanomaterials to synthetic (e.g., polyethylene glycol (PEG)) or natural (e.g., folic acid) ligands.12 In particular, aptamers have found increasing applications in such bioconjugated systems. Aptamers are single-stranded oligonucleotides generated from an in vitro process known as SELEX (Systematic Evolution of Ligands by Exponential Enrichment).13 By folding into distinct secondary or tertiary structures, aptamers can specifically bind to their targets with dissociation constants in the micromolar to picomolar range. Targets range in scope from small molecules14 to proteins,15 whole cells16 and even tissues.17 Our lab has developed a cell-based SELEX strategy that can produce a panel of aptamers for diseased cells, especially cancer cells.18 Some aptamers can be internalized into living cells,19 thus making them good candidates for drug delivery.
Because of the molecular recognition property of aptamers, they are sometimes called chemical antibodies. However, compared to antibodies, aptamers possess several unique advantages. First, the in vitro selection process of aptamers eliminates the need for the in vivo immunization of animals to generate antibodies, thus greatly widening the range of targets for aptamers, to include toxic or non-immunogenic materials.20 Second, aptamers can be easily prepared via a solid-phase oligonucleotide synthesis with good reproducibility.21 Compared to antibodies, aptamers are much more stable to heat, pH and organic solvents.22 Third, the chemical modification of aptamers with organic fluorophores, redox labels, functional groups or even nanomaterials is relatively simple.23 Finally, other than the specific molecular recognition toward their targets, aptamers are capable of recognition through base-pairing, enabling tailorable designs for desired applications.24
For a fair comparison, however, the disadvantages of aptamers should also be mentioned. One problem is that aptamers are vulnerable to nuclease digestion in cells or biological fluids, such as sera, blood and urine.25 In addition, some aptamers have relatively weak binding affinities to their targets.26 Thus, the question becomes whether these disadvantages can be successfully overcome. Regarding the stability issue, modification of aptamers with 2′-aminopyrimidine has generated anti-bFGF aptamer that can survive in urine for up to 17 h.27 In addition, modification of atpamers with 2′-fluoropyrimidine28 or 2′-O-methyl nucleotides29 has also achieved similar success. As for weak binding, many molecular engineering strategies have greatly improved the affinities of aptamers toward their targets.30
On balance, the myriad of nanomaterials available for the construction of aptamer-based bioconjugates will still result in structures capable of precise molecular recognition and enhanced target specificity according to pre-defined biochemical and biomedical applications. To demonstrate this point, the synthesis and properties of various nanomaterials, such as silica nanoparticles, gold nanoparticles, carbon nanotubes, gold nanorods, liposomes and micelles, are reviewed. Then the integration of these nanomaterials with aptamers is discussed. Finally the applications of these aptamer-conjugated systems as sensing platforms for bioanalysis and delivery vehicles are given.
Aptamer-conjugated nanomaterials as sensing platforms for bioanalysis
Understanding basic biological phenomena with the aim of exploring potential therapeutic applications depends on the identification and characterization of biomolecules and biological interactions. To achieve these objectives, suitable biosensing platforms are required. Two essential characteristics of biosensors are molecular recognition and signal transduction. Nanomaterials add signal transduction, as well as signal amplification. Meanwhile, aptamers have excellent molecular recognition ability. Thus, the integration of nanomaterials and aptamers will lead to a potentially promising construct for biosensing platforms. Today, rapid development in nanotechnology has resulted in the successful synthesis of numerous nanomaterials for bioanalysis. We will focus on silica nanoparticles, gold nanoparticles, carbon nanotubes and gold nanorods.Silica nanoparticles
Silica nanoparticles, especially dye-doped silica nanoparticles, are currently used in many areas of bioanalysis. There are two standard methods for synthesizing dye-doped silica nanoparticles: the Stöber method31 and the reverse micro-emulsion method.32 The former method is relatively simple, and either organic or inorganic dyes can be incorporated. However, it produces dye-doped silica nanoparticles with a relatively large size distribution because of the hydrolysis procedure. The latter method can generate uniform dye-doped silica nanoparticles, but only inorganic dyes can be incorporated. Modifications and optimizations have been applied to make both methods easier and more universal. Apart from single-dye doping, multiple dyes have also been incorporated into the silica matrix.33Compared to common fluorophores, dye-doped silica nanoparticles are superior in three respects:34 (1) Dye molecules doped inside are immune to degradation and photo-bleaching due to the protection of the silica matrix; (2) No further signal amplification scheme is needed, since thousands of dye molecules are trapped inside one single nanoparticle; (3) Various biomolecules can be introduced onto the surface of a single nanoparticle by the versatility of silicon chemistry. These advantages make dye-doped silica nanoparticles increasingly attractive for use in sensitive bioanalysis.
Using dye-doped silica nanoparticles as reporters, many detection methods have been developed for small molecules35 and proteins.36 In recent work by Wang et al., a sensitive and selective aptasensor was developed using Ru(bpy)32+-doped silica nanoparticles as DNA tags for the detection of thrombin, based on target-induced strand displacement.36 As a consequence of aptamers' high binding affinity and excellent binding specificity, they rival antibodies for molecular recognition. This makes aptamer-conjugated dye-doped silica nanoparticles increasingly used in the area of early diagnostics. For instance, sensitive detection of CEM cells by Sgc8 conjugated dye-doped silica nanoparticles has been achieved.37 Inspired by the successful synthesis of dye-doped silica nanoparticles with three different types of dyes with efficient fluorescence resonance energy transfer (FRET),38 Chen et al. realized multiplexed cancer cell detection by conjugating this triad of dye-doped silica nanoparticles with three different aptamers specific for three individual cancer cell types [Fig. 1].39 FAM-doped silica nanoparticles were conjugated with T1 aptamer for Toledo cells; FAM- and R6G-doped silica nanoparticles were conjugated with Sgc8 aptamer for CEM cells; and FAM-, R6G-, and ROX-doped silica nanoparticles were conjugated with Td05 aptamer for Ramos cells. Both flow cytometry histograms and confocal microscopy images showed the selectivity of this assay, demonstrating the high specificity of using aptamers for molecular recognition. Because of the FRET phenomenon between FAM, R6G and ROX dyes, different emissions can be produced by only one single excitation. When compared to single dye-doped silica nanoparticles, these quantum dot-like, multiple dye-doped silica nanoparticles greatly simplify the instrumental setup for multiplexed bioanalysis, since only one laser source is needed.
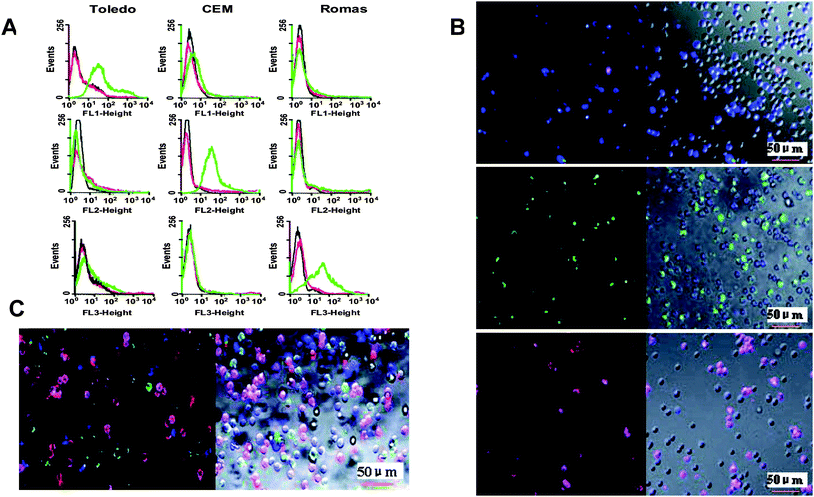 | ||
| Fig. 1 (A) Flow cytometry histograms of aptamer-conjugated dye-doped silica nanoparticles with three different cells: Top row, T1 aptamer conjugated to FAM-doped particles; middle row, Sgc8 aptamer conjugated to FAM- and R6G-doped particles; bottom row, Td05 aptamer conjugated to FAM-, R6G- and ROX-doped particles. On each histogram, the black curve corresponds to cells only; red curve corresponds to dye-doped silica nanoparticles with cells; and green curve corresponds to aptamer-conjugated dye-doped silica nanoparticles with cells. (B) Confocal microscopy images of aptamer-conjugated dye-doped silica nanoparticles with a mixture of three different cells: Top row, T1 aptamer conjugated to FAM-doped particles; middle row, Sgc8 aptamer conjugated to FAM- and R6G-doped particles; bottom row, Td05 aptamer conjugated to R6G- and ROX-doped particles. (C) Confocal microscopy images of all the aptamer-conjugated dye-doped silica nanoparticles with a mixture of three different cells. Adapted from ref. 39. | ||
Dye-doped silica nanoparticles can greatly increase measurement sensitivity because of their inherent signal amplification. Estévez et al. have demonstrated this point by using aptamer-conjugated dye-doped silica nanoparticles for cancer cell monitoring on a flow cytometer [Fig. 2].37 Enhanced sensitivity arises from (1) thousands of dye molecules trapped inside one single nanoparticle and (2) millions of cell membrane receptors for aptamer binding on one cancer cell. In order to further increase sensitivity and simplify sample preparation, magnetic nanoparticles have been introduced into the dye-doped silica nanoparticle based bioanalysis.40,41 Smith et al. developed a novel two-nanoparticle assay for the rapid collection and detection of leukemia cells.40 In this assay, aptamer-conjugated magnetic nanoparticles were used to simplify sample preparation and realize target cell extraction from complex mixtures, including whole blood samples. Aptamer-conjugated dye-doped silica nanoparticles were used to signal enriched cancer cells. As a consequence of magnetic enrichment, as well as the high affinity and specificity of aptamers, this assay is highly sensitive and selective. Later, by employing a similar principle, Smith et al. further expanded this assay for multiplexed cancer cell monitoring.41
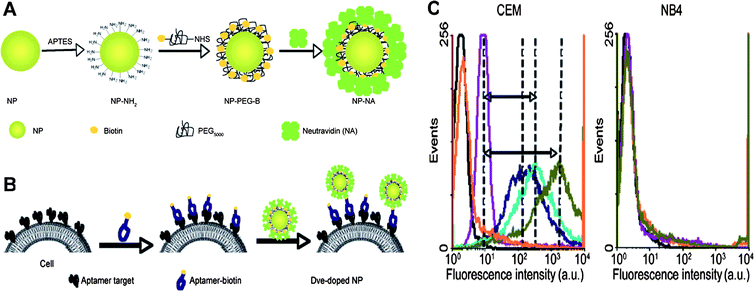 | ||
| Fig. 2 (A) Surface modification scheme for dye-doped silica nanoparticles. (B) Incubation procedures of aptamer-conjugated dye-doped silica nanoparticles with cells. (C) Flow cytometry histograms of increasing amounts of aptamer-conjugated dye-doped silica nanoparticles with CEM cells (target cells) and NB4 cells (control cells). Adapted from ref. 37. | ||
Gold nanoparticles
Gold nanoparticles, commonly known as gold colloid or colloidal gold, are also widely used for bioanalysis.42 Generally, they are produced in a liquid by reduction of chloroauric acid (HAuCl4) and their size can be easily controlled according to specific applications.43 Standard gold-thiol chemistry allows a monolayer of biomolecules having thiol end groups to be anchored on the gold nanoparticle surface through a ligand exchange process. As a consequence, combination of gold nanoparticles with thiol bearing ligands (e.g., small molecules,44proteins45 and DNA strands46) gives rise to a large number of functionalized gold nanoparticles with different sizes, different recognition moieties and inherent polyvalent abilities, satisfying the need for a wide range of biological applications.Due to the spatial length scale reduction of electronic motion and the coherent oscillation of the conduction band electrons, gold nanoparticles possess a strong surface plasmon resonance (SPR) band.47 In addition, the SPR band of gold nanoparticles has strong distance-dependent properties.48 That is, when gold nanoparticles come into close proximity with each other, their absorption spectra and scattering profiles change correspondingly, producing a color change in the sample that can be observed by the naked eye. Taking advantage of this phenomenon, a novel method of using aptamer-conjugated gold nanoparticles for colorimetric detection of cancer cells was developed.49,50 In this assay, gold nanoparticles were assembled and brought together via the recognition between aptamers and their target proteins on the cell membrane. This, in turn, caused the SPR bands of the gold nanoparticles to overlap, providing a direct visualization of CEM cells by using Sgc8-conjugated gold nanoparticles. Although slight non-specific binding was observed, this assay was sufficiently sensitive and selective. Experiments on Ramos cells with Td05-conjugated gold nanoparticles showed that this method could also be used for visualizing Ramos cells.49 Adapting a similar scheme, Liu et al. fabricated a strip biosensor for sensitive detection of circulating tumor cells, using Ramos cells as proof of principle [Fig. 3].50 They showed that 4000 Ramos cells can be seen by the naked eye, and as low as 800 Ramos cells can be detected by a portable strip reader.
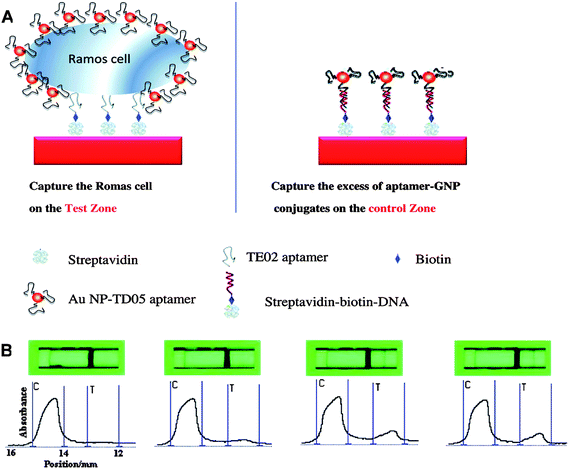 | ||
| Fig. 3 (A) Schematic illustration of detecting Ramos cells on an aptamer-conjugated gold nanoparticle strip biosensor. Ramos cells are captured on the test zone through specific aptamer-cell interaction, while excess aptamer-conjugated gold nanoparticles are captured on the control zone through aptamer-DNA hybridization. (B) Typical photo images (top) and corresponding responses (bottom) of aptamer-conjugated gold nanoparticle strip biosensor with samples containing different amounts of Ramos cells (target cells) and CLL cells (control cells). From left to right: 0 Ramos cells; 8*104CCL cells; 8*104 Ramos cells; 8*104CLL cells and 8*104 Ramos cells. C = Capture zone; T = Test zone. The large signal in the capture zone is due to excess aptamer-conjugated Au nanoparticles. Adapted from ref. 50. | ||
Apart from having SPR bands, gold nanoparticles also have the ability to quench the fluorescence of fluorophores through FRET or an electron-transfer mechanism.51 By careful choice of gold nanoparticles, fluorophores, and ligands to assemble into a complex, target-induced fluorescence signal turn-on can be observed, thus leading to versatile and simple platforms for bioanalysis. Based on this strategy, considerable work has been done using aptamer-conjugated gold nanoparticles.52–54 In one paper,52 Huang et al. constructed platelet-derived growth factor (PDGF) binding aptamer-conjugated gold nanoparticles for sensitive detection of PDGF, using N,N-dimethyl-2,7-diazapyrenium dication (DMDAP) as a reporter. Mirkin's group found that polyvalent DNA functionalized gold nanoparticles, composed of a gold nanoparticle core (2–250 nm) and a dense shell of synthetic oligonucleotides (including aptamers), were capable of entering a variety of cell types with enhanced stability towards enzymatic digestion.55 This finding rendered aptamer conjugated gold nanoparticles useful for a variety of challenging investigations, such as intracellular detection. By using nano-flares (gold-nanoparticle functionalized thiol-terminated aptamers hybridized with a short complementary Cy5-labeled reporter strand), specific intracellular ATP detection was achieved.54 When the nano-flares met ATP inside living cells, the binding between ATP and the corresponding aptamers caused a conformational change in the aptamers, displacing the Cy5-labeled reporter strand to restore the fluorescence signal.
Carbon nanotubes
Apart from these spherical nanomaterials, nanomaterials with other shapes also have great significance for bioanalysis. Among them, carbon nanotubes (CNTs), including single and multi-walled CNTs, have been extensively studied since their discovery in the 1990s.56 They are mosaics of carbon atoms that form graphene sheets and curl into seamless tubules. This review focuses on single-walled carbon nanotubes (SWNTs). Recent research has found that SWNTs can act cooperatively as effective quenchers for a variety of fluorophores, even nearby quantum dots, through an energy or electron transfer process, resulting in low background and high signal-to-noise ratio for their use in biosensor development.57,58 Researchers also found that SWNTs can interact with a variety of biomolecules covalently or non-covalently, such as proteins59,60 and nucleic acids.61,62With respect to nucleic acids, single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) interact with individual SWNTs by adopting different patterns.63,64 Through π–π stacking interactions between the nucleotide bases and the SWNT sidewalls, ssDNA, including aptamers, wrap around SWNTs with nearly complete DNA coverage to form stable complexes. On the other hand, dsDNA interacts relatively weakly with SWNTs, and the interaction is highly dependent on the surface charge of the SWNTs. That is, towards uncharged SWNTs, dsDNA interacts with either end because of the exposed hydrophobic bases. In contrast, for positively charged SWNTs, dsDNA is adsorbed with a roughly parallel configuration. As such, both modes are distinguished from the wrapping mode for ssDNA. In addition, target binding can also interrupt the interaction between aptamers and SWNTs.
A number of sensing events based on the assembly of fluorophore-labeled functional ssDNA and SWNTs have been reported.65,66 In the absence of target (either DNA or other biomolecules), the strong interaction between ssDNA and SWNTs brings the fluorophore and SWNTs into close proximity, quenching the fluorescence signal of the fluorophore, which, in turn, causes low background. However, in the presence of target, specific binding to the functional ssDNA, either through DNA hybridization or formation of an ssDNA-target complex, leads to the dissociation of functional ssDNA from the SWNTs, resulting in restoration of the fluorescence signal. This principle has opened new avenues in biosensor development. For example, Yang et al. harnessed this scheme for the detection of complementary DNAvia a molecular beacon-SWNT complex.65 With the help of SWNTs to reduce background and enhance the signal-to-background ratio, a detection limit of 4.0 nM, 8-fold lower than that of a normal molecular beacon, was achieved. The probe also had the ability to differentiate perfectly matched and single-base mismatched targets. In addition, compared to a normal molecular beacon, this molecular beacon-SWNT complex had much higher thermal stability. The method is cost effective, since no quencher is needed to prepare the molecular beacon, and further modification of the beacon is easy.
Zhu et al. engineered a novel photosensitizer-aptamer-SWNT complex to achieve singlet oxygen generation (SOG) regulation for photodynamic therapy (PDT) [Fig. 4].66 In their demonstration, ATP and thrombin were used as small molecule and protein targets, respectively. In the absence of target, the photosensitizer Ce6 was brought into close proximity of the SWNT surface via normal aptamer wrapping, as described above. Since the fluorescence process and SOG share a similar photophysical mechanism, SOG is efficiently quenched at this stage. However, upon target binding, the conformation of the probe changes, thus disturbing the DNA interaction with the SWNT to remove the aptamer. This leads to the restoration of SOG upon illumination for PDT applications. In this manner, the highly specific regulation of SOG can be directly attributed to the high specificity of aptamers. Moreover, by simply changing the aptamer-target pair, this approach can be used as a general method for SOG regulation.
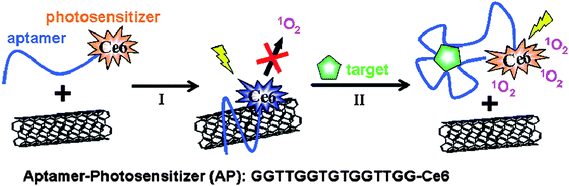 | ||
| Fig. 4 Schematic illustration of aptamer-photosensitizer-SWNT complex construction and the regulation of SOG upon target binding. Adapted from ref. 66. | ||
Gold nanorods
Gold nanorods comprise another type of non-spherical nanomaterial widely used for biosensing. Gold nanorods are mainly synthesized by either the seed-mediated growth method67,68 or the lithographic method,69,70 although electrochemical71 and catalytic72 methods are also used. By conjugating with specific biomolecules, such as enzymes,73,74proteins,75 or DNAs,76–78gold nanorods can be equipped with molecular recognition modules for biosensing. Changes in Raman spectra, bioconjugate stability79 and zeta potential are commonly used to verify the successful attachment of biomolecules to gold nanorods.Compared to gold nanoparticles, gold nanorods are especially intriguing in that they have two distinctive features. One is their relatively larger surface area.80 The other is their dimensional anisotropy, giving rise to a transverse as well as a longitudinal surface plasmon resonance (LSPR) band.81 By reason of their large surface areas, it is easy to functionalize gold nanorods with multiple biomolecules. Huang et al. showed that up to 80 fluorophore-labeled aptamers can be conjugated onto a 12 nm * 56 nm gold nanorod.82 This resulted in a much stronger fluorescence signal and a much higher binding affinity toward target cells compared to an individual fluorophore-labeled aptamer. According to their results, this strategy is versatile, working not only for aptamers with strong binding capability (e.g., Sgc8 for CEM cells), but also for aptamers with weak binding capability (e.g., KK1H08 for K-562 cells). As a result of the enhanced fluorescence signal and improved binding affinity associated with multivalent binding, aptamer conjugated gold nanorods can be good candidates for more sensitive and specific bioanalyses. Regarding the LSPR band of gold nanorods, the wavelength can be tuned with ease from near infrared (NIR) to the infrared (IR) region by controlling the aspect ratio of the gold nanorods.83 Recent theoretical84,85 and experimental results86,87 have shown that an enhanced Raman scattering signal can be produced from the large electromagnetic field generated by the SPR on metal nanostructures, making both gold nanoparticles and gold nanorods suitable candidates for surface enhanced Raman scattering (SERS) applications. This is attributed to the long-range electromagnetic field mechanism for SERS,88 as well as the short-range chemical enhancement mechanism.89
As for the electromagnetic field mechanism, proximity to the metal surface and particle shape both play an important role in enhancing the Raman scattering signal.88 An enhancement factor of 104–105 for adsorbed molecules on gold nanorods was observed, whereas no similar phenomenon was observed for gold nanoparticles.90 Therefore, higher sensitivity can be achieved by using gold nanorods for biosensing based on SERSin vitro and in vivo. For example, Wang et al. proposed a novel SERS aptasensor based on gold nanorod-gold nanoparticle junctions to detect thrombin [Fig. 5].91 In their work, gold nanorods were functionalized with thrombin-binding antibodies, while gold nanoparticles were conjugated with corresponding Raman reporter labeled aptamers. The interaction of thrombin with antibodies on gold nanorods and aptamers on gold nanoparticles brought the two nanostructures into close proximity, creating SERS hot spots for an enhanced Raman scattering signal. When there was no thrombin, no detectable signal was observed. By using this approach, 220 pM thrombin in Tris-HCl buffer and 887 pM thrombin in 5% human blood serum can be detected. By using different Raman reporters, this approach based on aptamer-protein recognition and hot spot formation can be used for multiplexed detection of rare proteins in blood samples.
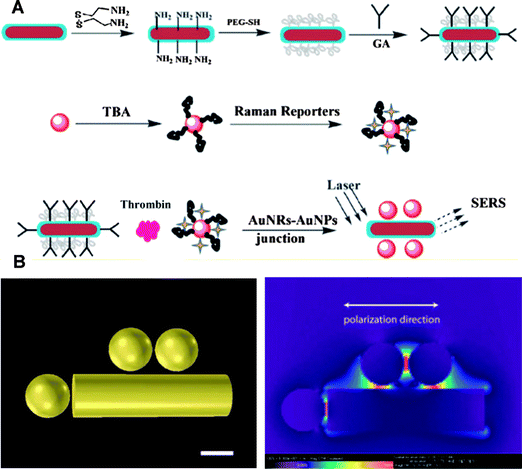 | ||
| Fig. 5 (A) Schematic illustration of gold nanorod-gold nanoparticle junction construction for thrombin detection, GA represents glutaraldehyde and TBA represents thrombin binding aptamer. (B) Finite difference time domain (FDTD) simulation and junction enhancement at the excitation laser wavelength of 632.8 nm. Adapted from ref. 91. | ||
Aptamer-conjugated nanomaterials as delivery vehicles
Apart from their use as sensing platforms for bioanalysis, nanomaterials can also be used for other biomedical purposes.92–95 It is these advances in bionanotechnology that are fueling the shift from traditional symptomatologic medicine to future prospective medicine, which embodies predictive, preventive, personalized and participatory (P4 Medicine TM) medicine. Nanomaterials are currently applied as delivery vehicles,96 regulation moieties97 and therapeutic agents.98 When used as carriers, nanomaterials are bound with cargo molecules, such as drugs or functional proteins, either covalently or non-covalently. Covalent modifications of cargoes are generally achieved through standard gold-thiol chemistry,99peptide bond formation100 or other methods.101 Electrostatic adsorption102 and hydrophobic interaction103 schemes are commonly employed to introduce cargo molecules non-covalently. Regardless of the methods for cargo molecule introduction, the water-solubility and biocompatibility of nanomaterials themselves as delivery vehicles are crucial to their in vivo applications.Compared with the solid nanomaterials described, soft nanomaterials (e.g., liposomes and micelles) have high water-solubility, prolonged circulation time in the blood and enhanced accumulation at the tumor site.104 In addition, most soft nanomaterials are prepared from the same or similar materials as cell membranes, and are therefore inherently biocompatible.105–107 By introducing PEG molecules and other targeting ligands, non-specific binding between building blocks of soft nanomaterials and the cell membrane can be greatly reduced. Since most recognition events with target cells are achieved by the specific interaction between ligands and cell membrane receptors, soft nanomaterials modified with aptamers generated from cell-based SELEX are superior in delivery.108,109
Using liposomes for drug delivery is already an established technology. Liposomes have been successfully used for the delivery of poorly soluble chemotherapeutics. However, targeted delivery with high specificity is still needed. To this end, Kang et al. linked aptamers to liposomes to build a multi-functional target specific delivery system [Fig. 6].110 Briefly, hydrogenated soy phosphatidyl choline (HSPC), cholesterol (Chol), methoxypoly-(ethylene glycol)-distearoyl-phosphatidyl-ethanolamine (MPEG-DSPE) and maleimide-terminated PEG-DSPE (MalPEG) with a ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.08
0.08![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.02 were mixed together to construct liposomes with MalPEG as the dock for Sgc8 aptamer covalent conjugation. To serve as a model drug as well as a reporter, FITC-Dextran was encapsulated into the Sgc8 aptamer-conjugated liposomes. Specific binding of the FITC-Dextran-loaded liposomes to their targeted cells was demonstrated by both flow cytometry histograms and confocal microscopy images.
0.02 were mixed together to construct liposomes with MalPEG as the dock for Sgc8 aptamer covalent conjugation. To serve as a model drug as well as a reporter, FITC-Dextran was encapsulated into the Sgc8 aptamer-conjugated liposomes. Specific binding of the FITC-Dextran-loaded liposomes to their targeted cells was demonstrated by both flow cytometry histograms and confocal microscopy images.
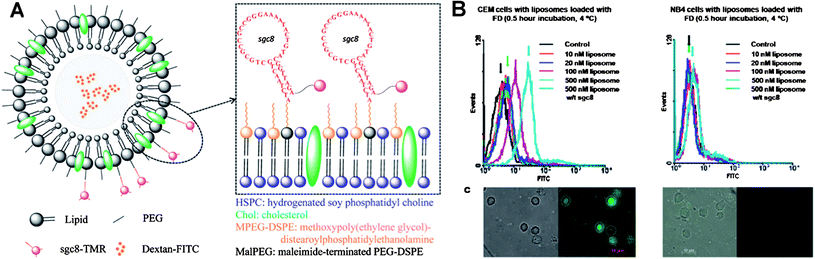 | ||
| Fig. 6 (A) Design scheme of multifunctional liposome nanostructure construction for targeted delivery. (B) Flow Cytometry histograms (top) and corresponding confocal microscopy images (bottom) of aptamer-conjugated liposome with target cells (left) and control cells (right). Adapted from ref. 110. | ||
Apart from liposomes, micelle aggregates composed of hydrophilic single-stranded DNA and hydrophobic polymers have also emerged as new delivery vehicles with good biocompatibility and high stability.111–113 However, the coupling between the DNA and hydrophobic polymer is often inefficient, and the micelle sizes are not well controlled.111 Liu et al. reported the easy synthesis of a well-defined DNA-diacyllipid micelle [Fig. 7].114 The diacyllipid tail could be efficiently incorporated at the 5′-end of DNA through solid-phase oligonucleotide synthesis to construct the conjugate. In aqueous solution, the diacyllipid-DNA conjugates spontaneously self-assembled into mono-dispersed micelles with low critical micelle concentration (CMC) and high stability. When incubated with cells, these micelles dissociated, allowing the diacyllipid-DNA conjugates to enter living cells through endocytosis. They also found that the ability of micelles to permeate cells was size-dependent. Based on these features and properties, they envisioned that these DNA-micelles would be attractive for applications in bionanotechnology, cell biology and drug delivery. Based on their work, Wu et al. further explored the application of these superior DNA-micelles, especially as detection/delivery vehicles to cancer cells.115 Instead of random ssDNA, the Td05 aptamer was introduced. In addition, they added a PEG linker between the lipid tail and the aptamer. It was found that the Td05 aptamer specifically binds to Ramos cells at 4 °C, but loses its binding affinity at 37 °C. However, the DNA-micelle composed of Td05-PEG-lipid conjugates showed strong binding to Ramos cells, even at 37 °C, with extremely low off rate (dissociation rate). Similar phenomena were also observed for other aptamer-micelles (e.g., KK-micelles and KB-micelles). By doping Td05-micelle with CellTracker™ Green that only fluoresces inside the cell, a strong fluorescence inside Ramos cells was observed after only 2 h. In contrast, fluorescence with similar intensity for free CellTracker™ Green required 12 h. The aptamer-micelles were also flushed through a channel containing immobilized cancer cells, as a tumor site mimic, demonstrating that micelles have good recognition ability in whole blood samples. Taken together, these results showed that the DNA-micelle with enhanced binding affinity and unique internalization allows rapid molecular recognition of cancer cells and in vivodrug delivery.
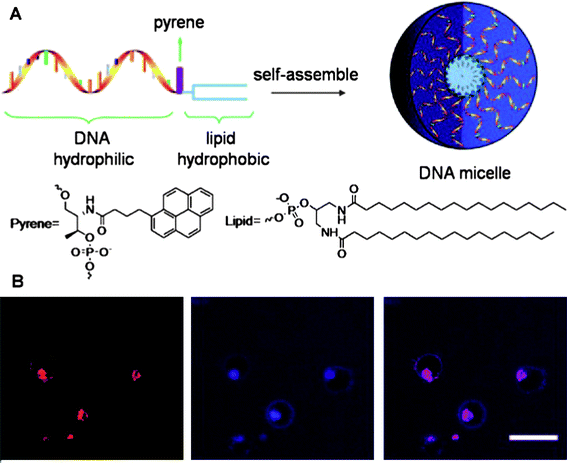 | ||
| Fig. 7 (A) Design scheme of DNA-micelle. (B) Localization and distribution of DNA-micelle inside CEM cells, from left to right: CEM cells treated with TAMRA-labeled DNA-micelles; CEM cells treated with Tf-Alexa 633; overlay of the first two channels. Adapted from ref. 114. | ||
Outlook
Aside from silica and gold nanoparticles, carbon nanotubes and gold nanorods, many other nanomaterials, including silver nanowires and biodegradable nanomaterials, such as chitosan nanotubes116 and chitosan dispersed carbon nanotubes,117 will carry nanotechnology one step further in satisfying the demands of biomedical applications. Bioconjugates integrating these nanomaterials with aptamers will further mediate the development of bioanalysis and biotechnology. Particularly, in terms of multiplexing, the combination of four DNA bases can, for example, produce millions of DNA strands with different sequences to meet the needs of specific recognition towards the desired targets with high affinity. In addition, advances in DNA engineering are expected to make the design of aptamer-conjugated nanomaterials more widely available.Other than their utilization as platforms for bioanalysis and carriers for drug delivery, nanomaterials will find more applications in the biomedical field. They have, for instance, been used as imaging agents for increased resolution and enhanced contrast.118–120 Future efforts will focus on using aptamer-conjugated nanomaterials to achieve localized enrichment in order to further improve imaging resolution and contrast. Nanomaterials have also been applied to three-dimensional cell culture and tissue engineering.121–123 For example, magnetic nanoparticles functionalized with appropriate ligands have been used to guide the direction of cell growth.124 This line of research is still in its infancy and aptamer-conjugated nanomaterials will pave the way for this field based on the specific interaction between aptamers and cell membrane receptors.
Acknowledgements
We thank NIH and NSF for the funding and support. We also acknowledge the support from the National Grand Program on Key Infectious Disease (2009ZX10004-312).References
- W. J. Parak, D. Gerion, T. Pellegrino, D. Zanchet, C. Micheel, S. C. Williams, R. Boudreau, M. A. Le Gros, C. A. Larabell and A. P. Alivisatos, Nanotechnology, 2003, 14, R15 CAS.
- J. Wang, Analyst, 2005, 130, 421 RSC.
- Q. A. Pankhurst, J. Connolly, S. K. Jones and J. Dobson, J. Phys. D: Appl. Phys., 2003, 35, R167 CrossRef.
- S. Cuenot, C. Fretigny, S. D. Champagne and B. Nysten, Phys. Rev. B: Condens. Matter Mater. Phys., 2004, 69, 165410 CrossRef.
- C. B. Murray, C. R. Kagan and M. G. Bawendi, Annu. Rev. Mater. Sci., 2000, 30, 545 CrossRef CAS.
- X. Chen, S. Xu, N. Yao and Y. Shi, Nano Lett., 2010, 10, 2133 CrossRef CAS.
- M. Victor, M. S. Fernando, K. Casimir, R. P. Alfonso and F. Martin, Pediatr. Res., 2010, 67, 481 CAS.
- N. Jha and S. Ramaprabhu, Nanoscale, 2010, 2, 806 RSC.
- F. A. Jaffer, M. Nahrendorf, D. Sosnovik, K. A. Kelly, E. Aikawa and R. Weissleder, Mol Imaging, 2006, 5, 85.
- I. I. Slowing, B. G. Trewyn, S. Giri and V. S. Y. Lin, Adv. Funct. Mater., 2007, 17, 1225 CrossRef CAS.
- I. Brigger, C. Dubernet and P. Couvreur, Adv. Drug Delivery Rev., 2002, 54, 631 CrossRef CAS.
- M. Shim, N. W. S. Kam, R. J. Chen, Y. Li and H. Dai, Nano Lett., 2002, 2, 285 CrossRef CAS.
- T. Sampson, World Pat. Inf., 2003, 25, 123 CrossRef CAS.
- D. E. Huizenga and J. W. Szostak, Biochemistry, 1995, 34, 656 CrossRef CAS.
- L. C. Bock, L. C. Griffin, J. A. Latham, E. H. Vermaas and J. J. Toole, Nature, 1992, 355, 564 CrossRef CAS.
- X. Fang and W. Tan, Acc. Chem. Res., 2010, 43, 48 CrossRef CAS.
- S. Li, H. Xu, H. Ding, Y. Huang, X. Cao, G. Yang, J. Li, Z. Xie, Y. Meng, X. Li, Q. Zhao, B. Shen and N. Shao, J. Pathol., 2009, 218, 327 CrossRef CAS.
- D. Shangguan, Y. Li, Z. Tang, Z. C. Cao, H. W. Chen, P. Mallikaratchy, K. Sefah, C. J. Yang and W. Tan, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11838 CrossRef CAS.
- Z. Xiao, D. Shangguan, Z. Cao, X. Fang and W. Tan, Chem.–Eur. J., 2008, 14, 1769 CrossRef CAS.
- J. Tang, T. Yu, L. Guo, J. Xie, N. Shao and Z. He, Biosens. Bioelectron., 2007, 22, 2456 CrossRef CAS.
- A. D. Keefe, S. Pai and A. Ellington, Nat. Rev. Drug Discovery, 2010, 9, 537 CrossRef CAS.
- J. H. Lee, M. V. Yigit, D. Mazumdar and Y. Lu, Adv. Drug Delivery Rev., 2010, 62, 592 CrossRef CAS.
- J. Zhou, M. R. Battig and Y. Wang, Anal. Bioanal. Chem., 2010 DOI:10.1007/S00216-010-3987-Y.
- Z. Tang, P. Mallikaratchy, R. Yang, Y. Kim, Z. Zhu, H. Wang and W. Tan, J. Am. Chem. Soc., 2008, 130, 11268 CrossRef CAS.
- S. E. Osborne, I. Matsumura and A. D. Ellington, Curr. Opin. Chem. Biol., 1997, 1, 5 CrossRef CAS.
- Y. Chen, A. C. Munteanu, Y. F. Huang, J. Phillips, Z. Zhu, M. Mavros and W. Tan, Chem.–Eur. J., 2009, 15, 5327 CrossRef CAS.
- L. S. Green, D. Jellinek, C. Bell, L. A. Beebe, B. D. Feistner, S. C. Gill, F. M. Jucker and N. Janjić, Chem. Biol., 1995, 2, 683 CrossRef CAS.
- L. Gold, B. Polisky, o. Uhlenbeck and M. Yarus, Annu. Rev. Biochem., 1995, 64, 763 CrossRef CAS.
- P. E. Burneister, S. D. Lewis, R. F. Silva, J. R. Preiss, L. R. Horwitz, P. S. Pendergrast, T. G. McCauley, J. C. Kurz, D. M. Epstein, C. Wilson and A. D. Keefe, Chem. Biol., 2005, 12, 25 CrossRef CAS.
- Y. Kim, D. M. Dennis, T. Morey, L. Yang and W. Tan, Chem.–Asian J., 2010, 5, 56 CrossRef CAS.
- W. Stober, A. Fink and E. Bohn, J. Colloid Interface Sci., 1968, 26, 62 CrossRef.
- F. J. Arriagada and K. O. Asare, Advances in Chemistry, American Chemical Society, 1994, Vol. 234, Ch. 5, pp. 113 Search PubMed.
- L. Wang and W. Tan, Nano Lett., 2006, 6, 84 CrossRef CAS.
- W. Lian, S. A. Litherland, H. Badrane, W. Tan, D. Wu, H. V. Baker, P. A. Gulig, D. V. Lim and S. Jin, Anal. Biochem., 2004, 334, 135 CrossRef CAS.
- Y. Wang, Y. Wang and B. Liu, Nanotechnology, 2008, 19, 415605 CrossRef.
- X. Wang, J. Zhou, W. Yun, S. Xiao, Z. Chang, P. He and Y. Fang, Anal. Chim. Acta, 2007, 598, 242 CrossRef CAS.
- M. C. Estévez, M. B. O'Donoghue, X. Chen and W. Tan, Nano Res., 2009, 2, 448 CrossRef CAS.
- L. Wang and W. Tan, Nano Lett., 2006, 6, 84 CrossRef CAS.
- X. Chen, M. C. Estévez, Z. Zhu, Y. F. Huang, Y. Chen, L. Wang and W. Tan, Anal. Chem., 2009, 81, 7009 CrossRef CAS.
- J. K. Herr, J. E. Smith, C. D. Medley, D. Shangguan and W. Tan, Anal. Chem., 2006, 78, 2918 CrossRef CAS.
- J. E. Smith, C. D. Medley, Z. Tang, D. Shangguan, C. Lofton and W. Tan, Anal. Chem., 2007, 79, 3075 CrossRef CAS.
- M. C. Daniel and D. Astruc, Chem. Rev., 2004, 104, 293 CrossRef CAS.
- T. K. Sau, A. Pal, N. R. Jana, Z. L. Wang and T. Pal, J. Nanoparticle Res., 2001, 3, 257 CrossRef.
- J. Liu and Y. Lu, Angew. Chem., Int. Ed., 2006, 45, 90 CrossRef CAS.
- S. Chah, M. R. Hammond and R. N. Zare, Chem. Biol., 2005, 12, 323 CrossRef CAS.
- R. Kanjanawarut and X. Su, Anal. Chem., 2009, 81, 6122 CrossRef CAS.
- T. A. El-Brolossy, T. Abdallah, M. B. Mohamed, S. Abdallah, K. Easawi, S. Negm and H. Talaat, Eur. Phys. J. Spec. Top., 2008, 153, 361 Search PubMed.
- L. He, E. A. Smith, M. J. Natan and C. D. Keating, J. Phys. Chem. B, 2004, 108, 10973 CrossRef CAS.
- C. D. Medley, J. E. Smith, Z. Tang, Y. Wu, S. Bamrungsap and W. Tan, Anal. Chem., 2008, 80, 1067 CrossRef CAS.
- G. Liu, X. Mao, J. A. Phillips, H. Xu, W. Tan and L. Zeng, Anal. Chem., 2009, 81, 10013 CrossRef CAS.
- U. H. F. Bunz and W. M. Rotello, Angew. Chem., Int. Ed., 2010, 49, 3268 CAS.
- C. Huang, S. Chiu, Y. Huang and H. Chang, Anal. Chem., 2007, 79, 4798 CrossRef CAS.
- J. Zhang, L. Wang, H. Zhang, F. Boey, S. Song and C. Fan, Small, 2010, 2, 201 CrossRef.
- D. Zheng, D. S. Seferos, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Nano Lett., 2009, 9, 3258 CrossRef CAS.
- D. A. Giljohann, D. S. Seferos, P. C. Patel, J. E. Millstone, N. L. Rosi and C. A. Mirkin, Nano Lett., 2007, 7, 3818 CrossRef CAS.
- S. Iijima, Nature, 1991, 354, 56 CrossRef CAS.
- R. B. Martin, L. Qu, Y. Lin, B. A. Harruff, C. E. Bunker, J. R. Gord, L. F. Allard and Y. P. Sun, J. Phys. Chem. B, 2004, 108, 11447 CrossRef CAS.
- D. Cui, B. Pan, H. Zhang, F. Gao, R. Wu, J. Wang, R. He and T. Asahi, Anal. Chem., 2008, 80, 7996 CrossRef CAS.
- H. M. So, K. Won, Y. H. Kim, B. K. Kim, B. H. Ryu, P. S. Na, H. Kim and J. O. Lee, J. Am. Chem. Soc., 2005, 127, 11906 CrossRef CAS.
- N. W. S. Kam and H. Dai, J. Am. Chem. Soc., 2005, 127, 6021 CrossRef CAS.
- M. Zheng, A. Jagota, E. D. Semke, B. A. Diner, R. S. Mclean, S. R. Lutig, R. E. RichardsonI and N. G. Tassi, Nat. Mater., 2003, 2, 338 CrossRef CAS.
- S. Wang, E. S. Humphreys, S. Y. Chung, D. F. Delduco, S. R. Lustig, H. Wang, K. N. Pprker, N. W. Rizzo, S. Subramoney, Y. M. Chiang and A. Jagota, Nat. Mater., 2003, 2, 196 CrossRef CAS.
- B. Gigliotti, B. Sakizzie, D. S. Bethune, R. M. Shelby and J. N. Cha, Nano Lett., 2006, 6, 159 CrossRef CAS.
- X. Zhao and J. K. Johnson, J. Am. Chem. Soc., 2007, 129, 10438 CrossRef CAS.
- R. Yang, J. Jin, Y. Chen, N. Shao, H. Kang, Z. Xiao, Z. Tang, Y. Wu, Z. Zhuand and W. Tan, J. Am. Chem. Soc., 2008, 130, 8351 CrossRef CAS.
- Z. Zhu, Z. Tang, J. A. Phillips, R. Yang, H. Wang and W. Tan, J. Am. Chem. Soc., 2008, 130, 10856 CrossRef CAS.
- N. R. Jana, L. Gearheart and C. J. Murphy, Adv. Mater., 2001, 13, 1389 CrossRef CAS.
- C. J. Murphy, T. K. Sau, A. M. Gole, C. J. Orendorff, J. Gao, L. Gou, S. E. Hunyadi and T. Li, J. Phys. Chem. B, 2005, 109, 13857 CrossRef CAS.
- J. Grand, S. Kostcheev, J. L. Bijeon, M. L. de la Chapelle, P. M. Adam, A. Rumyantseva, G. Lérondel and P. Royer, Synth. Met., 2003, 139, 621 CrossRef CAS.
- E. Cubukcu, E. A. Kort, K. B. Crozier and F. Capassoa, Appl. Phys. Lett., 2006, 89, 093120 CrossRef.
- Y. Y. Yu, S. S. Chang, C. L. Lee and C. R. C. Wang, J. Phys. Chem. B, 1997, 101, 6661 CrossRef CAS.
- N. Taub, O. Krichevski and G. Markovich, J. Phys. Chem. B, 2003, 107, 11579 CrossRef.
- Z. Ma and T. Ding, Nanoscale Res. Lett., 2009, 4, 1236 CrossRef CAS.
- X. Ren, D. Chen, X. Meng, F. Tang, A. Du and L. Zhang, Colloids Surf., B, 2009, 72, 188 CrossRef CAS.
- C. Yu and J. Irudayaraj, Anal. Chem., 2007, 79, 572 CrossRef CAS.
- E. Dujardin, L. B. Hsin, C. R. C. Wang and S. Mann, Chem. Commun., 2001, 1264 RSC.
- J. Li, J. Peng, W. Huang, Y. Wu, J. Fu, Y. Cong, L. Xue and Y. Han, Langmuir, 2005, 21, 2017 CrossRef CAS.
- T. S. Sreeprasad, A. K. Samal and T. Pradeep, Langmuir, 2008, 24, 4589 CrossRef CAS.
- T. Niidome, M. Yamagata, Y. Okamoto, Y. Akiyama, H. Takahashi, T. Kawano, Y. Katayama and Y. Niidome, J. Controlled Release, 2006, 114, 343 CrossRef CAS.
- H. Liao and J. H. Hafner, Chem. Mater., 2005, 17, 4636 CrossRef CAS.
- E. T. Castellana, R. C. Gamez, M. E. Gómez and D. H. Russell, Langmuir, 2010, 26, 6066 CrossRef CAS.
- Y. Huang, H. Chang and W. Tan, Anal. Chem., 2008, 80, 567 CrossRef CAS.
- L. Gou and C. J. Murphy, Chem. Mater., 2005, 17, 3668 CrossRef CAS.
- F. J. García-Vidal and J. B. Pendry, Phys. Rev. Lett., 1163, 77 Search PubMed.
- O. E. Rivera-betancourt, O. M. Primera-Pedrozo, L. C. Pacheco-Londoήo and S. P. Hernάndez-Rivera, IEEE Sens. J., 2010, 10, 699 CrossRef CAS.
- C. J. Orendorff, L. Gearheart, N. R. Jana and C. J. Murphy, Phys. Chem. Chem. Phys., 2006, 8, 165 RSC.
- G. V. Maltzahn, A. Centrone, J. Park, R. Ramanathan, M. J. Sailor, T. A. Hatton and S. N. Bhatia, Adv. Mater., 2009, 21, 3175 CrossRef.
- M. Moskovits, J. Chem. Phys., 1978, 69, 4159 CrossRef CAS.
- M. Moskovits, Rev. Mod. Phys., 1985, 57, 783 CrossRef CAS.
- B. Nikoobakht, J. Wang and M. A. El-Sayed, Chem. Phys. Lett., 2002, 366, 17 CrossRef CAS.
- Y. Wang, K. Lee and J. Irudayaraj, Chem. Commun., 2010, 46, 613 RSC.
- A. Joseph, L. Villaraza, A. Bumb and M. W. Brechbiel, Chem. Rev., 2010, 110, 2921 CrossRef CAS.
- S. Lin, X. Xie, M. R. Pate, Y. H. Yang, Z. Li, F. Cao, O. Gheysens, Y. Zhang, S. S Gambhir, J. H. Rao and J. C. Wu, BMC Biotechnol., 2007, 7, 67 CrossRef.
- M. Strømme, U. Brohede, R. Atluri and A. E. Garcia-Bennett, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol., 2009, 1, 140 Search PubMed.
- M. O. Riehle, Nanobiotechnology, 2005, 1, 308 CrossRef.
- J. Liu, S. Z. Qiao, S. B. Hartono and G. Q. (Max) Lu, Angew. Chem. Int. Ed, 2010, 49, 1.
- Z. Wu, B. Zhang and B. Yan, Int. J. Mol. Sci., 2009, 10, 4198 Search PubMed.
- K. Cho, X. Wang, S. Nie, Z. (Georgia) Chen and D. M. Shin, Clin. Cancer Res., 2008, 14, 1310 CrossRef CAS.
- Z. Li, R. Jin, C. A. Mirkin and R. L. Letsinger, Nucleic Acids Res., 2002, 30, 1558 CrossRef CAS.
- L. Xu, Y. Guo, R. Xie, J. Zhuang, W. Yang and T. Li, Nanotechnology, 2002, 13, 725 CrossRef CAS.
- E. Turos, J. Y. Shim, Y. Wang, K. Greenhalgh, G. S. K. Reddy, S. Dickey and D. V. Lim, Bioorg. Med. Chem. Lett., 2007, 17, 53 CrossRef CAS.
- S. Patil, A. Sandberg, E. Heckert, W. Self and S. Seal, Biomaterials, 2007, 28, 4600 CrossRef CAS.
- G. D. Bothun, J. Nanobiotechnol., 2008, 6, 13 CrossRef.
- K. Kataoka, A. Harada and Y. Nagasaki, Adv. Drug Delivery Rev., 2001, 47, 113 CrossRef CAS.
- T. Wang, V. A. Petrenko and V. P. Torchilin, Mol. Pharmaceutics, 2010 DOI:10.1021/mp1001125.
- A. S. Gobin, R. Rhea, R. A. Newman and A. B. Mathur, Int. J. Nanomed., 2006, 1, 81 CrossRef CAS.
- D. A. Eavarone, X. Yu and R. V. Bellamkonda, J. Biomed. Mater. Res., 2000, 51, 10 CrossRef CAS.
- J. Zhu, F. Yan, Z. Guo and R. E. Marchant, J. Colloid Interface Sci., 2005, 289, 542 CrossRef CAS.
- W. Y. Yu and N. Zhang, Current Nanoscience, 2009, 5, 123 CrossRef CAS.
- H. Kang, M. B. O'Donoghue, H. Liu and W. Tan, Chem. Commun., 2010, 46, 249 RSC.
- Z. Li, Y. Zhang, P. Fullhart and C. A. Mirkin, Nano Lett., 2004, 4, 1055 CrossRef CAS.
- F. E. Alemdaroglu, N. C. Alemdaroglu, P. Langguth and A. Herrmann, Adv. Mater., 2008, 20, 899 CrossRef CAS.
- J. H. Jeong and T. G. Park, Bioconjugate Chem., 2001, 12, 917 CrossRef CAS.
- H. Liu, Z. Zhu, H. Kang, Y. Wu, K. Sefah and W. Tan, Chem.–Eur. J., 2010, 16, 3791 CrossRef CAS.
- Y. Wu, K. Sefah, H. Liu, R. Wang and W. Tan, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5 CrossRef CAS.
- Y. Yang, Q. He, L. Duan, Y. Cui and J. Li, Biomaterials, 2007, 28, 3083 CrossRef CAS.
- S. Ozarkar, M. Jassal and A. K. Agrawal, Fibers Polym., 2008, 4, 410 Search PubMed.
- Z. Liu, F. Kiessling and J. Gatjens, J. Nanomater., 2010 DOI:10.1155/2010/894303.
- Z. Liu and R. Peng, Eur. J. Nucl. Med. Mol. Imaging, 2010 DOI:10.1007/s00259-010-1452-y.
- M. Ogawa, C. A. S. Regino, B. Marcelino, M. Williams, N. Kosaka, L. H. Bryant, Jr., P. L. Choyke and H. Kobayashi, Bioconjugate Chem., 2010, 21, 955 CrossRef CAS.
- M. O. Riehle, NanoBiotechnology, 2005, 1, 308 CrossRef.
- A. C. A. Wan and J. Y. Ying, Adv. Drug Delivery Rev., 2010, 62, 731 CrossRef CAS.
- V. Jayawarna, M. Ali, T. A. Jowitt, A. F. Miller, A. Saiani, J. E. Gough and R. V. Ulijn, Adv. Mater., 2006, 18, 611 CrossRef CAS.
- J. H. Lee, E. S. Kim, M. H. Cho, M. Son, S. I. Yeon, J. S. Shin and J. Cheon, Angew. Chem. Int. Ed., 2010, 49, 1.
| This journal is © The Royal Society of Chemistry 2011 |
